Abstract
The goose parvovirus (GPV) Rep 1 and Rep 2 proteins are encoded by P9-generated mRNAs that are either unspliced or spliced within the rep gene region, respectively. These mRNAs are present in an approximately equal ratio. The translation of Rep 1 was initiated from the first AUG in unspliced P9-generated mRNA; however, this AUG was bypassed in spliced P9-generated RNA and Rep 2 translation initiated predominately at the next initiating AUG downstream. We show that the choice of the site of initiation of translation of GPV Rep-encoding mRNAs is governed both by the splicing process itself and by the nature of the excised intron.
Goose parvovirus (GPV) has identical hairpin termini, is most similar in both nucleotide sequence and protein homology to adeno-associated virus 2 (AAV2), and has been classified as a member of the Dependovirus genus (10-12); however, unlike the AAVs, GPV can replicate efficiently without the aid of a helper virus (12). The RNA expression profile of GPV is a surprising hybrid of features of the Parvovirus and Dependovirus genera of the Parvovirinae (7). Similar to the Dependovirus AAV5, RNAs transcribed from the GPV upstream P9 promoter, which encode the viral Rep protein(s), are polyadenylated at high efficiency at a polyadenylation [(pA)p] site located within the small intron in the center of the genome (7). No promoter analogous to the Dependovirus P19 promoter has been detected; however, similar to minute virus of mice (MVM) and other members of the Parvovirus genus, approximately half of the pre-mRNAs generated from the P9 promoter are additionally spliced within the putative GPV Rep coding region between a donor site located at nucleotide (nt) 814 and an acceptor site at nt 1198 (7). The GPV RNA profile has been shown to be the same in both human 293T and goose CGBQ cells (7). Thus, the mechanism that GPV uses for the expression of its nonstructural gene is more like that used by members of the autonomous Parvovirus group.
In this report, we describe the coding strategy for the nonstructural proteins of GPV. We demonstrate that the large Rep 1 protein is encoded uninterruptedly in open reading frame 1 (ORF 1) from the unspliced P9-generated mRNA using an initiating AUG codon at nt 537. The smaller Rep 2 protein is encoded by the spliced P9-generated mRNA; it initiates in ORF 2 at an AUG at nt 650 and continues in ORF 1 after the splice. Strikingly, the first upstream AUG at nt 537 is not utilized in spliced P9-generated mRNA. We show that the choice of initiation site is governed by the splicing process itself and by the nature of the excised intron.
AUG 1 is utilized efficiently in unspliced but not spliced P9-generated mRNA, while AUG 2 is utilized efficiently only in spliced P9-generated mRNA.
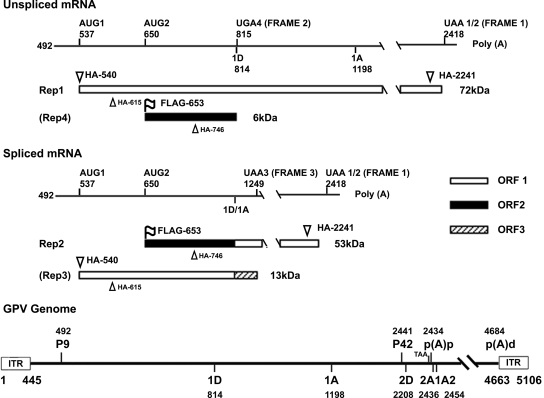
Figure 1 shows a diagram of the Rep coding region of GPV. This region is expressed from mRNAs initiated at nt 492 by the P9 promoter (7). Greater than 95% of the mRNAs generated by P9 are polyadenylated at a (pA)p site in the center of the genome at nt 2434 within the small central intron (7). P9-generated mRNAs accumulate as two predominant species at an approximate steady-state ratio of 1:1 (7). The first is unspliced across its length, while the second is additionally spliced between nt 814 to 1198 (7). The AUG at nt 537 (designated AUG 1) is the first AUG present in P9-generated RNA downstream of the initiation site (7). A sequence analysis suggested that AUG 1 could initiate, in unspliced P9-generated mRNA, the production of a large, 627-amino-acid protein (designated Rep 1 in Fig. 1) of approximately 72 kDa in ORF 1 which extends until a termination codon at nt 2418 shortly upstream of the (pA)p site (Fig. 1). In P9-generated RNA spliced between nt 814 and 1198, translation initiated in ORF 1 at AUG 1 would be shifted to ORF 3 after the splice and terminate shortly after at nt 1249. This would generate a small protein of approximately 13 kDa (designated Rep 3 in Fig. 1). However, if the translation of spliced P9-generated RNA initiated at a downstream AUG at nt 650 (designated AUG 2) in ORF 2, it would continue in ORF 1 after the splice to generate a protein of approximately 53 kDa (designated Rep 2 in Fig. 1) which would share its carboxyl-terminal region with Rep 1. Translation initiated in ORF 2 at nt 650 in unspliced RNA would terminate at nt 815, generating a small protein of approximately 6 kDa (designated Rep 4 in Fig. 1).
