Abstract
The broadly conserved AvrE-family of type III effectors from Gram-negative plant pathogenic bacteria includes important virulence factors, yet little is known about the mechanisms by which these effectors function inside plant cells to promote disease. We have identified two conserved motifs in AvrE-family effectors: a WxxxE motif and a putative C-terminal endoplasmic reticulum membrane retention/retrieval signal (ERMRS). The WxxxE and ERMRS motifs are both required for the virulence activities of WtsE and AvrE1, which are major virulence factors of the corn pathogen Pantoea stewartii subsp. stewartii and the tomato/Arabidopsis pathogen Pseudomonas syringae pv. tomato, respectively. The WxxxE and the predicted ERMRS motifs are also required for other biological activities of WtsE, including elicitation of the hypersensitive response in nonhost plants and suppression of defense responses in Arabidopsis. A family of type III effectors from mammalian bacterial pathogens requires WxxxE and sub-cellular targeting motifs for virulence functions that involve their ability to mimic activated G-proteins. The conservation of related motifs and their necessity for the function of type III effectors from plant pathogens indicates that disturbing host pathways by mimicking activated host G-proteins may be a virulence mechanism employed by plant pathogens as well.
Additional keywords: Zea mays, Stewart's wilt, bacterial speck, type III secretion system, Hrp
Introduction
Many Gram-negative bacterial pathogens of plants and animals utilize type III secretion systems (TTSSs) to deliver bacterial virulence proteins into eukaryotic host cells (He et al., 2004). The translocated proteins, which are called type III effectors, serve to promote infection by modulating host cellular processes such as cytoskeleton dynamics, host immune responses, and cell death. A plant pathogenic bacterium usually produces and secretes more than 20 different effector proteins, presumably to block or avoid eliciting host resistance responses and to release nutrients from host tissues in order to support the rapid increase of bacterial populations in infected plants. With a few exceptions, the virulence functions of most type III effectors in plant pathogenic bacteria remain poorly understood (da Cunha et al., 2007; Speth et al., 2007).
AvrE-family effector proteins are among the most important virulence factors in plant pathogenic bacteria and are unique in several respects. First, AvrE-family effectors are widely conserved in the divergent genera Pseudomonas, Pantoea, Erwinia, Dickeya, and Pectobacterium. These effectors are encoded by genes that are physically adjacent to the Hrp TTSS gene cluster as part of a large pathogenicity island (PAI). Since acquisition of this PAI is believed to play an essential role in the evolution of many bacterial plant pathogens, the virulence function of AvrE-family effectors is likely to be central to pathogenesis. Second, while it has been difficult to demonstrate a role of many type III effectors in virulence using mutational approaches, the role of AvrE-family effectors in virulence is well recognized. Most significantly, DspE (alternatively named DspA) of the fire blight pathogen Erwinia amylovora (Ea) and WtsE of Pantoea stewartii subsp. stewartii (Pnss) are essential pathogenicity factors because the virulence reduction of the dspE and wtsE mutants is comparable to that of TTSS-defective mutants (Bogdanove et al., 1998; Frederick et al., 2001). An important role for AvrE-family effectors in virulence has also been shown in strains of Pseudomonas syringae pv. tomato (Pto) and, interestingly, the function of AvrE1 appears to be redundant to that of HopM1 in Pto strain DC3000 (Lorang and Keen, 1995; DebRoy et al., 2004; Badel et al., 2006). Third, AvrE-family effectors are very large proteins (most are > 200 kDa) with dual functions in promoting symptoms associated with host cell death and in suppressing basal defenses (DebRoy et al., 2004; Boureau et al., 2006; Ham et al., 2008). Despite their importance, the molecular mechanisms by which AvrE-family effectors carry out their virulence functions are not understood. In this study, we investigated WtsE from Pnss, which causes Stewart's bacterial wilt and leaf blight of sweet corn and maize, and AvrE1 from Pto, which causes bacterial speck of tomatoes and a leaf spot on Arabidopsis thaliana. Although WtsE and AvrE1 are each members of this large family (Fig. 1), the two proteins are fairly divergent with only 27.1% aa identity.
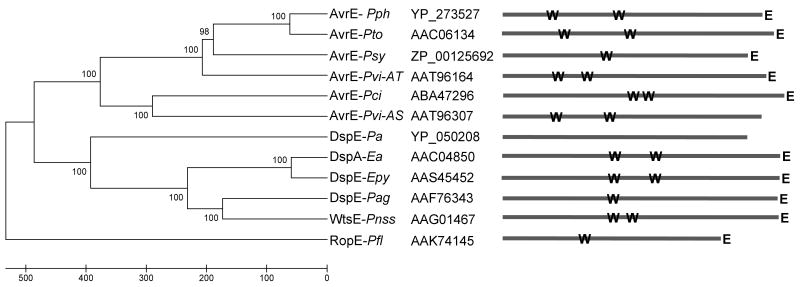
Fig. 1.
