Abstract
Background
The global regulatory system ArcAB controls the anaerobic growth of E. coli, however, its role in aerobic conditions is not well characterized. We have previously reported that ArcA was necessary for Salmonella to resist reactive oxygen species (ROS) in aerobic conditions.
Results
To investigate the mechanism of ROS resistance mediated by ArcAB, we generated deletion mutants of ArcA and ArcB in E. coli. Our results demonstrated that both ArcA and ArcB were necessary for resistance to hydrogen peroxide (H2O2), a type of ROS, and their function in this resistance was independent from H2O2 scavenge. Mutagenesis analysis of ArcA indicated that ROS resistance was mediated through a distinct signaling pathway from that used in anaerobic conditions. An abundant protein flagellin was elevated at both the protein and mRNA levels in the ΔarcA mutant as compared to the wild type E. coli, and deletion of flagellin restored the resistance of the ΔarcA mutant to H2O2. The resistance of the ΔarcA mutant E. coli to H2O2 can also be restored by amino acid supplementation, suggesting that a deficiency in amino acid and/or protein synthesis in the mutant contributed to its susceptibility to H2O2, which is consistent with the notion that protein synthesis is necessary for ROS resistance.
Conclusion
Our results suggest that in addition to its role as a global regulator for anaerobic growth of bacteria, ArcAB system is also important for bacterial resistance to ROS in aerobic conditions, possibly through its influence on bacterial metabolism, especially amino acid and/or protein assimilation and synthesis.
Background
Aerobic bacteria use oxygen as a terminal electron acceptor in oxygen-containing environments for their metabolism. Although aerobic growth has its obvious advantages (e. g. high energy efficiency, abundance of oxygen in the atmosphere, etc), bacteria must deal with the undesired consequences from exposure to oxygen and oxidative environments. Oxygen and its derivatives, such as superoxide and hydrogen peroxide, are often highly reactive and pose a threat to many macromolecules, such as enzymes with iron-sulfur centers, nucleic acids, and lipids. Therefore, bacteria undergoing aerobic growth must be able to sense, respond to, and detoxify reactive oxygen species (ROS), and maintain their structural and functional integrities.
The principle mechanism through which bacteria respond to environmental signals is through two-component and other regulatory systems [1,2]. At least four global regulatory systems -OxyRS, SoxRS, Fnr and ArcAB – are identified to respond to oxygen and its derivatives [3,4]. OxyRS and SoxRS systems control the response of bacteria to hydrogen peroxide and superoxide, respectively [3-12]. Fnr (fumarate and nitrate reduction) controls the transition from aerobic growth to anaerobic growth [13-17]. Fnr is believed to directly sense oxygen [18-20] and regulate at least 100 operons [21-23]. In addition to Fnr, the two-component regulatory system ArcAB also regulates the transition of bacteria from aerobic to anaerobic growth and is active under microaerobic conditions. It controls at least 100 operons that are involved in the TCA cycle and energy metabolism [16,24-29]. The sensor kinase ArcB undergoes auto-phosphorylation at His292 under anaerobic conditions, and this activation is negatively regulated by the oxidized quinones under aerobic conditions [25]. Activated ArcB undergoes a phosphorelay of His292 to Asp576 to His717, and subsequently activates its cognate transcriptional regulator ArcA by phosphorylating ArcA at Asp54 to repress genes contributing to aerobic metabolism (e.g. citrate synthase and isocitrate lyase) and activates genes necessary for anaerobic metabolism (e.g. pyruvate formate lyase and hydrogenase) [23,25,30-34].
Although the function of the ArcAB system in the anaerobic growth of E. coli has been well characterized, its function is unlikely to be limited to those required for the anaerobic growth of bacteria. For example, the ArcAB system has been reported to be involved in chromosomal replication, stress responses and aging of bacteria [35-37]. We have previously reported that ArcA of Salmonella enterica is necessary for its resistance to reactive oxygen and nitrogen species (ROS and RNS) [38]. More recently, ArcA is implicated in the ROS stress response of Haemophilus influenzae [39]. In this report, we analyzed the role of ArcAB in reactive oxygen resistance of E. coli and investigated the mechanism of ROS resistance mediated by the ArcAB two-component system.
Results
ArcAB system is necessary for E. coli to resist hydrogen peroxide (H2O2)
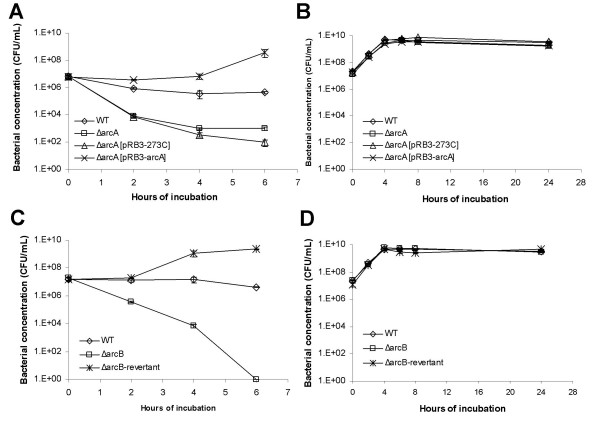
To determine if the ArcAB global regulatory system plays a role in the survival of E. coli under stress by reactive oxygen species (ROS), we generated deletion mutants of ArcA (the global regulator) and ArcB (the cognate sensor-kinase of ArcA) in E. coli (Table 1). Both ΔarcA and ΔarcB mutant E. coli formed smaller colonies than their parental E. coli, but otherwise showed similar colony morphology. The ΔarcA and ΔarcB mutant E. coli were tested for their growth properties in complete (Luria Bertani broth) or minimal (M9) medium with glucose as carbon source. Overnight culture of each bacterial strain was diluted 1:100 in LB or M9 medium, and the growth of bacteria was measured by the optical density of the culture at 550 nm (OD550 nm) every 2 hours for 8 hours and then at 24 hours. This incubation period includes both log phase of growth and stationary phase of bacteria. We found that OD550 nm of both ΔarcA and ΔarcB mutants appeared to be lower than that of the wild type E. coli during the log phase of growth. However, both mutants had similar bacterial concentrations and growth curves to those of the wild type E. coli when their growth was quantified by plating (Figure 1B and 1D). Therefore, no gross defect was observed in ΔarcA and ΔarcB mutants in spite of lower OD550 nm of their cultures. The anaerobic growth of the ΔarcA and ΔarcB mutant E. coli was also tested and compared to that of the wild type E. coli. No defect was detected (data not shown). Similar results were obtained with LB broth and M9 minimal medium, results obtained with LB broth are shown (Figure 1).
Table 1.
Bacterial strains, plasmids and oligonucleotides used for mutagenesis.
| Bacterial strains and plasmids | Characteristics | Source or reference | |
|---|---|---|---|
| E. coli strains | K12 | Isolate MG1655 | Dr. Sydney Kustu, University of California |
| ΔarcA | ΔarcA::kan derivative of K12 | This study | |
| ΔarcB | ΔarcB::cm derivative of K12 | This study | |
| arcB::kan | derivative of K12 in which Kanr was inserted adjacent to arcB while maintaining the function of arcB | This study | |
| ΔarcB-rev | kan derivative of ΔarcB with arcB::cm replaced by wild type arcB | This study | |
| ΔfliC | fliC non-polar deletion mutant of K12 | This study | |
| ΔarcA/ΔfliC | ΔarcA::kan/ΔfliC derivative of K12 | This study | |
| Plasmids | pRB3-273C | Apr, low to medium copy number plasmid | [40] |
| pRB3-arcA | derivative of pRB3-273C containing arcA | [38] | |
| pRB3-arcD2A | derivative of pRB3-arcA containing Asp54 → Ala mutation | This study | |
| Oligonucleotides | Used for | Sequence | |
| arcA5KO | mutagenesis of arcA | 5'-tcttatcgttgaagacgagttggtaacacgcaacacgttg aaaagtattttcgaagcggagtgtaggctggagctgcttc-3' |
|
| arcA3KO | mutagenesis of arcA | 5'-tcttccagatcaccgcagaagcgataaccttcaccgtgaa tggtggcgatgatttccggccatatgaatatcctccttag-3' |
|
| arcB5KO | mutagenesis of arcB | 5'-gccctcgtcgttcttgccattgtggtacaaatggcggtaaccatggtgct gcatggtcaggtcgaaagcattgatgttatgtgtaggctggagctgcttc-3' |
|
| arcB3KO | mutagenesis of arcB | 5'-gtggcttttgccacccacgctttcagcacttctacgtcgtgacgccactc ttctttcatctcttcaatccattcaccgaccatatgaatatcctccttag-3' |
|
| arcB-rev5 | generation of arcB::kan | 5'-cacattaatttttttaataaaaatggtacgcatcacacatttaactgattcatgtaacaa atcatttaagttttgctatcttaactgcgtcatatgaatatcctccttag-3' |
|
| arcB-rev3 | generation of arcB::kan | 5'-gcgaatactgcgccaacaccagggaaatcttggctgcgccgtaaattattatgatga gttacaagggcacagcactgtttttcaggccgcgtgtaggctggagctgcttc-3' |
|
| fliC5KO | mutagenesis of fliC | 5'-tcgctgatcactcaaaataatatcaacaagaaccagtctgcgctgtcgag ttctatcgagcgtctgtcttctggcttgcggtgtaggctggagctgcttc-3' |
|
| fliC3KO | mutagenesis of fliC | 5'-ctgcggtacctggttagcttttgccaacacggagttaccggcctgctgga tgatctgcgctttcgacatattggacacttcatatgaatatcctccttag-3' |
|
kan, kanamycin resistance cassette; cm, chloramphenicol resistance cassette. Sequences in bold in the table indicate those that are homologous to plasmids pKD3 and pKD4 [50], which were used as PCR templates for mutagenesis.
