Abstract
Barrier-to-autointegration factor (BAF) is a protein that has been proposed to compact retroviral DNA, making it inaccessible as a target for self-destructive integration into itself (autointegration). BAF also plays an important role in nuclear organization. We studied the mechanism of DNA condensation by BAF using total internal reflection fluorescence microscopy. We found that BAF compacts DNA by a looping mechanism. Dissociation of BAF from DNA occurs with multiphasic kinetics; an initial fast phase is followed by a much slower dissociation phase. The mechanistic basis of the broad timescale of dissociation is discussed. This behavior mimics the dissociation of BAF from retroviral DNA within preintegration complexes as monitored by functional assays. Thus the DNA binding properties of BAF may alone be sufficient to account for its association with the preintegration complex.
Keywords: autointegration, BAF (Banf1), DNA condensation, DNA looping, retrovirus
Barrier-to-autointegration factor (BAF) is an 89 aa protein (1) that is highly conserved among multicellular eukaryotes. It was identified as a protein factor that blocks self-destructive autointegration of retroviral DNA (1, 2). NMR studies have revealed that BAF is a dimer with the monomer composed of a bundle of helices (3). Crystallographic studies refined the dimer interface and identified a helix-hairpin-helix (HHH) motif as a putative DNA binding surface (4). DNA binding to the HHH motif (5, 6) lacks sequence specificity because the protein-DNA contacts are between amide groups on the protein backbone and DNA phosphates. This proposal was fully consistent with both bridging of double stranded DNA by BAF and the sequence nonspecific nature of binding of BAF to DNA (1, 7). Confirmation came from the structure of BAF in complex with DNA determined by x-ray crystallography (8). One DNA duplex bound to each monomer within the BAF dimer, with the two DNAs forming an angle close to 90° with one another. However, local bending of a single DNA duplex around the dimer to occupy both binding sites appears energetically unfavorable because of the preponderance of negative charges on the protein surface between the two DNA binding sites.
BAF was also identified as an interacting partner of lamina-associated-polypeptide 2 (LAP2) (9), a member of the inner nuclear membrane protein family that includes Lamin-associated-polypeptides, Emerin, and MAN1 (10). These proteins share a common “LEM” domain that interacts with BAF. NMR studies (11) show that the LEM domain binds to a surface of the BAF dimer that is distinct from the DNA binding site. BAF has been reported to be located in many cellular compartments including the nucleoplasm, cytoplasm, the nuclear membrane, the “core” region of teleophase chromosomes, and its cellular distribution varies according the cell cycle stage (9, 12–14). RNA interference (RNAi) or genetic inactivation of BAF demonstrates that it is an essential protein. Knockdown or knockout of BAF results in a lethal phenotype of aberrant chromatin organization as evidenced by a segregation defect during mitosis (7, 12, 13). BAF is also directly involved in nuclear envelope organization independent of the chromatin organization phenotype (15). In vitro, BAF has profound effects on chromatin decondensation and nuclear expansion in the in vitro Xenopus system for reconstitution of nuclear envelopes around DNA promoted by oocyte extracts in the presence of an energy regenerating system (16).
The function of BAF in nuclear organization involves interaction with multiple proteins of the LEM family and likely many indirect interaction partners including components of chromatin and DNA. BAF is also a substrate for vaccinia-related-kinase (VRK), which phosphorylates BAF and abrogates its DNA binding activity (17, 18). Cell-cycle–dependent phosphorylation by VRK is important for regulating the association of BAF with chromatin and the nuclear envelope (15). In addition to its normal cellular functions, which are still not well understood and involve many co-factors, BAF is likely to have a direct impact on the fate of DNA that enters the cytoplasm through its DNA binding activity. Although cytoplasmic DNA other than that within certain organelles is not known to be involved in normal cellular function, DNA is frequently introduced into the cytoplasm during viral infection; in fact, BAF was first identified through its binding to retroviral DNA (1). BAF has also been implicated in impacting vaccinia virus replication. Vaccinia replicates its DNA in the cytoplasm and encodes an essential kinase, the function of which has been unclear. Recent work indicates that its role is to inactivate the DNA binding activity of BAF which otherwise binds to vaccinia DNA and blocks its replication (17, 19). Thus the DNA binding activity of BAF in the cytoplasm can be beneficial for viral replication as in the case of retroviruses, or severely detrimental as for vaccinia. Compaction of DNA by BAF can potentially explain both phenomena; compaction of retroviral DNA blocks access of the integration machinery, thus blocking autointegration; similarly, compaction of vaccinia DNA interferes with replication. We have used total internal reflection fluorescence microscopy (TIRFM) to establish that BAF does indeed compact DNA and that this compaction results from the formation of intramolecular loops rather than collapse because of charge neutralization or wrapping of DNA around the protein.