Presence of WxxxE and ERMRS motifs in AvrE-family effector proteins of plant associated bacteria. Phylogenetic tree of the AvrE-family effector proteins from plant pathogenic bacteria. Genus and species abbreviations after each protein are as follows; Pph (P. syringae pv. phaseolicola strain 1448A), Pto (P. syringae pv. tomato strain DC3000), Psy (P. syringae pv. syringae strain B728a), Pvi-AT (Pseudomonas viridiflava strain LP23.1a), Pci (Pseudomonas cichorii strain 83-1), Pvi-AS (Pseudomonas viridiflava strain RMX23.1a), Pa (Pectobacterium atrosepticum strain SCRI1043), Ea (Erwinia amylovora strain Ea321), Epy (Erwinia pyrifoliae strain WT3), Pag (Pantoea agglomerans pv. gypsophilae strain Ehg284-1), Pnss (Pantoea stewartii subsp. stewartii strain SS104), and Pfl (Pseudomonas fluorescens strain SBW25). Also shown are the Genbank accession number for each protein. This neighbor-joining tree is built based on the observed number of amino acid differences. Bootstrap values (%) are shown beside each branch and the units at the bottom indicate the number of amino acid differences. To the right are stick diagrams showing the position of WxxxE motifs (W) and putative ERMRSs (E).
Recently, Alto et al. (2006) reported that members of a family of type III effectors from animal pathogens, including Escherichia coli, Shigella, and Salmonella, perturb host functions by mimicking constitutively active Ras-like G-proteins. Despite producing the same effects as activated G-proteins, these effectors do not bind guanine nucleotides or possess apparent GTPase domains. Moreover, they all contain a WxxxE motif that is essential for their function as G-protein mimics. In addition, these effectors also possess various C-terminal sub-cellular localization signals. In this study, we report that nearly all of the AvrE-family members contain one or two WxxxE motifs and a putative C-terminal endoplasmic reticulum membrane retention/retrieval signal (ERMRS). We used site-directed mutagenesis to inactivate both of the WxxxE motifs and/or the predicted ERMRS in Pnss WtsE and Pto AvrE1 and then tested their biological activities in host and nonhost plants. Our results demonstrate that both the WxxxE and predicted ERMRS motifs are required for multiple virulence and avirulence activities of AvrE-family effector proteins, which suggests that the AvrE-family effectors might mimic activated plant G-proteins. This would be a new mode of action for TTSS effector proteins from plant pathogenic bacteria.
Results
AvrE-family type III effectors contain WxxxE motifs
Recently, type III effector proteins from several animal pathogenic bacteria were shown to alter host cells by mimicking activated Ras-like GTPases (Alto et al., 2006; Arbeloa et al., 2008) and this activity depends on invariant WxxxE motifs and eukaryotic sub-cellular localization signals. We examined the available sequences of AvrE-family proteins and found that they form two major clades (Fig. 1), representing the erwinias and the pseudomonads. Moreover, the amino-terminal halves of members of both clades contain WxxxE motifs. AvrE homologs from Pnss, Ea, Erwinia pyrifoliae, Pto, Pseudomonas syringae pv. phaseolicola 1448A (Pph) and Pseudomonas cichorii possess two WxxxE motifs, while those from Pantoea agglomerans pv. gypsophilae, P. syringae pv. syringae B728a, and Pseudomonas fluorescens SBW45 (RopE) have only one WxxxE. Recently, Araki et al. (2007) released the sequences of 96 AvrE homologs from strains of Pseudomonas viridiflava, a pathogen of A. thaliana. These strains contained either of two different Hrp PAIs, each with a slightly diverged avrE allele. All 96 AvrE proteins from P. viridiflava contained two WxxxE motifs.
Arbeloa et al. (2008) reported that EspM2 and EspM3, WxxxE-containing mammalian effectors from Citrobacter rodentium, retained function when their WxxxE motifs were mutated with conservative W to Y or E to D substitutions, whereas changing either W or E to A disrupted their function. We have determined that the AvrE-family proteins contain seven highly conserved Ws (Supplementary Fig. 1). Those sites with substitutions of Y or F for W and D or other positively charged residues for E may be functional variants of the WxxxE motif.