Figure 1.
Resistance of the ΔarcA and ΔarcB mutant of E. coli to H2O2. (A and B) Growth and survival of wild type E. coli (diamond), ΔarcA mutant E. coli (square), ΔarcA mutant E. coli transformed with plasmid pRB3-273C (triangle) and ΔarcA mutant E. coli transformed with plasmid pRB3-arcA (cross) in LB broth with 1.5 mM H2O2 (A) or LB broth alone (B). (C and D) Growth and survival of wild type E. coli (diamond), ΔarcB mutant E. coli (square) and ΔarcB revertant mutant E. coli (cross) in LB broth with 1.5 mM H2O2 (C) or LB broth alone (D). Bacterial concentration was determined by plating and plotted against the indicated incubation time period. At least three experiments were performed, and results from a representative experiment performed in triplicates are shown. Error bars indicate standard deviation and sometimes fall within the data label.
We assayed the resistance of the ΔarcA mutant E. coli to hydrogen peroxide (H2O2). Overnight culture of the ΔarcA mutant E. coli was exposed to H2O2, and its survival was compared to that of the wild type E. coli. The ΔarcA mutant E. coli was more susceptible than the wild type E. coli (Figure 1A). Plasmid pRB3-arcA, which carries a wild type allele of arcA in plasmid pRB3-273C [38,40], complemented the survival defects in H2O2. This indicates that the susceptible phenotype of the ΔarcA mutant E. coli was likely due to the deletion of the arcA allele (Figure 1A). Assays performed with log-phase culture of the ΔarcA mutant E. coli yielded similar results (data not shown). Similar results were obtained with LB broth and M9 minimal medium, results obtained with LB broth are shown (Figure 1).
The same analysis was carried out for ArcB, the cognate sensor-kinase of the ArcAB system. The ΔarcB mutant E. coli survived less than the wild type parental strain (Figure 1C). We had attempted to clone a wild type allele of arcB into plasmid pRB3-273C to complement the ΔarcB mutant E. coli. However, the cloning efficiency was unusually low as compared to similar cloning attempts we had conducted with the plasmid vector. Of a total of 7 recombinant plasmids we eventually obtained from several transformations, 5 contained mutations at the start codon of arcB and the remaining 2 had mutations that produced truncations early in the ORF (data not shown). This indicates that an over-expression of arcB from a plasmid is probably toxic to E. coli. As an alternative, we constructed a revertant of the ΔarcB mutant E. coli, in which a wild type arcB allele replaced the deleted arcB allele (see Materials and Methods). The revertant mutant of ΔarcB was shown to have the same resistance to H2O2 as the wild type E. coli (Figure 1C).
The ArcAB system is dispensable for H2O2 scavenge
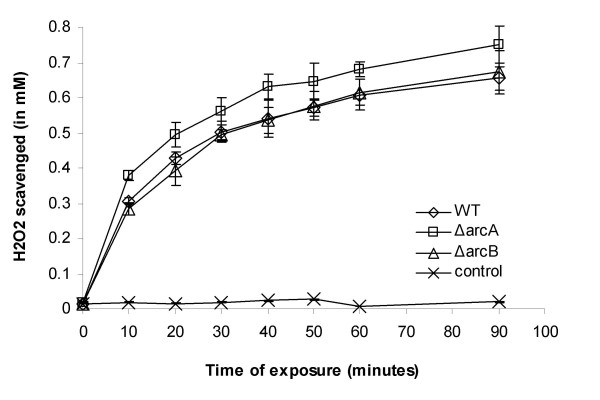
To determine the mechanism of how the ArcAB system is involved in H2O2 resistance, we analyzed the H2O2 scavenging activity of the ΔarcA and ΔarcB mutant of E. coli K12, since a defect in H2O2 scavenging activity may lead to the susceptibility to H2O2. The overnight culture was diluted in LB containing 2 mM of H2O2, and the concentration of the residual H2O2 was measured after various incubation period. The scavenge of H2O2 was measured as the reduction in H2O2 concentration over the incubation period. Our results indicate that both ΔarcA and ΔarcB mutants scavenged H2O2 normally as compared to the wild type E. coli K12., and no deficiency was observed (Figure 2).
Figure 2.
The ArcAB system is dispensable for H2O2 scavenge. The ΔarcA (square), ΔarcB (triangle) mutant and the wild type E. coli K12 (diamond) was cultured in LB broth supplemented with 2 mM of H2O2 at 37°C with shaking. Bacterial concentration and the H2O2 concentration were measured at various time points. The H2O2 scavenge was measured as the decrease of H2O2 concentration per 107 c.f.u. bacteria. A control sample without bacteria (cross) was included to monitor any possible spontaneous degradation of H2O2. The experiment was repeated at least three times, and data from one representative assay performed in duplicates were shown. Error bars indicate standard deviation and sometimes fall within the data label.
Phosphorylation at Asp54 is dispensable for H2O2 resistance mediated by ArcA
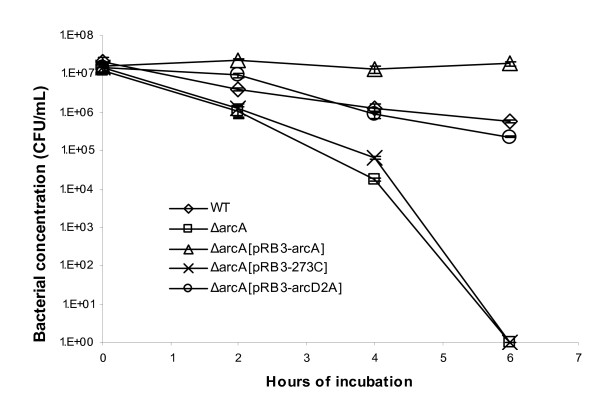
Under anaerobic conditions, ArcB is activated by reduced quinones, undergoes auto-phosphorylation, and transfers its phosphorylation to ArcA [25,32,41-43]. It is not known if ArcA is phosphorylated under aerobic conditions or if unphosphorylated ArcA has any function. To test if phosphorylation is necessary for H2O2 resistance mediated by ArcA, we generated an Asp54 → Ala mutation in ArcA in plasmid pRB3-arcA [38] and used the resulting plasmid pRB3-arcD2A to complement the ΔarcA mutant E. coli. In H2O2 resistance assays, plasmid pRB3-arcD2A rescued the ΔarcA mutant E. coli and the resistance of the mutant to H2O2 was restored to the wild type level (Figure 3). However, unlike the original plasmid pRB3-arcA, plasmid pRB3-arcD2A did not render the complemented ΔarcA mutant E. coli more resistant to H2O2 than the wild type E. coli (Figure 3).
Figure 3.
Plasmid containing phosphorylation-deficient arcA complements the ΔarcA mutant E. coli in resistance to H2O2. The wild type E. coli (diamond), ΔarcA mutant E. coli (square), ΔarcA mutant E. coli transformed with plasmid vector pRB3-273C (cross), ΔarcA mutant E. coli transformed with plasmid pRB3-arcA (triangle) and ΔarcA mutant E. coli transformed with plasmid pRB3-arcD2A which contains a phosphorylation-deficient arcA allele (circle) were incubated with LB medium containing 1.5 mM H2O2 at 37°C. The survival of bacteria was determined by plating and plotted against the indicated incubation time period. At least three experiments were performed, and results from a representative experiment performed in triplicates are shown. Error bars indicate standard deviation and sometimes fall within the data label.