Results
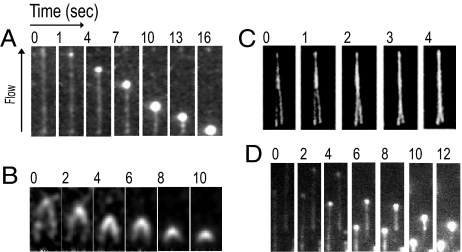
To study the interaction of BAF with DNA by total internal reflection fluorescence microscopy (TIRFM), we first used non-labeled BAF protein and phage λ DNA molecules fluorescently stained with YOYO-1 and attached to the flow–cell surface at one biotinylated end. In the absence of external force, the DNA molecules are not visualized because they are not confined within the penetration depth of the evanescent illumination field. By means of buffer flow in the microfluidic cell (10 mM Tris·HCl pH 7.5, 0.5 mM EDTA containing NaCl as noted), DNA molecules attached at one end are extended to near full length of 16 μm within the evanescent field (Fig. 1A). Next, the protein-free buffer flow is replaced with buffer containing BAF. Shortly after BAF injection, DNA condensation initiates from the free DNA end (Fig. 1A). This DNA end collapses into a BAF–DNA condensate that then appears to roll back along the extended DNA molecule until the entire DNA is condensed into a tight ball. At the flow rate of ≈20 μl/min (cross-section averaged velocity of 3.3 mm · s−1) that we typically used in these experiments, folding of the λ DNA occurred on a time scale of several seconds.
Fig. 1.
Condensation of λ-DNA by BAF. (A) λ-DNA, labeled with YOYO-1 was attached to the flow cell surface at one end and extended by buffer flow. Successive time frames are shown from left to right. (B) Same as in panel (A) except that both ends of the λ-DNA were attached to the flow cell surface. (C) Two separate λ-DNA molecules were attached in close proximity at one end. The molecules progressively “zippered up.” (D) Same as in (A) except that BAF was labeled with Alexa-488 and the DNA was unlabeled. All experiments were carried out in 100 mM NaCl buffer.
DNA condensation starts at the free DNA end, consistent with DNA loop formation involving the two DNA binding sites on the BAF dimer. Although the hydrodynamic tensions on different regions along the DNA in these experiments have not been measured, DNA near the free end is clearly under lowest tension. Thus, loop formation would take place more readily near the free end. Since we did not observe condensation starting from internal points along the DNA, we examined whether it requires a free DNA end. For this purpose we attached DNA molecules to the glass at both ends through biotin. The double-anchored DNA molecules were subjected to buffer flow, and BAF protein was injected into the flow cell (Fig. 1B). The DNA condensed as evidenced by shortening of the DNA contour length between the two attachment points, demonstrating that condensation does not require a free DNA end. In addition to intramolecular loop formation, a single BAF dimer should be able to bind to separate DNA molecules, forming intermolecular bridges. As predicted, Fig. 1C shows that when two DNA molecules are in close proximity, the addition of BAF results in the two molecules “zipping” together. The “zipped” double duplex appears to resist further extensive condensation, presumably because of quick saturation of the interduplex interface by BAF and the stiffness of the resulting structure.
Fluorescent Labeling of BAF with Alexa-488.
Although the condensation of DNA by BAF can be visualized with unlabeled BAF and fluorescently labeled DNA, study of the kinetics of BAF binding and dissociation requires direct visualization of the BAF protein. We first constructed GFP-BAF fusions for this purpose, but these proteins exhibited aberrant DNA binding properties compared with the wild-type BAF. Therefore we instead labeled BAF with Alexa-488 maleimide (see SI Text). Bulk DNA binding experiments confirmed that Alexa-488–BAF exhibits DNA binding properties that are indistinguishable from those of wild-type BAF. Alexa-488–BAF also condenses λ DNA in TIRFM experiments in the same way as wild-type BAF (compare Fig. 1D with Fig. 1A), and “zippers up” adjacent DNA molecules. We conclude that the labeling of BAF with Alexa-488 does not significantly perturb its DNA-binding properties. Binding experiments showed that DNA is saturated at BAF concentrations greater than 100 nM with half-saturation at ≈10 nM (Fig. S1).
Dissociation of BAF from DNA Suggests Multiple Modes of DNA Binding.
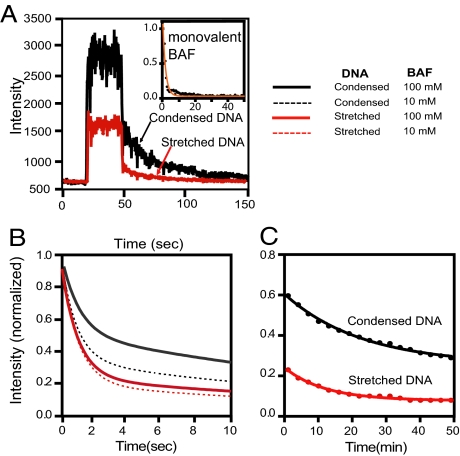
The kinetics of BAF dissociation from condensed DNA may be complicated because of the possible “caging” effect of the locally high concentration of DNA that can facilitate re-binding. In contrast, if the BAF dimer binds to DNA in a single mode, dissociation from stretched DNA would be expected to follow a simple exponential decay. To test these models, we studied dissociation from the stretched part of the DNA in parallel with dissociation from the condensed part within the same DNA molecule by performing the BAF-binding step at higher flow rates; under these conditions, many molecules are only partially “rolled back,” and the condensed part is attached to the flow cell surface. After binding, dissociation was initiated by replacing the flow of Alexa-488–BAF solution by buffer without protein. Fig. 2 shows the Alexa-488–BAF fluorescence signal of the stretched DNA and the condensed ball during one cycle of binding and dissociation. The results are qualitatively similar for both the condensed ball and the stretched DNA. Fluorescence reaches a plateau within seconds during the binding phase. Upon initiation of dissociation by switching from flow of protein solution to 100 mM NaCl buffer, a rapid decrease in fluorescence occurs over the first few seconds, followed by a slower decrease over several hundred seconds. The fast dissociation phase accounts for ≈50% of the initial fluorescence of the condensed ball and ≈80% of the initial fluorescence of the stretched DNA. As this apparently biphasic dissociation is observed with both stretched DNA and the condensed ball, it cannot be explained by caging-type effects that might be invoked to explain complex dissociation behavior in the latter case. For the dissociation rate estimation, photobleaching of Alexa-488 must be taken into consideration, especially for slow dissociation observed with high frame rate acquisition. The correction for photobleaching is described in SI Text and is discussed below.