Similar to the G-protein mimicking mammalian effectors, most AvrE-family proteins also contain a putative sub-cellular localization motif at their C-termini. In our previous studies, we found a motif at the C-terminus of WtsE (KKEGFEMK) that is predicted by the PSORT II algorithm to function as an ERMRS (Fig. 1) and we demonstrated that this motif is essential for the virulence and avirulence activities of WtsE (Ham et al., 2006; 2008). Among AvrE-family members examined, PSORT II predicted a C-terminal ERMRS (KKxGxExK) for all of the effectors except DspE from Pectobacterium atrosepticum and the AvrE alleles from P. viridflava strains containing one of the two alternative Hrp PAIs. (Representative AvrE homologs from P. viridiflava with and without an ERMRS are included in Fig. 1.). It is interesting to note that the WxxxE and predicted ERMRS motifs are found in all of the AvrE-family members that have been shown to make large virulence contributions, such as those from Pnss, Pto, and Ea. It is possible that the absence of the WxxxE or ERMRS motifs in P. atrosepticum and some strains of P. viridiflava is somehow related to their different pathogenic lifestyles. Pnss, Pto, and Ea are hemi-biotrophic pathogens, whereas P. atrosepticum and some strains of P. viridiflava are necrotrophic, soft-rot pathogens, which rely on cell wall-degrading enzymes more than type III effectors as major virulence factors. Mutant studies have shown that the virulence contribution of DspE in P. atrosepticum appears to be less obvious than in Pnss, Pto, and Ea (Holeva et al., 2004).
The WxxxE motifs are required for WtsE-induced cell death in host and nonhost plants
To study the cell death activity of WtsE in the absence of any other TTSS effector proteins produced by Pnss, we employed E. coli MC4100 carrying the entire Dickeya dadantii (Dda) hrp gene cluster in plasmid pCPP2156 as a delivery system for WtsE, which was encoded by derivatives of plasmid pJH021 wtsEF+. The Dda TTSS has been shown to promiscuously translocate effectors from many different plant pathogenic bacteria (Ham et al., 1998) and it has worked well to introduce WtsE into sweet corn, tobacco and N. benthamiana (Ham et al., 2006; 2008). Since changing either the W or E residue to A in the WxxxE motifs of mammalian effectors resulted in loss of G-protein mimicking activity (Alto et al., 2006; Arbeloa et al., 2008), we changed the W residue in each of the two WxxxE motifs of WtsE to A; W694A and W840A mutations were constructed by site-directed mutagenesis in plasmid pJH021. The single mutants were designated WtsE-wxe1 and WtsE-wxe2, respectively, and the double mutant was designated WtsE-wxe12. We previously deleted aa 1831-34 (FEMK) within the putative ERMRS (Ham et al., 2008) and designated this mutant ΔFEMK.
We first tested the ability of the wtsE-wxe mutants to cause disease-associated cell death in susceptible plants. Corn leaves were vacuum-infiltrated with E. coli MC4100 (pCPP2156) strains delivering the WtsE derivatives and water-soaking (Wts) and necrosis were both rated visually in corn seedlings and quantified by measuring electrolyte leakage (EL) from corn leaf discs. Wild-type WtsE caused severe Wts and substantial EL (Fig. 2A). Disruption of either of the two WxxxE motifs alone did not significantly reduce the amount of Wts, although the wxe1 mutant did reduce EL. However, the wxe12 and ΔFEMK mutants both failed to cause cell death or EL (Fig. 2A).
Fig. 2.
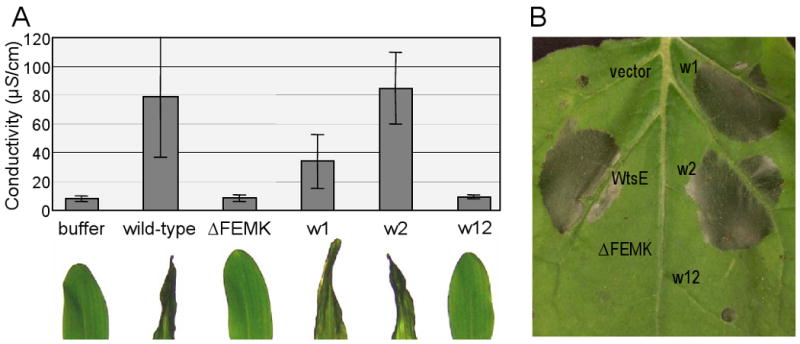
Cell death phenotypes induced by delivery of WtsE from E. coli into host and nonhost plants. A, photographs of corn seedling leaves (below) and corresponding ion leakage data (above) following infiltration with E. coli MC4100 (pCPP2156) strains delivering wild-type WtsE or the indicated derivatives. Each WtsE plasmid also encodes the chaperone WtsF. Conductivity values are means ± SD for three replications. B, photograph of the HR in N. benthamiana following delivery of WtsE or the indicated derivatives from E. coli. Photographs were taken at 15-18 h after infiltration and conductivity assays were done at 18 h after infiltration. Results are representative of three independent experiments
In addition to causing disease-associated cell death in susceptible plants, WtsE also causes a rapid hypersensitive response (HR) in nonhost plants (Ham et al., 2008). To determine if the WxxxE motifs and the putative ERMRS are also required for HR elicitation, we infiltrated the E. coli delivery strains into N. benthamiana (Fig. 2B) and tobacco leaves (Supplementary Fig. 2). Again, wild-type WtsE and the wxe1 and wxe2 single mutants elicited a strong HR, whereas the wxe12 and ΔFEMK mutants did not. Similar results were obtained when WtsE or the derivatives were delivered into N. benthamiana by agro-infiltration (Supplementary Fig. 3A) or by Pnss (Supplementary Fig. 3B). Collectively, these results indicate that the WxxxE motifs are functionally redundant and that both the C-terminal ERMRS and the WxxxE motifs within WtsE are required for both HR elicitation in nonhost plants and disease-associated cell death in susceptible plants.