Response of flagellin, OppA and GltI to H2O2 is altered in the ΔarcA mutant E. coli
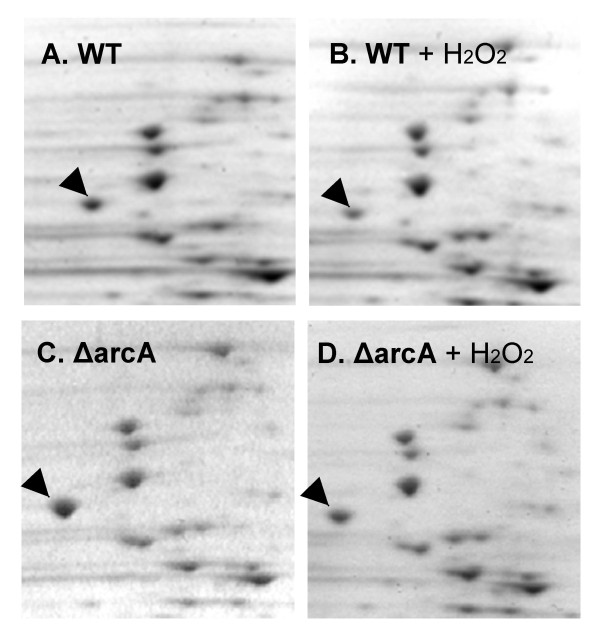
To investigate the mechanisms of H2O2 resistance mediated by ArcA, we performed two-dimensional gel electrophoresis to examine the protein profiles in the ΔarcA mutant E. coli in the presence or absence of H2O2, and compared to those of the wild type E. coli. While most proteins either were not altered by H2O2 treatment, or responded similarly to H2O2 treatment in the wild type and ΔarcA mutant E. coli, the levels of three proteins were observed to respond to H2O2 differently, the most abundant of which is shown in Figure 4. By peptide mass fingerprinting using MALDI-TOF, the prominent protein in Figure 4 was identified as flagellin encoded by fliC, while the other two less abundant proteins were identified as oligopeptide ABC transporter substrate-binding protein (OppA) and glutamate and aspartate transporter subunit (GltI) (data not shown). The levels of these proteins were quantified in the H2O2-treated and control untreated samples of the wild type and ΔarcA mutant E. coli (Table 2).
Figure 4.
Two-dimensional gel electrophoresis analysis of whole cell proteins of the wild type and ΔarcA mutant E. coli. The wild type (WT, A and B) and the ΔarcA (ΔarcA, C and D) mutant E. coli were exposed to H2O2 and total proteins from H2O2-exposed (+H2O2, B and D) and unexposed bacteria (A and C) were electrophoresed on 2-D gels. Arrows point to the flagellin protein.
Table 2.
Relative levels of differentially regulated proteins in the wild type and ΔarcA mutant of E. coli K12.
| Bacterial strain | Wild type | ΔarcA | |||
|---|---|---|---|---|---|
| Treatment | -H2O2 | + H2O2 | -H2O2 | + H2O2 | |
| Protein | FliC | 100 | 37.9 ± 16.7† | 188.9 ± 29.8† | 139.9 ± 57.8§ |
| GltI | 100 | 2555.5 ± 1343.1† | 892.0 ± 555.8† | 440.3 ± 202.2 | |
| OppA | 100 | 717.5 ± 390.5† | 205.2 ± 127.3 | 183.1 ± 67.9 | |
The level of each protein in the untreated wild type E. coli is arbitrarily set as 100, and levels of proteins in other samples are expressed as relative to the level in the untreated E. coli. Results are the average of three to five independent experiments (biological repeats) with standard deviation. † Level differs significantly from that of untreated wild type E. coli; and § level differs significantly from that of wild type E. coli treated with H2O2 (p < 0.05, Student's t-test).
Flagellin is the only one among the 10 most abundant proteins that responded to H2O2 treatment. In the wild type, un-treated E. coli flagellin was detected at a lower level than in the ΔarcA mutant E. coli, and H2O2 treatment further decreased the flagellin level (p < 0.05, Student's t-test, Table 2 and Figure 4). In the ΔarcA mutant E. coli H2O2 treatment also decreased flagellin level, however, the decrease was not statistically significant (Table 2). Therefore, compared to the wild type the E. coli, ΔarcA mutant displayed higher flagellin levels both constitutively and following H2O2 treatment, and its flagellin level did not respond to H2O2 treatment as that in the wild type E. coli.
The response of OppA and GltI expression was different from that of flagellin. In the untreated bacteria levels of both GltI and OppA appeared to be higher in the ΔarcA mutant than in the wild type E. coli (p < 0.05, Student's t-test for GltI, Table 2). Following H2O2 treatment the levels of OppA and GltI in the wild type E. coli became higher (p < 0.05, Student's t-test), while neither protein displayed a statistically significant change in the ΔarcA mutant E. coli (Table 2). This results in a lower GltI and OppA level in the H2O2 treated ΔarcA mutant than the wild type E. coli.
Flagellin messenger RNA is over-expressed in the ΔarcA mutant E. coli
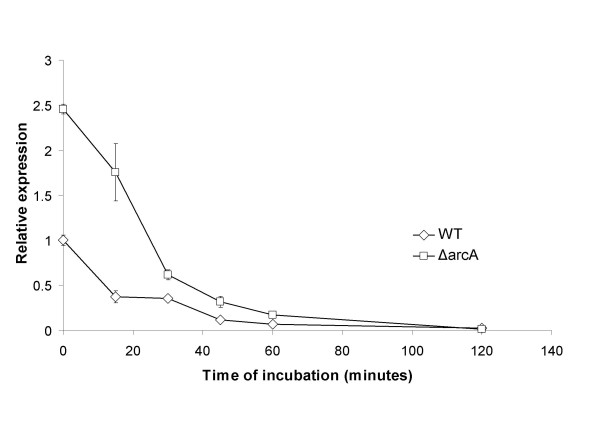
Since flagellin was one of the most abundant proteins in E. coli and the only abundant protein whose level was altered in response to H2O2, we decided to investigate the influence of flagellin on the survival of the ΔarcA mutant E. coli in the presence of H2O2. To determine if the higher protein levels of flagellin in the ΔarcA mutant E. coli was due to higher levels of mRNA, we examined the expression of the fliC transcripts by Real-Time Reverse Transcriptase PCR analysis (RT-PCR). RNA was prepared from the wild type and ΔarcA mutant E. coli before and after exposure to H2O2, and subjected to RT-PCR analysis. Similar to protein levels, the ΔarcA mutant E. coli had higher levels of fliC mRNA than the wild type E. coli both constitutively and after exposure to H2O2. In both strains, H2O2exposure reduced the fliC mRNA level progressively (Figure 5). The difference in fliC mRNA levels between the wild type and ΔarcA mutant E. coli decreased with longer exposure periods and no difference could be detected by 120 minutes of exposure (Figure 5). To determine if ArcA directly regulates fliC expression, we expressed and purified recombinant ArcA from aerobic cultures of E. coli and carried out electrophoretic mobility shift assay of the fliC upstream sequence. No specific binding was detected (data not shown).
Figure 5.
Expression of fliC messenger RNA is regulated in response to H2O2 exposure. Expression of fliC messenger RNA is regulated in response to H2O2 exposure. The wild type and the ΔarcA mutant E. coli was exposed to H2O2, and the fliC messenger RNA in wild type (diamond) and the ΔarcA mutant E. coli (square) was quantified by Real-time Reverse Transcriptase PCR after various periods of exposure. The level of the fliC messenger RNA in the unexposed wild type E. coli (at 0 hour) was arbitrarily set as 1, and levels of fliC messenger RNA in other samples were expressed as relative expression levels and plotted against the exposure time. At least three experiments were performed, and results from a representative experiment performed in triplicates are shown. Error bars indicate standard deviation.