Fig. 2.
Kinetics of BAF dissociation. (A) Alexa-488–BAF was first bound to λ-DNA in 100 mM NaCl buffer. Dissociation was initiated by switching to protein-free buffer and the fluorescence of the stretched (red line) and condensed (black line) DNA segments was monitored as a function of time. An initial fast dissociation phase was followed by a slower phase. When BAF was made monovalent by inactivating one of the two DNA binding sites within the dimer, only the fast phase dissociation was observed (Top Right Inset; orange line shows a single exponential fit of the data). (B) The first 10 s of BAF dissociation from stretched DNA (red lines) and condensed DNA (gray lines) assembled at 100 nM (solid lines) and 10 nM BAF (dashed lines). Shown here are double exponential fits of actual data, which are shown in Fig. S3. The first dissociation phase of all curves fits koff,fast of 1 s−1. (C) To avoid a significant contribution from photo-bleaching, the slow dissociation phase was monitored with a frame rate of 1 frame per 3 min instead of 1 per 0.3 s, which was used to monitor the fast dissociation phase. Slow phase dissociation data are fitted to the single exponential decay functions with rate koff,slow ≈ 5 × 10−4 · s−1.
Monovalent BAF Exhibits Only Fast Phase Dissociation.
We reasoned that biphasic dissociation of BAF from DNA might involve binding of DNA to one or to both binding sites in the BAF dimer. It follows that BAF dimers with only a single DNA binding site should exhibit only the fast dissociation phase. To test this prediction, we generated fluorescently labeled BAF dimers with only one functional DNA binding site. Briefly, this was accomplished by mixing Alexa-488–BAF dimers with the DNA binding site inactivated by phosphorylation with wild-type BAF dimers. The mixture was then denatured and refolded to randomly assort the monomers within the dimers (see SI Text). In the resulting dimer mixture, the only fluorescently labeled dimers that can bind DNA have a functional DNA binding site on only one of the monomers. These monovalent BAF dimers exhibited only the fast phase dissociation kinetics (Fig. 2A Inset), demonstrating that biphasic dissociation requires two functional DNA binding sites on the BAF dimer.
Kinetics of BAF Dissociation.
To measure the rates of the fast and slow BAF dissociation components, we conducted washing experiments monitoring fluorescence decay from folded and stretched DNA segments after binding Alexa-488–BAF at 10 nM and 100 nM (data in Fig. S3, curve fits shown in Fig. 2B). The fluorescence intensity at time 0 refers to apparent saturation binding, which we estimate to correspond to one BAF dimer per 6 bp (Fig. S2). The dissociation kinetics fit a double-exponential decay model. The fast decay component, koff,fast obtained from the fitting curve shown in Fig. 2B corresponds to 1 s−1 regardless of the protein concentration during the binding step and regardless of whether dissociation from the stretched or condensed DNA is measured. Photobleaching does not contribute significantly to this measurement (see SI Text). However, this represents a lower limit of the rate for the fast dissociation phase, because buffer exchange in our system takes close to 1 s.
The slow decay in the experiment of Fig. 2B is mainly caused by bleaching (SI Text). Thus, we repeated the experiment using a decreased frame rate of one 0.1-s exposure every 3 min to mitigate bleaching effects. The data (Fig. 2C) are fitted to a single exponential decay with koff,slow ≈ 5 × 10−4 s−1 after correction for the bleaching effect (SI Text). Strikingly, this is three orders of magnitude slower than the fast component for both the stretched and condensed DNA. Furthermore, the single exponential fitting left a fraction of BAF that has the appearance of irreversible binding.
DNA Condensation by BAF Is Reversed at High Ionic Strength.
Fitting the dissociation data in Fig. 2 as a double exponential decay requires additive offsets corresponding to ≈25% and 8% of the BAF irreversibly bound to the condensed DNA and stretched DNA segments, respectively. At the end of the experiment, the condensed DNA did not show any extension, indicating that this residual BAF was sufficient to maintain compaction.