The WxxxE motifs in WtsE are required for full virulence of P. stewartii in host plants
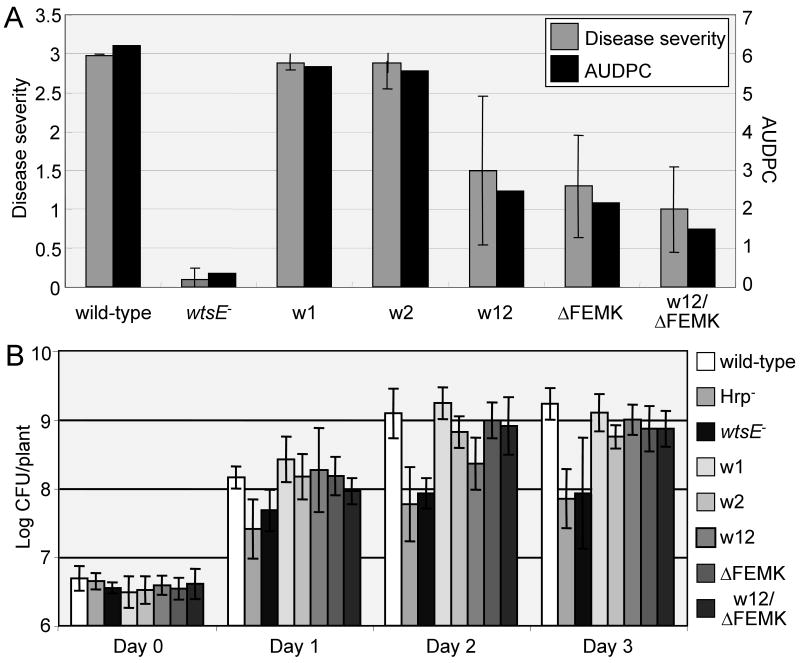
The wxe1, wxe2, wxe12, and ΔFEMK mutations, as well as a triple wxe12/ΔFEMK mutation, were introduced into the Pnss DC283 genome by homologous recombination. Sweet corn seedlings were inoculated with these mutants by placing bacterial suspensions into the whorls and then disease severity was rated daily for 5 days. Disrupting either one of the WxxxE motifs did not significantly alter the ability of Pnss to cause water-soaked lesions, as shown by both the final disease severity rating and the area under the disease progress curve (AUDPC) in Fig. 3A. The wxe12 mutation resulted in significant loss of virulence, again indicating that the two WxxxE motifs are functionally redundant. A comparable reduction was observed for the ΔFEMK mutant, but this level was not reduced any further in the triple wxe12/ΔFEMK mutant. The virulence of the wxe12, ΔFEMK, and triple mutants was fully restored by complementation with plasmid pAA007 wtsEF+, with disease severity ratings of 2.8 to 3.0±0.3 for all mutants and the DC283 parent strain at 5 days after inoculation. These results indicate that the WxxxE and putative ERMRS motifs likely contribute to the same virulence function of WtsE and that the wxe12 or ΔFEMK mutations each fully disrupt this activity.
Fig. 3.