Deletion of flagellin increased the survival of the ΔarcA mutant E. coli
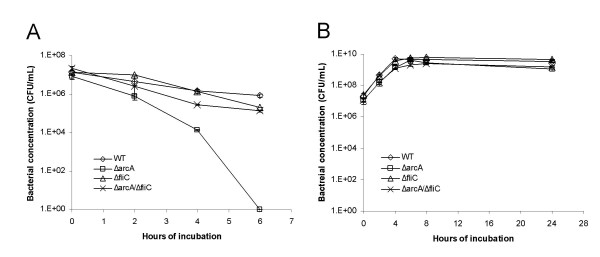
Flagellin is one of the most abundant proteins in E. coli, and we have shown that its level was higher in the ΔarcA mutant E. coli both constitutively and upon H2O2 exposure (Figure 4 and Table 2). We reasoned that expressing an abundant protein such as flagellin at a higher level might be a burden to the ΔarcA mutant E. coli, especially under stress conditions such as those caused by H2O2. We hypothesize that a deletion of flagellin encoded by fliC may facilitate the survival of the ΔarcA mutant E. coli exposed to H2O2. To test this hypothesis, we generated a non-polar ΔfliC mutant and an ΔarcA/ΔfliC double mutant E. coli. The non-polar deletion of fliC itself had no obvious effect on the survival of E. coli in the presence of H2O2 (Figure 6). However, the fliC deletion improved the survival of the ΔarcA mutant E. coli, and the survival of the ΔarcA/ΔfliC double mutant E. coli was close to that of the wild type E. coli (Figure 6). This indicates that elimination of flagellin in the ΔarcA mutant E. coli enhanced its survival under H2O2 stress.
Figure 6.
Deletion of fliC increased the resistance of the ΔarcA mutant E. coli to H2O2. Growth and survival of wild type E. coli (diamond), ΔarcA mutant E. coli (square), ΔfliC mutant E. coli (triangle) and ΔarcA/ΔfliC double mutant E. coli (cross) in LB medium containing 1.5 mM H2O2 (a) or LB broth alone (b). The survival of bacteria was determined by plating and plotted against the indicated incubation time period. At least three experiments were performed, and results from a representative experiment performed in triplicates are shown. Error bars indicate standard deviation and sometimes fall within the data label.
In addition to flagellin, we have also attempted to delete other abundant proteins to determine if such deletions would improve the survival of the arcA mutant E. coli. Our efforts were not successful, however, because most abundant proteins such as elongation factors, 30 s ribosomal proteins, and chaperone proteins are either essential or important for E. coli, and such deletions would be detrimental to E. coli. We successfully deleted D-ribose periplasmic binding protein (RbsB) encoded by rbsB, a protein which is as abundant as or more abundant than flagellin. The ΔrbsB mutant itself was found to be susceptible to H2O2, therefore could not be used to test the effect of RbsB on the H2O2 resistance of the arcA mutant E. coli (data not shown).
Amino acid supplementation improved the survival of the ΔarcA mutant E. coli under H2O2 stress
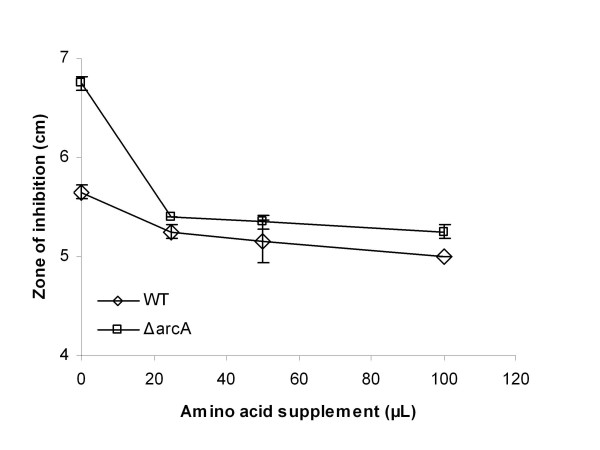
We described above that a deletion of flagellin in E. coli improved the survival of the ΔarcA mutant E. coli in the presence of H2O2. Our analysis of the proteome of the wild type and ΔarcA mutant E. coli indicated that levels of glutamine/aspartate periplasmic binding protein (GltI) and oligopeptide binding protein precursor (OppA) increased in the ΔarcA mutant as compared to the wild type E. coli (Table 2). In addition, the ΔarcA mutant E. coli failed to increase GltI and OppA protein levels in response to H2O2 as the wild type E. coli. This suggests that E. coli may have an increased need for amino acids under H2O2 stress and the ΔarcA mutant E. coli may benefit from amino acid supplementation. To test this hypothesis, we determined the effect of amino acid supplementation on the survival of the ΔarcA mutant E. coli in the presence of H2O2. To facilitate a direct comparison between the resistance of the wild type and ΔarcA mutant E. coli to H2O2 with or without amino acid supplementation, we carried out a disc diffusion assay, and bacterial resistance to H2O2 was measured by the diameter of the zone of inhibition (ZOI). Without amino acid supplementation the ZOI of the ΔarcA mutant E. coli was significantly larger than that of the wild type E. coli (Figure 7). With amino acid supplementation, sizes of the ZOI reduced for both the wild type and the ΔarcA mutant E. coli, and the difference in the sizes of the ZOI between wild type and ΔarcA mutant E. coli diminished with amino acid supplementation (Figure 7). We tested single amino acids and combinations of various amino acids, and none of the combinations tested was able to complement the susceptibility of the ΔarcA mutant E. coli as the total amino acids (data not shown).
Figure 7.
Amino acid complementation increased the resistance of E. coli to H2O2 and reduced the difference in H2O2 resistance between the wild type and ΔarcA mutant E. coli. Resistance of wild type (diamond) and the ΔarcA mutant E. coli (square) to H2O2 was assayed by the ability to grow in the presence of H2O2 and more resistant bacteria show a smaller diameter of inhibition. Various volumes of 20 mM amino acid solution was spread onto each M9 minimal medium plate containing approximately 1 × 106 c.f.u. wild type or ΔarcA mutant E. coli and a paper disc of 1/4" with 10 μl of 30% H2O2 was added to the center of each plate. Zone of inhibition was measured after overnight incubation and plotted against the volume of amino acid supplementation. At least three experiments were performed, and results from a representative experiment performed in triplicates are shown. Error bars indicate standard deviation and sometimes fall within the data label..
Antibiotic that inhibits protein synthesis increased susceptibility of E. coli to H2O2
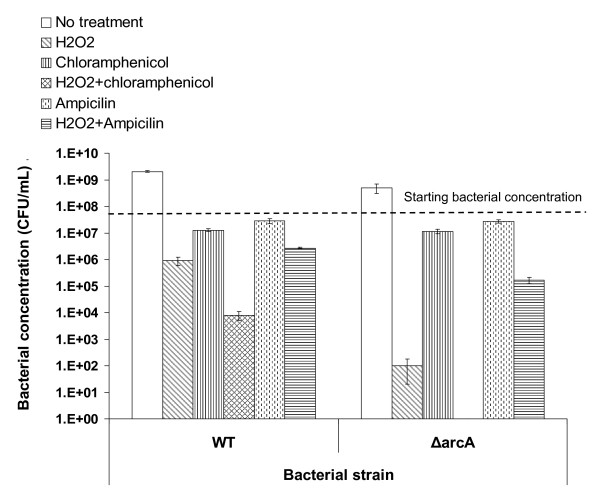
To test if protein synthesis is important for bacterial survival and if protein synthesis inhibition is detrimental to bacteria under reactive oxygen stress, we assayed the resistance of E. coli to H2O2 in the presence of chloramphenicol, an antibiotic that inhibits peptide bond formation and hence protein synthesis. Without H2O2 or antibiotic, wild type E. coli grew approximately 2log10 during 6 hours of incubation (Figure 8, left half, open bar). Hydrogen peroxide was bactericidal and the bacterial concentration decreased for over 1log10 (Figure 8, left half, diagonally-hatched bar). Supplementation of chloramphenicol alone prohibited bacterial proliferation and the bacterial concentration decreased slightly (Figure 8, left half, vertically-hatched bar). Incubation in the presence of both H2O2 and chloramphenicol was more detrimental to E. coli than either H2O2 or chloramphenicol alone, and the bacterial concentration decreased by nearly 4log10 (Figure 8, left half, cross-hatched bar). This indicates that chloramphenicol enhanced the bactericidal activity of H2O2. To determine if this enhanced bactericidal activity is due to the bacteriostatic activity of chloramphenicol, we tested the effect of ampicillin, an antibiotic that inhibits the bacterial cell wall synthesis, in the same assay. When added alone, ampicillin had similar effect on bacterial growth as chloramphenicol did (Figure 8, left half, dotted line-filled bar). However, in contrast to chloramphenicol that enhanced the bactericidal effect of H2O2 (Figure 8, left half, cross-hatched bar), the addition of ampicillin reduced the bactericidal activity of H2O2 for unknown reasons (Figure 8, left half, compare horizontally hatched bar to diagonally-hatched bar). This indicates that the synergistic effect of chloramphenicol on the bactericidal activity of H2O2 is not due to its bacteriostatic effect and suggests that protein synthesis is important for E. coli to resist the killing by H2O2.