FIG. 1.
The putative genetic map of unspliced and spliced P9-generated mRNAs of the GPV rep gene. The P9-generated pre-mRNA initiates at nt 492 and is polyadenylated in the middle of the genome at nt 2434. About 50% of these RNAs are further spliced between nt 814 and 1198. The ORFs encoding the potential GPV Rep proteins are shown. Parentheses surrounding the Rep 3 and Rep 4 labels indicate the lack of evidence of their production. Also shown are the locations of the various initiating AUGs, termination codons, and HA and FLAG tags described in the text. A diagram of the GPV genome with relevant landmarks is shown at the bottom.
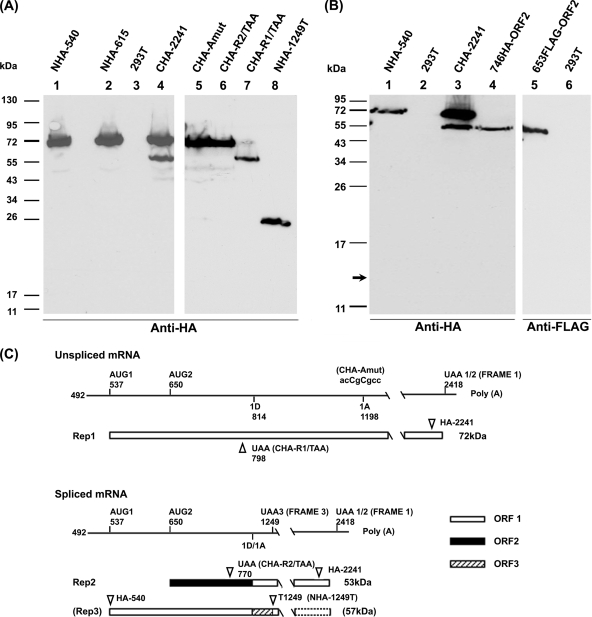
To determine the rep gene coding strategy utilized by GPV, we initially inserted either hemagglutinin (HA) or FLAG tags in various ORFs at various positions within the gene (Fig. 1) and analyzed protein production following the transfection of 293T cells using linear polyethylenimine as previously described (8). The GPV-RepCap plasmid used as the parent for all mutant constructs described in this report has been previously described (7), and all nucleotide numbers refer to GenBank accession number U25749. The insertion of an HA epitope in ORF 1 upstream of the rep gene intron at either nt 540 (NHA-540) or nt 615 (NHA-615) tagged primarily a single protein of approximately 72 kDa (Fig. 2A, lanes 1 and 2) as detected by immunoblotting, performed as previously described (6) using anti-HA antibody (HA-7; Sigma, St. Louis, MO). This demonstrated that AUG 1 was used to initiate a large Rep protein (Rep 1) from unspliced P9-generated mRNA. The absence of a smaller band in these lanes, as well as its absence when samples were run on 15% polyacrylamide gel electrophoresis (PAGE) gels (Fig. 2B, lane 1), suggested that AUG 1 was not used to initiate a protein from spliced P9-generated mRNA (which, as described above, would be terminated shortly after the splice at UAA 3, generating a protein of approximately 13 kDa [Fig. 1]). The insertion of an HA epitope in the C-terminal shared ORF 1 at nt 2241 (CHA-2241), tagged both a protein of approximately 72 kDa (Rep 1) and a protein of approximately 53 kDa, which is the size predicted for a protein (Rep 2) initiated by AUG 2 at nt 650 in spliced P9-generated RNA (Fig. 2A, lane 4). Consistent with this prediction, the insertion in ORF 2 of either HA at nt 746 (746HA-ORF2), or FLAG at nt 653 (653FLAG-ORF2), downstream of AUG 2 tagged a single detectable protein of 53 kDa (Fig. 2B, lanes 4 and 5; the FLAG epitope was detected by anti-FLAG M2 antibody F3165 [Sigma, St. Louis, MO]). Together, these results demonstrated that AUG 2 was used to initiate an additional Rep protein (Rep 2) from spliced P9-generated mRNA. These results also confirmed that the 53-kDa protein is a primary translation product and not the result of the degradation of Rep 1. Similar results were obtained following the transfection of the goose embryonic kidney cell line CGBQ (AATC CCL-169), using the Amaxa Nucleofector device (Amaxa, Inc., Gaithersburg, MD) with solution V and program A23, as described previously (7; data not shown). It is not yet clear why Rep 2 is seen to accumulate at lower levels than Rep 1; however, we are currently examining the relative stabilities of both proteins.