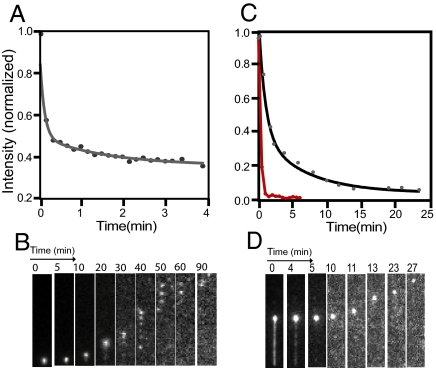
We next examined dissociation of Alexa-488–BAF from condensed DNA in the presence of 1 M NaCl (Fig. 3 A and B). ≈60% of the bound BAF dissociated with fast kinetics similar to that observed when washing with 100 mM NaCl buffer. This was followed by a slower dissociation phase with about 30% of the BAF remaining bound after 5 min. This dissociation was not accompanied by decondensation, indicating that the residual bound BAF was sufficient to maintain the condensed state. By 10 min, the fluorescence dropped to less than 10% of the original level, and at about the same time the DNA began to decondense as evidenced by movement of the DNA ball away from the attachment point. The opening process consists of a series of events (Fig. 3B). The DNA condensate breaks up into several clusters, each of which independently fragments further into smaller clusters. The process continues until, after about 90 min, the DNA extends to near its original length.
Fig. 3.
(A and B) Dissociation of BAF from DNA in 1 M NaCl. (A) Decrease in fluorescence of the BAF–DNA condensate assembled in 100 mM NaCl buffer after switching to 1 M NaCl buffer without BAF. Data are fitted with a double exponential decay function. (B) Extension of the DNA occurs in discrete steps as the condensate progressively fragments into smaller clusters. (C and D) BAF dissociation at 100 mM NaCl in the presence of 100 ng/μl sonicated salmon sperm competitor DNA. (C) Kinetics of dissociation from the stretched (red line) and the condensed (black line) DNA segments. Dissociation from the condensed DNA was fitted to a double exponential. (D) Time course of extension. In (B) and (D), the gray scale was adjusted to make the weak signal visible for later time points.
Competitor DNA Induces Rapid Dissociation of BAF.
Complete dissociation of BAF from DNA is slow even at high ionic strength. However, the dissociation rate is profoundly elevated by the presence of competitor DNA even at 100 mM NaCl. Figure 3 C and D depict the dissociation of Alexa-488–BAF from DNA in the presence of 100 mM NaCl and 100 ng/μl sonicated DNA fragments. Dissociation from the condensed DNA is biphasic, with a slow phase that is much faster relative to that with 100 mM NaCl and is comparable to the rates with 1 M NaCl buffer. Unlike with high salt buffer, opening in the presence of competitor DNA occurs smoothly without large jumps and fragmentation of condensed balls. This suggests that competitor DNA sequentially removes BAF molecules that are exposed near the surface of the condensed balls, in contrast to high salt, which seems to destabilize the entire condensate. Figure 3C shows that dissociation of BAF from stretched DNA is complete within 1 min in the presence of competitor DNA.
Condensation of DNA by BAF Monitored Using Magnetic Tweezers.
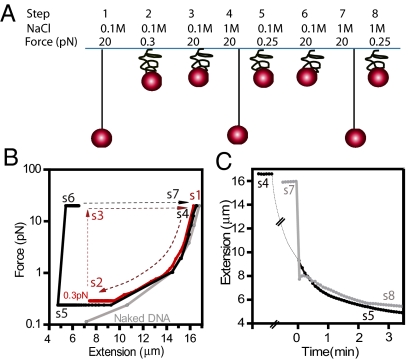
To probe BAF binding and DNA condensation under conditions in which stretching force is known and constant along the molecule contour, we conducted experiments with magnetic tweezers. A single λ-DNA molecule was attached to the slide glass of a flow cell and was stretched using force generated by a magnetic field gradient acting on a paramagnetic bead coupled to its free end. DNA was stretched in the vertical direction and its length measured by tracking the bead. We measured the length of the BAF-DNA complex under different tensile forces and compared it to the well-known force–extension curve of naked DNA (20–22).
A schematic diagram of the experiment is shown in Fig. 4A. Naked DNA was first stretched by applying a force of 20 pN in 100 mM NaCl buffer; the resulting force/extension curve is shown in Fig. 4B. Next, unlabeled BAF was introduced into the flow cell in 100 mM NaCl (step 1), and the force was gradually decreased to 0.3 pN. The force extension curve is shown in Fig. 4B. When the force was reduced to 0.3 pN, the DNA rapidly condensed (step 2), consistent with the formation of DNA loops, which form by Brownian motion with appreciable rate only at stretching forces of less than 1 pN (23–25). To prevent the BAF/DNA condensate and the bead from sticking to the glass, condensation was then stopped by increasing the force to 20 pN and replacing the BAF solution with 100 mM NaCl. The DNA–BAF remained condensed under these conditions (Fig. 4B, step 3). The 100 mM NaCl buffer was then replaced by 1 M NaCl buffer while maintaining the force at 20 pN (Fig. 4B, step 4). At 1 M NaCl, the DNA extended to its original length over several minutes. The NaCl concentration was then lowered back to 100 mM, and the force was slowly decreased (Fig. 4B, step 5). The force–extension curve shows that the DNA again condensed at 0.25 pN, apparently indicating that some BAF remained bound during step 4 even though the DNA was extended in buffer without added BAF, consistent with the slow dissociation of BAF in the presence of 1 M NaCl observed in the TIRFM experiments. We caution that complete removal of BAF from the flow cell during steps 3 and 4 was not experimentally verified. Finally, a similar force extension behavior was observed when steps 3–5 were repeated, this time maintaining the high NaCl concentration during step 8 (steps 6–8). The extension as a function of time for steps 5 and 8 is shown in Fig. 4C.