Figure 8.
Chloramphenicol enhanced the bactericidal activity of H2O2. The wild type E. coli (WT) and the ΔarcA mutant E. coli (ΔarcA) were incubated in M9 minimal medium containing 1.5 mM H2O2 for 6 hours at 37°C. The survival of bacteria was determined by plating. Bacterial concentration following each treatment (open bars, no treatment; diagonally-hatched bars, H2O2; vertically-hatched bars, 25 μg ml-1 of chloramphenicol; cross-hatched bars, H2O2 and 25 μg ml-1 of chloramphenicol; dotted line-hatched bars, 50 μg ml-1 ampicillin and horizontally-hatched bars, H2O2 and 50 μg ml-1 ampicillin) was plotted on the graph. The horizontal dashed line indicates the starting concentration of bacteria.
Similar assays were carried out with the ΔarcA mutant E. coli and the results were consistent with those of the wild type E. coli. While incubation with H2O2 alone reduced the concentration of the ΔarcA mutant E. coli by over 5log10 after 6 hours of incubation (Figure 8, right half, diagonally-hatched bar), the addition of chloramphenicol to the assay eliminated all E. coli (Figure 8). The synergistic effect of the bactericidal activity of H2O2 and chloramphenicol on the ΔarcA mutant E. coli is not because it is more susceptible to chloramphenicol (Figure 8, vertically-hatched bars). Similarly to that observed with wild type E. coli, ampicillin reduced the bactericidal activity of H2O2, and the ΔarcA mutant E. coli survived better in the presence of both ampicillin and H2O2 than H2O2 alone (1.7 × 105 CFU/ml vs. 1.0 × 102 CFU/ml) (Figure 8).
Discussion
Although the ArcAB system has been extensively investigated for its role as the global control system of E. coli in anaerobic growth, its role, if any, in aerobic growth is much less understood. We have previously reported that ArcA is necessary for the pathogenic bacterium Salmonella enterica to resist reactive oxygen and nitrogen species under aerobic conditions [38]. In this report, we used E. coli as our model to further explore the role of both ArcA and ArcB in ROS resistance, and to investigate the mechanism of ROS resistance mediated by the ArcAB two-component system. Here we demonstrate that deletion mutants of ArcA and ArcB were more susceptible to H2O2, suggesting that both ArcA and ArcB were necessary for E. coli to resist the stress caused by H2O2 (Figure 1), and that their functions were not limited to anaerobic growth of bacteria. Interestingly, we have not detected any growth defects of ΔarcA or ΔarcB mutant E. coli under anaerobic conditions (data not shown) and to our knowledge no such defect has been reported in the literature. In addition, an ΔarcA mutant of Salmonella enterica grew normally in anaerobic medium [38]. This further indicates that ArcAB has wider roles in the physiology and metabolism of enteric bacteria besides its well-characterized regulation of anaerobic growth of bacteria.
The signaling pathway of the ArcAB system under anaerobic conditions has been extensively characterized [25-28,30-34,42,44]. The membrane-bound sensor-kinase ArcB is activated by reduced quinones under anaerobic conditions, and subsequently activates its cognate transcriptional regulator ArcA by phosphorylating ArcA at Asp54 [30,42,25]. Matsushika and Mizuno previously reported that ArcB can also phosphorylate ArcA directly through His292 under aerobic conditions [45], however, its physiological relevance to E. coli has not been reported. Our results on the role of ArcAB in ROS resistance suggest that ArcAB can be activated by novel signals other than reduced quinones and anaerobic conditions, and the activation is independent of phosphorylation at Asp54 of ArcA as demonstrated under anaerobic conditions [41,42,46], since phosphorylation-defective ArcA expressed from a plasmid fully complemented an ΔarcA mutant E. coli for its susceptibility to H2O2 (Figure 3). We would like to point out that our analysis was conducted using a phosphorylation-mutant ArcA (Asp54 → Ala) expressed from a plasmid. It is yet to be determined if a mutant carrying a corresponding mutation of arcA in the chromosome is susceptible to H2O2. (Our attempts to generate a mutant arcA encoding an Asp54 → Ala mutation in the chromosome were unsuccessful due to technical difficulties. Similar to what we observed for arcB, plasmids carrying arcA were prone to mutations during cloning.) We have also noticed that the wild type ArcA expressed from a plasmid confers a stronger H2O2 resistance phenotype than the phosphorylation-defective ArcA. The ΔarcA mutant E. coli complemented in trans with a wild type arcA allele demonstrated higher H2O2 resistance than the wild type E. coli (Figure 1 and 3), while the same mutant E. coli complemented with a phosphorylation-defective arcA allele has the same H2O2 resistance as the wild type E. coli (Figure 3).
In addition to novel signals and signaling pathways that may mediate the function of the ArcAB system in the ROS resistance, the ArcAB system may also regulate a distinct set of genes under aerobic conditions. Under anaerobic conditions ArcA mostly negatively regulates genes involved in the TCA cycle and electron transport [26-28]. Under aerobic conditions, a microarray study by Oshima et al. demonstrated that expression of a large number of genes in the ΔarcA or ΔarcB mutant E. coli was altered [23]. Our results suggest that levels of at least three proteins (flagellin, GltI and OppA) were altered in the ΔarcA mutant E. coli both constitutively and in response to H2O2 treatment (Figure 4 and Table 2). Our further analysis on the messenger RNA level of fliC indicates that the RNA levels are higher in the ΔarcA mutant E. coli and corresponded to the protein levels, suggesting that the regulation is likely on the transcriptional or post-transcriptional level (Figure 5). Oshima et al. did not detect a significant alteration in the expression of fliC in their microarray analysis, although flagellar synthesis was identified as a system that was affected in the ΔarcA mutant but not the ΔarcB mutant E. coli [23]. The discrepancy is possibly due to the differences in experimental conditions (shaking bacterial cultures at 120 rpm vs. 225 rpm) and detection methods (microarray vs. Real-Time Reverse Transcriptase PCR and 2-D gel electrophoresis). Since we detected an elevation of both mRNA and protein levels of flagellin in the ΔarcA mutant E. coli (Figures 4 and 5), we believe that our observation is valid. The regulation of ArcA on flagellin is likely to be indirect, as we did not detect specific binding of recombinant ArcA protein to the upstream sequence of fliC (data not shown).
Given that the ArcAB system regulates a large number of genes in E. coli, its role in the ROS resistance is likely to be complex. We have demonstrated that mutation of ArcA or ArcB did not alter the H2O2 scavenging ability of E. coli (Figure 2), however, the precise molecular mechanism on how ArcA regulates ROS resistance in E. coli is yet to be elucidated. ArcA was reported to be necessary for the ROS resistance of Haemophilus influenzae due to its regulation of Dps, a ferritin-like small protein that was previously reported to be involved in ROS resistance of Salmonella [39,47]. The mechanism of the ROS resistance mediated by ArcA is likely to be different in E. coli, since dps is expressed close to the wild type level in the ΔarcA or ΔarcB mutant (84% and 99% respectively), and our preliminary microarray analysis with Salmonella ΔarcA mutant indicated that dps responded normally to H2O2 in the ΔarcA mutant (unpublished results). One possible clue on the mechanism of how ArcAB contributes to the ROS resistance of E. coli came from our proteomic analysis that showed altered expression of flagellin, GltI and OppA between the wild type and ΔarcA mutant E. coli (Table 2). The constitutive GltI and OppA levels are higher in the ΔarcA mutant than in the wild type E. coli, suggesting that the mutant may have a higher need for amino acid transport. In contrast to the GltI and OppA levels in the wild type E. coli that increased 6- and 24-fold respectively in response to H2O2 exposure (possibly due to a higher need for amino acid transport under ROS stress), the level of neither protein in the ΔarcA mutant increased under the same condition (Table 2). A higher level of flagellin in the ΔarcA mutant likely put further constraint on the protein synthesis, and as a result the ΔarcA mutant E. coli might have become less fit under H2O2 stress. Our genetic study demonstrating that deletion of fliC "rescued" the survival defect of the ΔarcA mutant E. coli under H2O2 stress (Figure 6) supports the hypothesis.