FIG. 2.
Genetic organization of the GPV rep gene. (A) Immunoblot analysis of GPV proteins run on 10% PAGE gels, using anti-HA antibody. Protein samples were prepared 48 h following transfection of 293T cells by the constructs listed at the top of each lane. Untransfected 293T cell control samples are labeled 293T (lane 3). The constructs are described in the text. Marker migration shown to the left. (B) Immunoblot analysis of GPV proteins run on 15% PAGE gels, using either anti-HA antibody, lanes 1 to 4, or anti-FLAG antibody, lanes 5 to 6. Protein samples were prepared 48 h following the transfection of 293T cells by the constructs listed at the top of each lane. Untransfected 293T cell control samples are labeled 293T (lanes 2 and 6). The constructs are described in the text. Marker migration is shown to the left. (C) Diagram showing the position of the mutations in the CHA-Amut, CHA-R2/TAA, CHA-R1/TAA, and NHA-1249T mutant constructs, explained further in the text. The transcript landmarks are the same as shown in Fig. 1. The box with a stippled border indicates a frameshift of putative Rep 3 at nt 1249.
Additional experiments were performed to confirm the coding strategy of Rep 2. When the splicing of the rep gene intron was prevented (as confirmed by RNase protection analysis, done as previously described [7] using the P1D probe [7] across the rep gene splice donor [data not shown]) by the introduction of an acceptor site mutation (CAACAG/AGCG to CAACCG/CGCC [changes shown in italics]) in the parent CHA-2241 construct tagged in the shared C-terminal ORF 1 region (CHA-Amut), the expression of the 53-kDa protein was abolished, while the 72-kDa Rep 1 was unaffected (Fig. 2A, lane 5). Furthermore, insertion in the CHA-2241 parent construct of a stop codon at nt 798 which terminated only ORF 1 (CHA-R1/TAA) resulted in the loss of Rep 1 while allowing the expression of the 53-kDa Rep 2 protein, confirming the independent translation of Rep 2 in an alternate ORF in this region (Fig. 2A, lane 7). Conversely, the insertion of a termination codon in ORF 2 at nt 770 (CHA-R2/TAA) resulted in the loss of the 53-kDa Rep 2, while allowing the translation of Rep 1 (Fig. 2A, lane 6). Similar results were also obtained following the transfection of CGBQ cells as described above (data not shown).