Fig. 4.
DNA condensation by BAF monitored using magnetic tweezers. (A) λ DNA was tethered to the glass slide of a flow cell and stretched with a magnetic force applied to a paramagnetic bead coupled to the free DNA end. The flow cell buffer was changed sequentially as indicated. The buffer conditions, applied force, and the final resulting DNA extension are indicated for each of the sequential steps. (B) Force/extension curves during the steps depicted in (A). The curve for naked λ DNA in 100 mM NaCl buffer is shown in gray. DNA condensation with BAF is observed at 0.3 pN and 0.25 pN during steps 2 and 5. (C) Time trajectory of DNA condensation during steps 5 and 8. Time 0 is when the force was reduced to and held constant at 0.25 pN in steps 5 and 8.
State of BAF-Induced DNA Condensation Probed by Sedimentation in High and Low Salt Sucrose Gradients.
To further probe the stability of BAF-DNA complexes, we conducted velocity sedimentation experiments in sucrose gradients at 100 mM NaCl and 1M NaCl. Alexa-488–BAF was bound to φX174RF DNA at 150 nM BAF and a stoichiometry of one dimer per 10 bp DNA. The complex was sedimented through the gradient, and after fractionation the DNA was detected by Southern blotting.
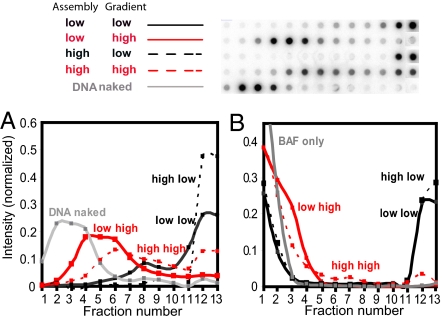
As shown in Fig. 5A, complexes assembled at either low (black solid line) or high salt (black dashed line) sedimented to the bottom of the gradient when the gradient contained low salt, indicative of a high degree of DNA compaction regardless of the ionic strength of the binding buffer. Complexes assembled at low-salt (red solid line) or high-salt (red dashed line) sedimented faster than naked DNA (blue) when sedimented in a high-salt gradient. The finding that complexes assembled and sedimented in the presence of 1M NaCl sediment faster than naked DNA confirms our conclusion that BAF can bind and compact DNA even at high ionic strength, although the degree of compaction is less than at low ionic strength.
Fig. 5.
BAF–DNA complex assembly and stability assayed by velocity sedimentation at high and low ionic strengths. φX174RF DNA was incubated with 150 nM Alexa-488–BAF at a ratio of one BAF dimer per 10 bp in either 100 mM (low) or 1 M NaCl (high) buffer, and the mixture was layered onto a sucrose gradient containing either low or high salt. (A) After centrifugation, the gradients were fractionated and the DNA content of each fraction was quantified by Southern blotting (top). The DNA distributions in the gradient are plotted below. Labels refer to the salt concentration during assembly (first) and in the sucrose gradient (second). (B) The Alexa-488–BAF content of each fraction was monitored by fluorescence. Curves are labeled as in (A).
We also measured the BAF protein present in each gradient fraction (Fig. 5B). The gray curve represents the fluorescence of 150 nM Alexa-488–BAF applied to the sucrose gradient in the absence of DNA. When the gradient contained 100 mM NaCl, DNA and a fraction of the BAF sedimented together near the bottom of the gradient regardless of whether complex assembly was carried out under low- or high-salt conditions, indicating that DNA and BAF remain stably associated in the gradient. In contrast, when complexes were sedimented in gradients containing 1M NaCl, a significant fraction of the BAF sedimented between the positions expected for free BAF and the DNA signal, indicating that dissociation of BAF occurred gradually during centrifugation. However, a small but detectable amount of BAF was found in the fractions containing DNA. Although most of the BAF dissociated from the DNA during centrifugation at high salt, the sedimentation velocity of the DNA shows that this residual bound BAF significantly compacts the DNA.
Discussion
BAF Condenses DNA by Looping.
We conclude that DNA looping by intersegmental bridging is responsible for DNA condensation by BAF. First, condensation requires low (< 1 pN) DNA tension, as shown by the tweezers experiments. When tension is unevenly applied to DNA molecules, condensation starts exclusively from the region of the lowest tension, i.e., the most downstream portion of the DNA molecule in the TIRFM experiments, again supporting a DNA looping mechanism. Second, intermolecular cross-bridging competes with and prevents condensation, supporting the notion that condensation is caused by intramolecular cross-bridging (Fig. 1C). Third, from the data in Fig. 3 and Fig. S2, we estimate that approximately one BAF dimer per 100 bp is sufficient to condense DNA, a stoichiometry below that required for condensation by a DNA wrapping mechanism. Fourth, decondensation initiated by washing at high ionic strength occurs in discrete steps of variable elongation length that can be explained by the sequential opening of large loops of variable sizes. Finally, BAF is an acidic protein with a calculated pI of 5.8, making condensation by net charge neutralization unlikely.
Multiphasic Dissociation of BAF from DNA.