ROS stress conditions induce growth arrest in E. coli. Chang et al. has reported that in growth arrest induced by either glucose-lactose diauxie, entry into stationary phase, or H2O2 treatment, genes involved in amino acid biosynthesis pathways are down-regulated except those of histidine and arginine biosynthesis [24]. Recently, Jang and Imlay have shown that H2O2 damages enzymes with iron-sulfur and impairs bacterial metabolism, especially the biosynthesis of leucine [48]. This down regulation of amino acid synthesis may cause a strain on the protein synthesis of bacteria. Our results indicate that protein synthesis is important for E. coli to survive H2O2 treatment. Chloramphenicol, an antibiotic inhibiting protein synthesis, reduced the survival of both the wild type and ΔarcA mutant E. coli after H2O2 treatment, while ampicillin did not (Figure 8). Consistently, amino acid supplementation enhanced the survival of E. coli after H2O2 treatment (Figure 7). This is in agreement with the report by Calioz and Touati that amino acid supplementation facilitates the survival of superoxide dismutase-deficient E. coli under aerobic conditions [49].
Although our results and results from other investigators suggest that protein synthesis and amino acid availability are important for E. coli to survive ROS stress and the global regulatory system ArcAB plays a role this aspect of ROS stress resistance, protein synthesis and amino acid availability may be only one aspect of the pleiotropic effect of ArcAB system on E. coli, since chloramphenicol-treated ΔarcA mutant was still more susceptible than the similarly treated wild type E. coli. Further studies are necessary to elucidate more molecular mechanisms that control the ROS resistance mediated by the ArcAB global regulatory system.
Conclusion
The global regulatory system ArcAB of E. coli regulate many important functions of bacteria including anaerobic growth, motility, and cell division. Here we demonstrate that ArcAB regulates ROS resistance under aerobic condition, and the signalling pathway of this regulation is distinct from that under anaerobic conditions. The ArcAB system may regulate protein and amino acid synthesis and transport that influence the fitness of E. coli under ROS stress.
Methods
Reagents
Growth media for bacteria were purchased from Becton Dickinson and Company (Franklin Lakes, NJ). Anaerobic peptone-yeast medium was obtained from Anaerobe Systems (Morgan Hills, CA). Chemicals and antibiotics were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO) unless otherwise indicated. Restriction and modifying enzymes for manipulating DNA were purchased from the New England Biolabs (Beverly, MA). Custom oligonucleotides were purchased from Sigma Genosys (The Woodlands, TX).
Bacterial strains and plasmids
E. coli strain K12 isolate MG1655 (gift from Dr. Sydney Kustu, University of California) was used as the parental strain in all analyses described in this report. Mutagenesis was carried out using the one-step mutagenesis method by Datsenko and Wanner [50]. Mutant bacterial strains and sequences of oligonucleotides used for mutagenesis are listed in Table 1. In the ΔarcA mutant, the wild type arcA allele was replaced by a kanamycin-resistance cassette (Kanr). In the ΔarcB mutant, the wild type arcB allele was replaced by a chloramphenicol-resistance cassette (Cmr). Each mutation was transduced into fresh E. coli by general transduction with phage P1 before further analysis. In the ΔfliC mutant, the wild type fliC allele was replaced by Cmr, which was subsequently removed to generate a non-polar mutant [50]. The ΔarcA/ΔfliC mutant was prepared by transducing arcA::kan from the ΔarcA mutant into the ΔfliC non-polar mutant E. coli. A revertant of ΔarcB mutant E. coli was generated through a two-step process. First, a mutant, arcB(Kanr), was generated in which Kanr was inserted downstream to the arcB coding sequence without affecting the arcB open reading frame. Subsequently, phage P1 was prepared from arcB(Kanr) and used to transduce the ΔarcB mutant E. coli. Kanamycin-resistant and chloramphenicol-sensitive colonies were selected, in which the deletion mutant arcB allele in the ΔarcB mutant E. coli was replaced by a wild type allele from arcB(Kanr). The genome structure surrounding the arcB allele was determined to verify that wild type arcB allele was restored. The resultant bacterial strain was referred to as ΔarcB-rev.
Plasmid pRB3-arcA used to complement the ΔarcA mutant E. coli was described previously [38]. Plasmid pRB3-arcD2A was constructed using megaprimer method as described [51]. Briefly, a 260-bp section of the arcA gene that included the Asp54 was amplified using mutagenesis primer 5'-CAACCTGGTGATCATGGCGATCAATCTGCC-3' and an arcA primer 5'-CAACGCTACGACGCTCTTC-3'. Sequence in bold in the mutagenesis primer introduced an aspartate to alanine mutation (Asp → Ala) at amino acid 54 in ArcA. The PCR product was used as a megaprimer to amplify plasmid pRB3-arcA together with a vector primer 5'-GTTTTCCCAGTCACGAC-3'. The PCR product was subsequently digested with KpnI and cloned into KpnI-digested plasmid pRB3-arcA to replace the wild type arcA gene with the corresponding sequence that introduced an Asp54 → Ala mutation. The resulting plasmid pRB3-arcD2A contained the same sequence as the original plasmid pRB3-arcA except that GAT which codes for Asp54 of ArcA was mutated to GCG which codes for Ala.
Survival assays of bacteria after exposure to oxidative and other stresses
Survival of E. coli after H2O2 and other stress conditions was assayed as described previously [38,52]. E. coli was cultured in 2 ml of Luria Bertani (LB) broth at 37°C overnight with shaking at 225 rpm. Antibiotics were added as appropriate. Twenty microliters of overnight cultures were added to 2 ml of LB containing one of the following chemicals: hydrogen peroxide, sodium chloride, or sodium dodecyl sulfate (SDS). Cultures in all assays were grown aerobically by shaking at 225 rpm. After exposure to H2O2 or other stresses, aliquots of cultures were diluted and plated in triplicates. Bacterial colonies were enumerated as colony-forming units (CFU) after overnight incubation to determine the bacterial concentration. Disc diffusion assay was carried out as described previously [52]. Briefly, approximately 1 × 106 cfu bacteria were plated onto M9 minimal agar plates and paper discs of 1/4" diameter loaded with 10 μl of 30% H2O2 were placed in the center of plates onto the bacterial lawn. Plates were incubated overnight at 37°C, and the diameter of the inhibitory zone on each plate was measured.
Scavenging of H2O2 by E. coli
Wild type, the ΔarcA and the ΔarcB mutant E. coli were cultured overnight in LB broth at 37°C with shaking at 225 rpm. Twenty microliters of overnight bacterial culture was diluted in 1 mL of fresh LB broth containing 2 mM of H2O2 that had been pre-warmed to 37°C. An aliquot of 100 μL was taken as the 0 minute sample, and rest of the cultures were incubated at 37°C with shaking. Subsequently, aliquots were taken at 10' intervals. Aliquots of bacterial cultures were used for plating to determine the bacterial concentration, and the rest of the samples were used to determine the concentration of H2O2. A control sample of LB supplemented with H2O2 that contained no bacteria was included in all assays for spontaneous degradation of H2O2.
The concentration of H2O2 in bacterial cultures was determined as described [53]. Briefly, bacterial cultures were spun down to remove bacteria and 40 μL of supernatant was diluted in 260 μL of 50 mM potassium phosphate (pH7.0). Diluted supernatant was mixed with 600 μL of a reaction mixture containing 500 nM H2O2, 2.5 mM phenol, 0.5 mM 4-aminoantipyrine, 40 μg horseradish peroxidase, and 1 mM potassium phosphate (pH 7.0) [53]. The reactions were incubated at room temperature for approximately 10' till color stabilized, and OD505 nm was measured for each sample. The concentration of H2O2 was determined by a standard curve generated with known concentrations of H2O2 in LB broth. The H2O2 scavenging was determined as (initial H2O2 concentration – residual H2O2 concentration) (in mM)/bacterial concentration (in 107 cfu/mL).
Real-Time Reverse Transcriptase Polymerase Chain Reaction (RT-PCR) analysis of gene expression
To analyze the expression of fliC messenger RNA, we cultured the wild type and ΔarcA mutant E. coli in LB broth to log phase and divided each culture into two aliquots. One of the aliquots was exposed to 5 mM H2O2 and samples were taken after different exposure periods. The other aliquot was used as an unexposed control. Total RNA was purified from E. coli using TRIzol reagent (Invitrogen, Carlsbad, CA) followed by digestion with DNase I (Qiagen, Valencia, CA) and purification by RNeasy kit (Qiagen, Valencia, CA). Subsequently, 1.5 μg RNA were reverse-transcribed using M-MLV reverse transcriptase (Promega, Madison, WI), and cDNA samples were used for Real-Time Reverse Transcriptase PCR analysis (RT-PCR). RT-PCR was performed using the iQ SYBR Green PCR supermix (Bio-Rad, Hercules, CA) in an iCycler (Bio-Rad, Hercules, CA). Primers 5'-GGCGGAACTAACCCAGCTTCA-3' and 5'-TGCTCCAGTCGCCATTGTCA-3' were used for the RT-PCR analysis of fliC expression. The 16S ribosomal RNA level was determined with primers 5'-GGGACCTTCGGGCCTCTTG-3' and 5'-ACCGTGTCTCAGTTCCAGTGTGG-3', and was used to normalize expression levels of fliC from different samples.