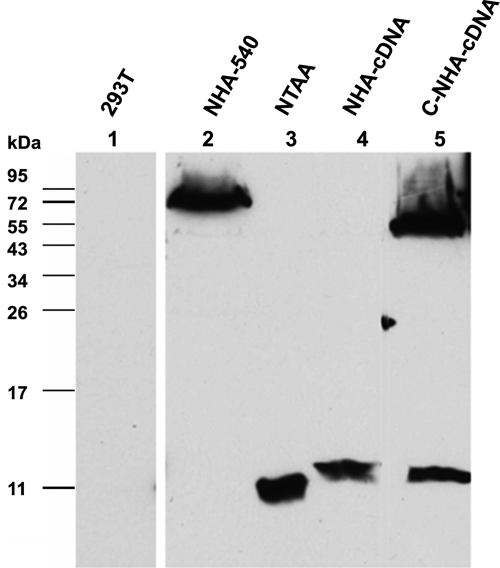
Our inability to detect a Rep 3-like product suggested that, surprisingly, AUG 1 was not used in spliced mRNA, although it was present as the first initiation signal, and functional, in unspliced RNA. To confirm that we could detect a 13-kDa product initiated at AUG 1 from spliced RNA if such a protein had in fact been generated by GPV, we examined the expression from a construct in which we introduced a termination signal in ORF 1 at nt 798 (NTAA). This construct, which was also NH2 terminally HA-tagged in ORF 1, was predicted to generate from AUG 1 an 87-amino-acid protein of approximately 10 kDa, 21 amino acids smaller than the approximate 13-kDa protein that would be generated if AUG 1 was used in spliced GPV mRNA. The expression of a protein of approximately this size was clearly detected following the transfection of this mutant construct (Fig. 3, lane 3), suggesting that our inability to detect the generation of a 13-kDa protein from wild-type GPV constructs was not due to technical considerations. Furthermore, as mentioned above, a putative 13-kDa Rep 3 protein initiated in ORF 1 from AUG 1 in spliced mRNA would be terminated by a TAA signal in ORF 3 at nt 1249, 50 nt after the intron acceptor at nt 1198 (Fig. 1). The insertion of a single nucleotide at position 1249 prior to the ORF 3-terminating TAA would shift ORF 3 to the open ORF 1 at this site and preclude the termination of an AUG 1-initiated Rep 3 product generated from spliced mRNA. Thus, if AUG 1 was used in spliced RNA generated from a construct with this insertion (NHA-1249T), it would produce a larger protein of approximately 57 kDa. Following the transfection of NHA-1249T, such a product was not detected, although as predicted, the insertion mutation shifted Rep 1 into ORF 2 in which it terminated 8 nt downstream of the insertion, generating a protein of approximately 24 kDa (Fig. 2A, lane 8). Similar results were seen with a mutant with a single nucleotide insertion at position 1221 (data not shown). Taken together, these results demonstrated that the 5′-most initiation codon AUG 1, which was the primary initiation codon used in unspliced P9-generated RNA, was not used at a detectable frequency from spliced P9-generated mRNA. Furthermore, none of our analyses detected a product initiated from AUG 2 in unspliced P9-generated RNA (the putative Rep 4 protein [Fig. 1 and data not shown]).
FIG. 3.
Both AUG 1 and AUG 2 are utilized in prespliced, intronless P9-generated RNA. Immunoblot analysis of GPV proteins run on 15% PAGE gels, using anti-HA antibody. Protein samples were prepared 48 h following the transfection of 293T cells by the constructs listed at the top of each lane. Untransfected 293T cell control samples are labeled 293T (lane 1). The constructs are described in the text. Marker migration is shown to the left.
Both AUG 1 and AUG 2 are utilized in prespliced, intronless P9-generated RNA and P9-generated RNA in which the GPV intron was replaced with a foreign intron.
Why was AUG 1 utilized in unspliced P9-generated mRNA but overlooked in spliced P9-generated mRNA? To determine whether the splicing process itself had an effect on subsequent translation initiation, we examined protein expression from constructs in which the intron was precisely removed to mimic a cDNA molecule in the rep gene region. In such constructs, which also contained an HA tag in frame in ORF 1 at nt 540 (NHA-cDNA), a protein of 13 kDa was clearly detected (Fig. 3, lane 4), and in constructs containing an HA tag in both the amino terminus and the shared C-terminal region (C-NHA-cDNA), both 13-kDa and 53-kDa proteins were apparent (Fig. 3, lane 5). These results indicated that both AUG 1 and AUG 2 were efficiently utilized from the prespliced, intronless RNA and suggested that the splicing process itself, rather than the structure of the mRNA molecules themselves, mediated the bypass of AUG 1 in spliced P9-generated mRNAs.