The kinetics of dissociation of BAF from both “stretched” and condensed DNA observed in the TIRFM experiments does not fit a single exponential decay. Completion of the initial fast dissociation phase within several seconds represents a lower limit for the rate because buffer exchange occurs over a similar time scale. This population may represent BAF bound to DNA through only one of the DNA binding sites within the dimer, consistent with the observation that dimers with only one subunit capable of DNA binding exhibit only fast dissociation kinetics. If this interpretation is correct, this fraction of BAF was inaccessible for divalent DNA binding. Thus, within a condensed DNA molecule, significant parts of the loops are too short to be further condensed by smaller internal loop formation and are also devoid of neighboring DNA segments to form cross-bridges, yet can accommodate univalent BAF binding.
The complexity of the slowly dissociating population of BAF does not allow us to estimate a meaningful Kd for this fraction. However, the slowly dissociating, hence more stably binding, fraction of BAF would saturate accessible loop bridging sites before saturation of the remaining DNA sites by monovalently binding BAF. Thus, from a titration experiment (Fig. S1), we could estimate the Kd for the fast dissociating fraction of BAF to be roughly 10 nM.
The slow dissociation phase can be fitted to a single exponential, but only if we assume that a fraction of the BAF remains irreversibly bound. If we assume that divalent BAF binding by intramolecular DNA looping can be treated as a pseudosecond order reaction and that the association rate constant for the tightly binding species and quickly dissociating species are similar, the Kd for the slow dissociation fraction is estimated to be 10 pM or lower, as judged by the rate of dissociation. Complex kinetics of dissociation of BAF from the condensed DNA ball may not be surprising if we consider the complexity of DNA compaction and the binding of free energy involved. From the binding constant for the monovalent BAF-DNA interaction, the monovalent binding energy is estimated to be roughly 11 kcal/mol. The same amount of energy would, in principle, be available for the binding to DNA at the other binding face of the BAF dimer. If there were no extra energetic cost for the second interaction, the Kd for the divalent binding could be below femtomolar (fM). However, there are additional free energy costs for the second interaction because of tight bending and compaction of DNA, and this cost increases as compaction gets tighter. The weakest of the divalently bound state in the highly compacted complex was estimated above to be roughly Kd of 10 pM, which translates to roughly 15.2 kcal/mol, leaving ≈6.8 kcal/mol as the extra energy cost for the second site binding at near-maximum compaction. The extra energy cost would be less at lower compaction. Then, binding should become tighter as BAF dissociates and the complex becomes less compact; and the Kd could approach, in theory, low fM, explaining the apparently “irreversible” binding in the absence of competitor DNA.
The apparently irreversibly bound fraction of BAF may just represent the slower dissociating states rather than a population of BAF that is bound in a fundamentally different way. This may also explain the apparently irreversible fraction of BAF bound to “stretched” DNA in the TIRFM experiments. Although this DNA is linear within the resolution limit of light microscopy, inhomogenities in the intensity of fluorescence observed along the DNA suggest the presence of microclusters of BAF that may exhibit dissociation behavior qualitatively similar to that of the condensed ball.
Influence of Competitor DNA on Dissociation of BAF from DNA.
Competitor DNA is frequently used as a “sink” in experiments involving protein–DNA interactions. The rationale is based on an assumption that the competitor does not influence the dissociation reaction but, rather, soaks up the free protein once dissociation has occurred. Our data on the effects of competitor DNA on BAF dissociation kinetics suggest the possibility that competitor DNA directly promotes dissociation of BAF from DNA, as previously suggested for HU and HMG-box proteins (26). This is easily understandable for a divalent DNA binding protein. Dissociation in the absence of competitor DNA requires simultaneous release of both binding sites, whereas protein transfer from the initially bound DNA to competitor could take place by sequential transfer of one binding site at a time with lower activation energy. This is equivalent to the process termed “intersegmental transfer” of DNA binding proteins (26, 27).
Implications for Organization of Retroviral Preintegration Complex.
Our results shed light on the interaction of BAF with the retroviral preintegration complex (PIC). The DNA within Moloney murine leukemia virus (MLV) PICs efficiently integrates into an intermolecular target in vitro; but after treatment with 750 mM or higher concentrations of KCl and removal of liberated factors, autointegration predominates (2). The preference for intermolecular integration is restored upon incubation with BAF at lower ionic strength (1). The high ionic strength required to strip BAF from the PIC has been puzzling. In a sucrose gradient assay similar to that reported here, binding of BAF to DNA was not observed at 400 mM KCl (28). This raised the question as to why greater than 500 mM KCl is required to strip BAF from the PIC. LAP2α, a known binding partner of BAF, was found to be present in MLV PICs; and BAF, together with LAP2α, formed a compacted DNA complex that was more salt stable in sucrose gradients than the complex with BAF alone. It was therefore suggested that LAP2α stabilizes the association of BAF with the PIC. However, knockdown of BAF with RNAi resulted in only a very modest replication defect that was observed at low multiplicity of infection (28), and later MLV was shown to efficiently infect a LAP2α knockdown cell line (29). It is possible that another protein may substitute for LAP2α in stabilizing the association of BAF with PIC; but the results reported here suggest a perhaps simpler explanation, that the compacted DNA structure within the PIC presents a kinetic barrier to BAF dissociation.