Q-Gene program and Relative Expression Software Tool (REST) were used for data analysis of threshold cycle numbers from the iCycler [54,55]. Mean values of normalized expression and standard error measurements were determined as described [54]. Comparisons of mean normalized expression were used to calculate expression ratios. REST was used to obtain statistical significance (p-value) as described [55].
Bacterial extracts and two-dimensional (2-D) gel electrophoresis
E. coli was cultured in LB broth overnight at 37°C with shaking. Overnight bacterial culture was diluted 1:100 in fresh LB and cultured for 4 hours at 37°C with shaking, and then split into two aliquots. Hydrogen peroxide was added to 5 mM to one of the aliquots, and both aliquots were further incubated for 2 hours at 37°C with shaking. Bacterial cultures were chilled on ice immediately and spun down. Bacterial pellets were then resuspended in 8 M urea and 4% CHAPS in 10 mM Tris 8.0 and sonicated. The insoluble fraction was removed by centrifugation, and soluble lysate was used for 2-D gel electrophoresis.
Two-dimensional gel electrophoresis of E. coli proteins was performed with the Zoom IPG Runner system following the manufacturer's instructions (Invitrogen, Carlsbad, CA). One hundred fifty micrograms of cellular proteins were diluted in rehydration buffer (8 M urea, 4% CHAPS and 0.5% pH 3–10 ampholytes) and loaded onto each pH 3–10 ZOOM strip (Invitrogen, Carlsbad, CA). The first dimension electrophoresis was carried out at 200 V for 20', 450 V for 15', 750 V for 15' and 2000 V for 60'. After isoelectric focusing, ZOOM strips were reduced and alkylated with 125 mM iodoacetamide and electrophoresed on NuPAGE Novex 4–12% Bis-Tris ZOOM gels (Invitrogen, Carlsbad, CA) at 100 V for 90'. Proteins were visualized by staining with ProteomIQ reagents (Proteome Systems, Woburn, MA), and then scanned with a HP Scanjet 5530 scanner (Hewlett-Packard, Palo Alto, CA). Individual proteins were quantified using ImageQuant (Amersham Biosciences, Piscataway, NJ) and normalized against the total protein content of the gel.
Mass spectrometry analysis of protein spots
Protein spots of interest were excised from gels and washed with 50% acetonitrile in 50 mM ammonium bicarbonate twice for 15' each. The gel spots were then dehydrated in acetonitrile for 30' and dried in a speed vac for 10'. Thirty microliters of 50 mM ammonium bicarbonate containing 0.3 μg of trypsin (Sigma-Aldrich, St Louis, MO) were added to each sample, and samples were incubated at 37°C for 16 hours. Digested peptides were extracted from gel spots by two washes of 50% acetonitrile/0.1% trifluoroacetic acid, and purified with Ziptips (Millipore, Billerica, MA). Purified peptides were eluted from Ziptips with 50% acetonitrile/0.05% trifluoroacetic acid with 10 mg/ml alpha-cyano-4-hydroxycinnamic acid, and spotted on a sample plate to obtain mass spectra using an Axima CFR Plus MALDI-ToF mass spectrometer (Shimadzu Biotech, Columbia, MD). Each spectrum was calibrated externally using the ProteoMass peptide MALDI-MS calibration kit (Sigma-Aldrich, St Louis, MO).
Peptide fingerprints obtained for each sample were used to search the databases at NCBI and SWISS-PROT using MASCOT search engine http://www.Matrixscience.com. Search parameters used were variable carbamidomethyl and propionamide modifications of cysteines and oxidation of methionines. A peptide tolerance window of 0.5 daltons was used for all searches. Once an identification was made with a statistically significant score, data were accepted when the peptide coverage of the protein was at least 20%, and the molecular weight and isoelectric point of the protein matched those observed on the 2D gel electrophoresis.
Authors' contributions
CL participated in the study design, carried out the microbiological studies and helped to draft the manuscript. AC carried out the microbiological studies. SL conceived of the study, participated in the study design, carried out the microbiological studies, performed the statistical analysis and drafted the manuscript. All authors read and approved the final manuscript..
Contributor Information
Cindy Loui, Email: csloui@berkeley.edu.
Alexander C Chang, Email: alex_c@berkeley.edu.
Sangwei Lu, Email: sangwei@berkeley.edu.
Acknowledgements
We thank Drs. Stuart Linn and Hiroshi Nikaido for insightful discussions. This work was supported by USDA CALR-2005-01892 (to S. L.).
References
- Hoch JA. Two-Component Signal Transduction. Washington, DC: American Society for Microbiology Press; 1995. [Google Scholar]
- Nixon BT, Ronson CW, Ausubel FM. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc Natl Acad Sci USA. 1986;83:7850–7854. doi: 10.1073/pnas.83.20.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Weiner L. Cellular and molecular physiology of Escherichia coli in the adaptation to aerobic environments. J Biochem (Tokyo) 1996;120:1055–1063. doi: 10.1093/oxfordjournals.jbchem.a021519. [DOI] [PubMed] [Google Scholar]
- Bauer CE, Elsen S, Bird TH. Mechanisms for redox control of gene expression. Annual Review of Microbiology. 1999;53:495–523. doi: 10.1146/annurev.micro.53.1.495. [DOI] [PubMed] [Google Scholar]
- Hidalgo E, Ding H, Demple B. Redox signal transduction via iron-sulfur clusters in the SoxR transcription activator. Trends Biochem Sci. 1997;22:207–210. doi: 10.1016/S0968-0004(97)01068-2. [DOI] [PubMed] [Google Scholar]
- Demple B. Study of redox-regulated transcription factors in prokaryotes. Methods. 1997;11:267–278. doi: 10.1006/meth.1996.0421. [DOI] [PubMed] [Google Scholar]
- Ding H, Demple B. Glutathione-mediated destabilization in vitro of [2Fe-2S] centers in the SoxR regulatory protein. Proc Natl Acad Sci USA. 1996;93:9449–9453. doi: 10.1073/pnas.93.18.9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunoshiba T, Hidalgo E, Amabile Cuevas CF, Demple B. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J Bacteriol. 1992;174:6054–6060. doi: 10.1128/jb.174.19.6054-6060.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha ER, Owens G Jr, Smith CJ. The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis. J Bacteriol. 2000;182:5059–5069. doi: 10.1128/JB.182.18.5059-5069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Storz G. Redox sensing by prokaryotic transcription factors. Biochem Pharmacol. 2000;59:1–6. doi: 10.1016/S0006-2952(99)00289-0. [DOI] [PubMed] [Google Scholar]
- Storz G, Altuvia S. OxyR regulon. Methods Enzymol. 1994;234:217–223. doi: 10.1016/0076-6879(94)34088-9. full_text. [DOI] [PubMed] [Google Scholar]
- Tao K, Makino K, Yonei S, Nakata A, Shinagawa H. Molecular cloning and nucleotide sequencing of oxyR, the positive regulatory gene of a regulon for an adaptive response to oxidative stress in Escherichia coli: homologies between OxyR protein and a family of bacterial activator proteins. Mol Gen Genet. 1989;218:371–376. doi: 10.1007/BF00332397. [DOI] [PubMed] [Google Scholar]
- Sawers G. The aerobic/anaerobic interface. Curr Opin Microbiol. 1999;2:181–187. doi: 10.1016/S1369-5274(99)80032-0. [DOI] [PubMed] [Google Scholar]
- Unden G, Schirawski J. The oxygen-responsive transcriptional regulator FNR of Escherichia coli: the search for signals and reactions. Mol Microbiol. 1997;25:205–210. doi: 10.1046/j.1365-2958.1997.4731841.x. [DOI] [PubMed] [Google Scholar]
- Unden G, Achebach S, Holighaus G, Tran HG, Wackwitz B, Zeuner Y. Control of FNR function of Escherichia coli by O2 and reducing conditions. J Mol Microbiol Biotechnol. 2002;4:263–268. [PubMed] [Google Scholar]
- Gunsalus RP, Park SJ. Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res Microbiol. 1994;145:437–450. doi: 10.1016/0923-2508(94)90092-2. [DOI] [PubMed] [Google Scholar]
- Spiro S. The FNR family of transcriptional regulators. Antonie Van Leeuwenhoek. 1994;66:23–36. doi: 10.1007/BF00871630. [DOI] [PubMed] [Google Scholar]
- Jordan PA, Thomson AJ, Ralph ET, Guest JR, Green J. FNR is a direct oxygen sensor having a biphasic response curve. FEBS Lett. 1997;416:349–352. doi: 10.1016/S0014-5793(97)01219-2. [DOI] [PubMed] [Google Scholar]