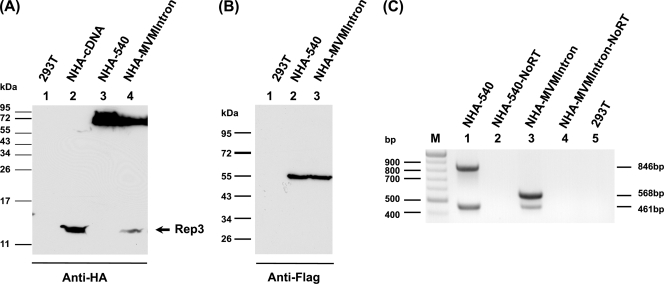
If the splicing of the GPV P9-generated pre-mRNA affected AUG usage in the spliced mRNA, was this specific to the splicing of the GPV rep gene intron? To address this question, we replaced the GPV rep gene intron with the small intron from the MVM (from D1-A1, 96 nt; MVM, nt 2280 to 2377). In order to restrict splicing to the natural D1-A1 pair, the MVM second small intron donor D2, internal to D1, was destroyed (by changing MVM nt 2315 to 2318 from AGGT to ACCA). The construct was designed such that splicing between the MVM D1-A1 would join the GPV upstream and downstream exons exactly as in the wild type. In addition, a single T nucleotide was inserted at position 2353 within the MVM intron to bypass termination signals in the MVM intron in frame with GPV ORF 1 that would terminate Rep 1. The T insertion allows the extension of a Rep 1-like molecule into ORF 1 until its native termination site at GPV nt 2418, thus preventing the generation of a potentially comigrating small product. This construct (NHA-MVMIntron) also contained an HA tag at nt 540 in frame in ORF 1 to detect AUG 1-initiated products, as well as a FLAG tag in ORF 2 at nt 653 to detect AUG 2-initiated products.
The semiquantitative reverse transcriptase PCR (RT-PCR) analysis shown in Fig. 4C indicated that the MVM small intron within the GPV background spliced somewhat less efficiently than the native GPV rep gene intron (lanes 3 and 1, respectively). The same-size (lower) bands, derived from spliced RNA, were produced from RNA generated by both the mutant and the wild type (461 nt), while the upper bands in this analysis, derived from unspliced RNA, reflected the different sizes of the introns present (568 nt for NHA-MVMIntron and 846 nt for NHA-540). RT-PCR was chosen for the initial analysis so that amplified bands could be sequenced; however, the relative ratios of spliced and unspliced RNAs were confirmed by RNase protection assays (data not shown). The sequencing of the bands derived from spliced RNA demonstrated that the GPV exons were joined exactly (data not shown). RNA used for the RT-PCR was isolated by the Paris kit (Ambion, Inc., Austin, TX), with additional treatment with DNase I. RT-PCR was performed for 35 cycles using the Titan-One RT-PCR kit (Roche, Mannheim, Germany) with primers F765 (AATCAGTTCAACCAGGACG) and R1610 (GCCATAGAAGGGTACAGCA). No bands were detected in the absence of RT or from untransfected control-cell RNA (Fig. 4C, lanes 2, 4, and 5).
FIG. 4.
Both AUG 1 and AUG 2 were utilized in spliced P9-generated RNA generated from a construct in which the GPV intron was replaced with the MVM small intron. Immunoblot analysis of GPV proteins run on 15% (A) or 10% (B) PAGE gels by using anti-HA antibody (A) or anti-FLAG antibody (B). Protein samples were prepared 48 h following the transfection of 293T cells by the constructs listed at the top of each lane. Untransfected 293T cell control samples are labeled 293T (lanes 1). The constructs are described in the text. Marker migration is shown to the left. Rep 3 is indicated with an arrow. (C) Semiquantitative RT-PCR, using primers described in the text, of RNA samples taken 48 h following the transfection of 293T cells by NHA-540 (lane 1) and NHA-MVMIntron (lane 3). Corresponding PCR controls in the absence of reverse transcriptase are shown in lanes 2 and 4. RT-PCR of untransfected 293T cells is shown in lane 5. M, markers. The bands at 461 nt are from spliced templates, while the larger bands are generated from unspliced templates of different sizes.