The major difference of our current results from the previously reported sedimentation experiments is the higher concentration of DNA and BAF in the binding mixture. The resulting complexes only partially dissociate during the time course of sedimentation even in gradients containing 1 M NaCl. This is consistent with the slow time scale for dissociation of BAF and DNA extension observed in the TIRFM experiments. The dissociation behavior of BAF from DNA may be sufficient to explain the dissociation of BAF from retroviral DNA within PICs as inferred from functional assays. This implies that the “salt-stripped PIC” likely still contains a limited amount of BAF that could partially condense viral DNA. Blocking of autointegration likely depends on the tight compaction of viral DNA by a relatively high stoichiometry of stably bound BAF/DNA in the complex. Different preparations of PICs from infected cells display different degrees of protection from autointegration, suggesting partial loss of BAF during preparation; however, full protection could be recovered by addition of BAF. These observations can easily be explained on the basis of the DNA binding properties of BAF reported here.
Materials and Methods
DNA and Protein Reagents.
Plasmid DNA, protein expression and purification are described in SI Text.
Total Internal Reflection Fluorescence Microscopy.
The instrumentation was essentially as described (30, 31). The methodology is detailed in SI Text.
Velocity Sedimentation of BAF-DNA Complexes in Sucrose Gradients.
Sucrose gradients (1.0 ml) were made in 11 × 34 mm centrifuge tubes by sequentially layering 200 μl of 30%, 25%, 20%, and 15% sucrose solutions in 20 mM Tris · HCl, pH 7.4, 0.5 mM EDTA containing either 100 mM or 1 M NaCl, and incubating at 4 °C overnight. Immediately before loading the sample, 100 μl of 50% CsCl was layered at the bottom of the gradient as a cushion. To assemble the complexes, 0.5 nM φX174RF DNA was mixed at room temperature with 150 nM Alexa-488–BAF in 200 μl of 10% glycerol, 20 mM Tris · HCl, pH 7.4, 0.5 mM EDTA, 2.5 mM DTT, and either 100 mM or 1 M NaCl. The mixture was immediately layered onto the top of the gradient and centrifuged at 30,000 rpm at 4 °C for 1 h in a Beckman TLS55 rotor. The gradient was fractionated from the top into 13 fractions using a MicroFractionator (Brandel Research & Development Laboratories). The fluorescently labeled BAF in each fraction was detected using a SpectraMax M5 microplate reader (Molecular Devices). The DNA in each fraction was detected by transfer to Zeta-Probe membrane (Bio-Rad) and Southern blotting using 32P-labeled φX174 DNA as the probe. The image was visualized and quantified by a PhosphorImager (FujiFilm BAS-2500).
Manipulation of BAF-DNA Complexes with Magnetic Tweezers.
λ DNA was tethered to the glass slide of a flow cell and stretched with a magnetic force applied to a paramagnetic bead coupled to the free DNA end, as described (26). All of the buffers used in tweezers experiments contained 0.5 mg/ml casein.
Supplementary Material
Acknowledgments.
We are grateful to Vassili Ivanov for the help with the microscope system. This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases intramural program and by the NIH Intramural Aids Targeted Antiviral Program, National Institutes of Health, Department of Health and Human Services, US Government. The work of B.X. and J.F.M. was supported by National Science Foundation Grants DMR-0715099, PHY-0852130 and by a Catalyst Award from the Chicago Biomedical Consortium.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909077106/DCSupplemental.
References
- 1.Lee MS, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc Natl Acad Sci USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee MS, Craigie R. Protection of retroviral DNA from autointegration: Involvement of a cellular factor. Proc Natl Acad Sci USA. 1994;91:9823–9827. doi: 10.1073/pnas.91.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai M, Huang Y, Zheng R, Wei SQ, Ghirlando R, et al. Solution structure of the cellular factor BAF responsible for protecting retroviral DNA from autointegration. Nat Struct Biol. 1998;5:903–909. doi: 10.1038/2345. [DOI] [PubMed] [Google Scholar]
- 4.Umland TC, Wei SQ, Craigie R, Davies DR. Structural basis of DNA bridging by barrier-to-autointegration factor. Biochemistry. 2000;39:9130–9138. doi: 10.1021/bi000572w. [DOI] [PubMed] [Google Scholar]
- 5.Thayer MM, Ahern H, Xing DX, Cunningham RP, Tainer JA. Novel DNA-binding motifs in the DNA-repair enzyme endonuclease-III crystal-structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doherty AJ, Serpell LC, Ponting CP. The helix-hairpin-helix DNA-binding motif: A structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng RL, Ghirlando R, Lee MS, Mizuuchi K, Krause M, Craigie R. Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc Natl Acad Sci USA. 2000;97:8997–9002. doi: 10.1073/pnas.150240197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradley CM, Ronning DR, Ghirlando R, Craigie R, Dyda F. Structural basis for DNA bridging by barrier-to-autointegration factor. Nat Struct Mol Biol. 2005;12:935–936. doi: 10.1038/nsmb989. [DOI] [PubMed] [Google Scholar]