- Becker S, Holighaus G, Gabrielczyk T, Unden G. O2 as the regulatory signal for FNR-dependent gene regulation in Escherichia coli. J Bacteriol. 1996;178:4515–4521. doi: 10.1128/jb.178.15.4515-4521.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiley PJ, Beinert H. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol Rev. 1998;22:341–352. doi: 10.1111/j.1574-6976.1998.tb00375.x. [DOI] [PubMed] [Google Scholar]
- Crack J, Green J, Thomson AJ. Mechanism of oxygen sensing by the bacterial transcription factor fumarate-nitrate reduction (FNR) J Biol Chem. 2004;279:9278–9286. doi: 10.1074/jbc.M309878200. [DOI] [PubMed] [Google Scholar]
- Constantinidou C, Hobman JL, Griffiths L, Patel MD, Penn CW, Cole JA, Overton TW. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J Biol Chem. 2006;281:4802–4815. doi: 10.1074/jbc.M512312200. [DOI] [PubMed] [Google Scholar]
- Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, Mori H, Mizuno T. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol Microbiol. 2002;46:281–291. doi: 10.1046/j.1365-2958.2002.03170.x. [DOI] [PubMed] [Google Scholar]
- Chang DE, Smalley DJ, Conway T. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol Microbiol. 2002;45:289–306. doi: 10.1046/j.1365-2958.2002.03001.x. [DOI] [PubMed] [Google Scholar]
- Georgellis D, Kwon O, Lin EC. Quinones as the redox signal for the Arc two-component system of bacteria. Science. 2001;292:2314–2316. doi: 10.1126/science.1059361. [DOI] [PubMed] [Google Scholar]
- Iuchi S, Lin EC. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci USA. 1988;85:1888–1892. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Cameron DC, Lin EC. A second global regulator gene (arcB) mediating repression of enzymes in aerobic pathways of Escherichia coli. J Bacteriol. 1989;171:868–873. doi: 10.1128/jb.171.2.868-873.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Matsuda Z, Fujiwara T, Lin EC. The arcB gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the arc modulon. Mol Microbiol. 1990;4:715–727. doi: 10.1111/j.1365-2958.1990.tb00642.x. [DOI] [PubMed] [Google Scholar]
- Liu X, De Wulf P. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J Biol Chem. 2004;279:12588–12597. doi: 10.1074/jbc.M313454200. [DOI] [PubMed] [Google Scholar]
- Georgellis D, Lynch AS, Lin EC. In vitro phosphorylation study of the Arc two-component signal transduction system of Escherichia coli. J Bacteriol. 1997;179:5429–5435. doi: 10.1128/jb.179.17.5429-5435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpica R, Sandoval GR, Rodriguez C, Franco B, Georgellis D. Signaling by the arc two-component system provides a link between the redox state of the quinone pool and gene expression. Antioxid Redox Signal. 2006;8:781–795. doi: 10.1089/ars.2006.8.781. [DOI] [PubMed] [Google Scholar]
- Iuchi S. Phosphorylation/dephosphorylation of the receiver module at the conserved aspartate residue controls transphosphorylation activity of histidine kinase in sensor protein ArcB of Escherichia coli. J Biol Chem. 1993;268:23972–23980. [PubMed] [Google Scholar]
- Iuchi S, Lin EC. Mutational analysis of signal transduction by ArcB, a membrane sensor protein responsible for anaerobic repression of operons involved in the central aerobic pathways in Escherichia coli. J Bacteriol. 1992;174:3972–3980. doi: 10.1128/jb.174.12.3972-3980.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Lee YS, Han JS, Kim JB, Hwang DS. Multimerization of phosphorylated and non-phosphorylated ArcA is necessary for the response regulator function of the Arc two-component signal transduction system. J Biol Chem. 2001;276:40873–40879. doi: 10.1074/jbc.M104855200. [DOI] [PubMed] [Google Scholar]
- Nystrom T, Larsson C, Gustafsson L. Bacterial defense against aging: role of the Escherichia coli ArcA regulator in gene expression, readjusted energy flux and survival during stasis. Embo J. 1996;15:3219–3228. [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Han JS, Jeon Y, Hwang DS. The arc two-component signal transduction system inhibits in vitro Escherichia coli chromosomal initiation. J Biol Chem. 2001;276:9917–9923. doi: 10.1074/jbc.M008629200. [DOI] [PubMed] [Google Scholar]
- Mika F, Hengge R. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of sigmaS (RpoS) in E. coli. Genes Dev. 2005;19:2770–2781. doi: 10.1101/gad.353705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Killoran PB, Fang FC, Riley LW. The global regulator ArcA controls resistance to reactive nitrogen and oxygen intermediates in Salmonella enterica serovar Enteritidis. Infect Immun. 2002;70:451–461. doi: 10.1128/IAI.70.2.451-461.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SM, Alugupalli KR, Ram S, Akerley BJ. The ArcA regulon and oxidative stress resistance in Haemophilus influenzae. Mol Microbiol. 2007;64:1375–1390. doi: 10.1111/j.1365-2958.2007.05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berggren RE, Wunderlich A, Ziegler E, Schleicher M, Duke RC, Looney D, Fang FC. HIV gp120-specific cell-mediated immune responses in mice after oral immunization with recombinant Salmonella. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;10:489–495. doi: 10.1097/00042560-199510050-00001. [DOI] [PubMed] [Google Scholar]
- Georgellis D, Kwon O, De Wulf P, Lin EC. Signal decay through a reverse phosphorelay in the Arc two-component signal transduction system. J Biol Chem. 1998;273:32864–32869. doi: 10.1074/jbc.273.49.32864. [DOI] [PubMed] [Google Scholar]
- Kwon O, Georgellis D, Lin EC. Phosphorelay as the sole physiological route of signal transmission by the arc two-component system of Escherichia coli. J Bacteriol. 2000;182:3858–3862. doi: 10.1128/JB.182.13.3858-3862.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpica R, Franco B, Rodriguez C, Kwon O, Georgellis D. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc Natl Acad Sci USA. 2004;101:13318–13323. doi: 10.1073/pnas.0403064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgellis D, Kwon O, Lin EC. Amplification of signaling activity of the Arc two-component system of Escherichia coli by anaerobic metabolites. An in vitro study with different protein modules. J Biol Chem. 1999;274:35950–35954. doi: 10.1074/jbc.274.50.35950. [DOI] [PubMed] [Google Scholar]
- Matsushika A, Mizuno T. A dual-signaling mechanism mediated by the ArcB hybrid sensor kinase containing the histidine-containing phosphotransfer domain in Escherichia coli. J Bacteriol. 1998;180:3973–3977. doi: 10.1128/jb.180.15.3973-3977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Lin EC. Purification and phosphorylation of the Arc regulatory components of Escherichia coli. J Bacteriol. 1992;174:5617–5623. doi: 10.1128/jb.174.17.5617-5623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsey TA, Vazquez-Torres A, Gravdahl DJ, Fang FC, Libby SJ. The ferritin-like Dps protein is required for Salmonella enterica serovar Typhimurium oxidative stress resistance and virulence. Infect Immun. 2004;72:1155–1158. doi: 10.1128/IAI.72.2.1155-1158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Imlay JA. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem. 2007;282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? Embo J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. PNAS. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Li M, Zhang X, Xing L. An improvement of the site-directed mutagenesis method by combination of megaprimer, one-side PCR and DpnI treatment. Anal Biochem. 2004;331:401–403. doi: 10.1016/j.ab.2004.04.019. [DOI] [PubMed] [Google Scholar]
- Lu S, Manges AR, Xu Y, Fang FC, Riley LW. Analysis of virulence of clinical isolates of Salmonella enteritidis in vivo and in vitro. Infect Immun. 1999;67:5651–5657. doi: 10.1128/iai.67.11.5651-5657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macomber L, Rensing C, Imlay JA. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J Bacteriol. 2007;189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PY, Janovjak H, Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques. 2002;32:1372–1374. [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]