In contrast to the GPV intron-containing construct (Fig. 4A, lane 3), the replacement of the GPV intron with the MVM intron led to the production of both a Rep 1-like protein (in this case approximately 62 kDa) and a 13-kDa Rep 3 protein (Fig. 4A, lane 4), as detected using antibody to the N-terminal HA tag present in ORF 1. The levels of the 13-kDa protein generated by NHA-MVMIntron were consistent with the level of splicing seen, and the size was indistinguishable from the AUG 1-derived small product generated by the prespliced, intronless construct-made cDNA across the rep gene intron (NHA-cDNA; Fig. 4A, lane 2). As predicted following the authentic splicing of the MVM intron, the MVM intron-containing construct also generated a Rep 2 protein of wild-type size from spliced RNA, as detected with antibody directed against the FLAG tag present at nt 653 in ORF 2 (Fig. 4B, compare lanes 2 and 3). Thus, initiation from AUG 1 was regained in spliced P9-generated mRNAs generated from a parent pre-mRNA containing the MVM intron. These results indicated that the initiation codon choice for GPV P9-generated mRNAs was governed by both the splicing process and the specific nature of the excised intron.
There is increasing recognition that nuclear RNA processing events, including splicing and export, can influence translation in the cytoplasm (2-3). Components of the exon-junction complex (1, 4), as well as shuttling serine-arginine-rich proteins ASF/SF2 and 9G8 (5, 9), loaded on RNA in the nucleus have been implicated in the modulation of mRNA translation, although the mechanism of their action in this regard is not yet fully clear. The GPV system may be a useful system to study this phenomenon, and further characterization of the cis-acting sequences within the intron, and potential trans-acting factors that mediate this effect, is currently being pursued.
Acknowledgments
We thank Lisa Burger for excellent technical help.
This work was supported by PHS grants AI46458 and AI56310 from NIAID to D.J.P.
Footnotes
Published ahead of print on 22 July 2009.
REFERENCES
- 1.Diem, M. D., C. C. Chan, I. Younis, and G. Dreyfuss. 2007. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat. Struct. Mol. Biol. 14:1173-1179. [DOI] [PubMed] [Google Scholar]
- 2.Le Hir, H., A. Nott, and M. J. Moore. 2003. How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 28:215-220. [DOI] [PubMed] [Google Scholar]
- 3.Le Hir, H., and B. Seraphin. 2008. EJCs at the heart of translational control. Cell 133:213-216. [DOI] [PubMed] [Google Scholar]
- 4.Ma, X. M., S. O. Yoon, C. J. Richardson, K. Julich, and J. Blenis. 2008. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell 133:303-313. [DOI] [PubMed] [Google Scholar]
- 5.Michlewski, G., J. R. Sanford, and J. F. Caceres. 2008. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol. Cell 30:179-189. [DOI] [PubMed] [Google Scholar]
- 6.Miller, C. L., and D. J. Pintel. 2001. The NS2 protein generated by the parvovirus minute virus of mice is degraded by the proteasome in a manner independent of ubiquitin chain elongation or activation. Virology 285:346-355. [DOI] [PubMed] [Google Scholar]
- 7.Qiu, J., F. Cheng, Y. Yoto, Z. Zádori, and D. J. Pintel. 2005. The expression strategy of goose parvovirus exhibits features of both the Dependovirus and Parvovirus genera. J. Virol. 79:11035-11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu, J., F. Cheng, and D. J. Pintel. 2007. Distance-dependent processing of adeno-associated virus type 5 RNA is controlled by 5′ exon definition. J. Virol. 81:7974-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swartz, J. E., Y. C. Bor, Y. Misawa, D. Rekosh, and M. L. Hammarskjold. 2007. The shuttling SR protein 9G8 plays a role in translation of unspliced mRNA containing a constitutive transport element. J. Biol. Chem. 282:19844-19853. [DOI] [PubMed] [Google Scholar]
- 10.Tattersall, P. 2006. The evolution of parvovirus taxonomy, p. 5-14. In J. Kerr et al. (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 11.Zádori, Z., R. Stefancsik, T. Rauch, and J. Kisary. 1995. Analysis of the complete nucleotide sequences of goose and Muscovy duck parvoviruses indicates common ancestral origin with adeno-associated virus 2. Virology 212:562-573. [DOI] [PubMed] [Google Scholar]
- 12.Zadori, Z., J. Szelei, I. Kiss, and P. Tijssen. 2006. Waterfowl parvoviruses, p. 447-456. In J. Kerr et al. (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.