- 9.Furukawa K. LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2-chromatin interaction. J Cell Sci. 1999;112:2485–2492. doi: 10.1242/jcs.112.15.2485. [DOI] [PubMed] [Google Scholar]
- 10.Wagner N, Krohne G. LEM-domain proteins: New insights into lamin-interacting proteins. Int Rev Cytol. 2007;261:1–46. doi: 10.1016/S0074-7696(07)61001-8. [DOI] [PubMed] [Google Scholar]
- 11.Cai ML, Huang Y, Suh JY, Louis JM, Ghirlando R, et al. Solution NMR structure of the barrier-to-autointegration factor-emerin complex. J Biol Chem. 2007;282:14525–14535. doi: 10.1074/jbc.M700576200. [DOI] [PubMed] [Google Scholar]
- 12.Furukawa K, Sugiyama S, Osouda S, Goto H, Inagaki M, et al. Barrier-to-autointegration factor plays crucial roles in cell cycle progression and nuclear organization in Drosophila. J Cell Sci. 2003;116:3811–3823. doi: 10.1242/jcs.00682. [DOI] [PubMed] [Google Scholar]
- 13.Margalit A, Segura-Totten M, Gruenbaum Y, Wilson KL. Barrier-to-autointegration factor is required to segregate and enclose chromosomes within the nuclear envelope and assemble the nuclear lamina. Proc Natl Acad Sci USA. 2005;102:3290–3295. doi: 10.1073/pnas.0408364102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haraguchi T, Kojidani T, Koujin T, Shimi T, Osakada H, et al. Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J Cell Sci. 2008;121:2540–2554. doi: 10.1242/jcs.033597. [DOI] [PubMed] [Google Scholar]
- 15.Gorjanacz M, Klerkx EPF, Galy V, Santarella R, Lopez-Iglesias C, et al. Caenorhabditis elegans BAF-1 and its kinase VRK-1 participate directly in post-mitotic nuclear envelope assembly. EMBO J. 2007;26:132–143. doi: 10.1038/sj.emboj.7601470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segura-Totten M, Kowalski AK, Craigie R, Wilson KL. Barrier-to-autointegration factor: Major roles in chromatin decondensation and nuclear assembly. J Cell Biol. 2002;158:475–485. doi: 10.1083/jcb.200202019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nichols RJ, Wiebe MS, Traktman P. The vaccinia-related kinases phosphorylate the N′ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol Biol Cell. 2006;17:2451–2464. doi: 10.1091/mbc.E05-12-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bengtsson L, Wilson KL. Barrier-to-autointegration factor phosphorylation on Ser-4 regulates emerin binding to lamin A in vitro and emerin localization in vivo. Mol Biol Cell. 2006;17:1154–1163. doi: 10.1091/mbc.E05-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiebel MS, Traktman P. Poxviral b1 kinase overcomes barrier to autointegration factor, a host defense against virus replication. Cell Host Microbe. 2007;1:187–197. doi: 10.1016/j.chom.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouchiat C, Wang MD, Allemand JF, Strick T, Block SM, Croquette V. Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophys J. 1999;76:409–413. doi: 10.1016/s0006-3495(99)77207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 22.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 23.Skoko D, Yan J, Johnson RC, Marko JF. Low-force DNA condensation and discontinuous high-force decondensation reveal a loop-stabilizing function of the protein Fis. Phys Rev Lett. 2005;95:208101. doi: 10.1103/PhysRevLett.95.208101. [DOI] [PubMed] [Google Scholar]
- 24.Yan J, Kawamura R, Marko JF. Statistics of loop formation along double helix DNAs. Phys Rev E. 2005;71 doi: 10.1103/PhysRevE.71.061905. 061905. [DOI] [PubMed] [Google Scholar]
- 25.Sankararaman S, Marko JF. Formation of loops in DNA under tension. Phys Rev E. 2005;71 doi: 10.1103/PhysRevE.71.021911. 021911. [DOI] [PubMed] [Google Scholar]
- 26.Skoko D, Wong B, Johnson RC, Marko JF. Micromechanical analysis of the binding of DNA-bending proteins HMGB1, NHP6A, and HU reveals their ability to form highly stable DNA-protein complexes. Biochemistry. 2004;43:13867–13874. doi: 10.1021/bi048428o. [DOI] [PubMed] [Google Scholar]
- 27.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki Y, Yang HF, Craigie R. LAP2 alpha and BAF collaborate to organize the Moloney murine leukemia virus preintegration complex. EMBO J. 2004;23:4670–4678. doi: 10.1038/sj.emboj.7600452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulky A, Cohen TV, Kozlov SV, Korbei B, Foisner R, et al. The LEM domain proteins emerin and LAP2 alpha are dispensable for human immunodeficiency virus type 1 and murine leukemia virus infections. J Virol. 2008;82:5860–5868. doi: 10.1128/JVI.00076-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greene EC, Mizuchi K. Direct observation of single MuB polymers: Evidence for a DNA-dependent conformational change for generating an active target complex. Mol Cell. 2002;9:1079–1089. doi: 10.1016/s1097-2765(02)00514-2. [DOI] [PubMed] [Google Scholar]
- 31.Tan X, Mizuuchi M, Mizuuchi K. DNA transposition target immunity and the determinants of the MuB distribution patterns on DNA. Proc Natl Acad Sci USA. 2007;104:13925–13929. doi: 10.1073/pnas.0706564104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.