Effects of mutations in the WxxxE and ERMRS motifs of WtsE on the virulence of Pnss. A, the whorls of sweet corn seedlings were inoculated and rated as described in Materials and Methods. Disease severity ratings, scoring for lesion formation, were taken daily and the area under the disease progress curve (AUDPC) was calculated for days 1 to 6 after inoculation. Disease severity ratings are shown for day 5 after inoculation. Mean ratings ± SD were calculated from 24-31 plants per strain and are representative of three trials. B, growth of wild-type Pnss and wtsE derivatives in sweet corn seedlings. Whorls were inoculated with 5 × 106 CFU/plant. At daily intervals, viable cell counts were done for each of four plants per strain. The experiment was done twice and bacterial populations ± SD are the combined means from both experiments. The following Pnss strains were tested; DC283 (wild-type), AA005 wtsE∷aphT, JH037 wtsE-wxe1, JH030 wtsE-wxe2, JH039 wtsE-wxe12, JH033 wtsE-ΔFEMK, and JH042 wtsE-wxe12/ΔFEMK.
The growth of the wild-type and mutant Pnss strains in sweet corn plants was also examined (Fig. 3B). In contrast to the reduction in symptoms, the growth of the wxe12, ΔFEMK, and wxe12/ΔFEMK mutant strains was not significantly compromised. Each of these mutants grew significantly better than the wtsE null mutant, AA005, and a TTSS-deficient mutant, DM711 hrpJ. Thus, the mutant WtsE proteins apparently retain an additional growth-promoting virulence function that does not require WxxxE or ERMRS motifs, at least under the experimental conditions used in this study. In addition to promoting Pnss growth, the wxe12 and ΔFEMK mutants retain other activities, including some slight residual cell death activity in N. benthamiana (Supplemental Figs. 3A and 3B) and production of a few lesions in sweet corn (Fig. 2A), which were not observed with a wtsE null mutant. The relationship of these functions to the variants of WxxxE motifs (Supplementary Fig. 1) and to G-protein mimicry is unknown.
To determine whether the altered virulence of the mutant strains is caused by changes in the production, stability or secretion of the WtsE derivatives from Pnss, we did Western blots of cell extracts and supernatants using antibodies to Ea DspE, which cross-react with WtsE. All of the WtsE derivatives exhibited normal production and secretion in culture (Supplementary Fig. 3C). Evidence that wild-type WtsE can be expressed and delivered from E. coli MC4100 (pCPP2156) and from Pph (see below) comes from its in planta activity following delivery by these systems. Since the studied mutations do not affect type III secretion from Pnss, they are similarly unlikely to affect type III secretion in the E. coli/hrpDda or Pph systems. Efforts to monitor in planta production and stability of WtsE following Agrobacterium-mediated transient expression failed to detect WtsE or its derivatives (data not shown). However, the ability of the wtsE-wxe and wtsE-ΔFEMK mutants to promote Pnss lesions and growth relative to the wtsE null mutant (Fig. 3) indicates that the mutant proteins accumulate and retain some function in corn cells.
WxxxE and predicted ERMRS motifs are responsible for cell death and defense suppression activities of WtsE in A. thaliana
Pph strain NPS3121 is a bean pathogen that is non-pathogenic in A. thaliana. Although Pph does not elicit an HR in A. thaliana, it does elicit strong basal defense responses, including PR-1 protein production and callose deposition in the cell wall (Ham et al., 2007). When Pph expresses HopM1, which is functionally redundant to AvrE1, these defenses are suppressed (Ham et al., 2007). Thus, Pph apparently lacks versions of HopM1 and AvrE1 that are functional in Arabidopsis. We previously reported that Pph translocates WtsE into A. thaliana, where it induces water-soaking and suppresses callose formation and the synthesis of PR-1 (Ham et al., 2008). Here, we tested the effects of the wxe12 and ΔFEMK mutations on these virulence activities of WtsE. As shown in Fig. 4A, Pph expressing either wild-type WtsE or HopM1 caused tissue collapse of A. thaliana at 24 h after infiltration with a bacterial suspension (1 × 108 CFU/ml), whereas Pph strains with the wxe12 or ΔFEMK derivatives of WtsE did not show any apparent cell death activity. To determine whether these motifs were also important for the suppression of Pph-induced defenses, we examined callose deposition and PR-1 protein accumulation after infiltration of Pph expressing WtsE or its derivatives. WtsE suppressed Pph-induced small callose deposits by ∼50% and large callose deposits by ∼90%. WtsE also suppressed Pph-induced accumulation of PR-1 by 50 to 90% in individual biological replicates. In contrast, the wxe12 and ΔFEMK mutant proteins failed to significantly suppress callose deposition (Fig. 4B) and PR-1 protein accumulation (Fig. 4C). These results indicate that, in addition to their role in the induction of disease-associated cell death, these motifs are also important for suppression of Pph-induced defenses by WtsE.
Fig. 4.
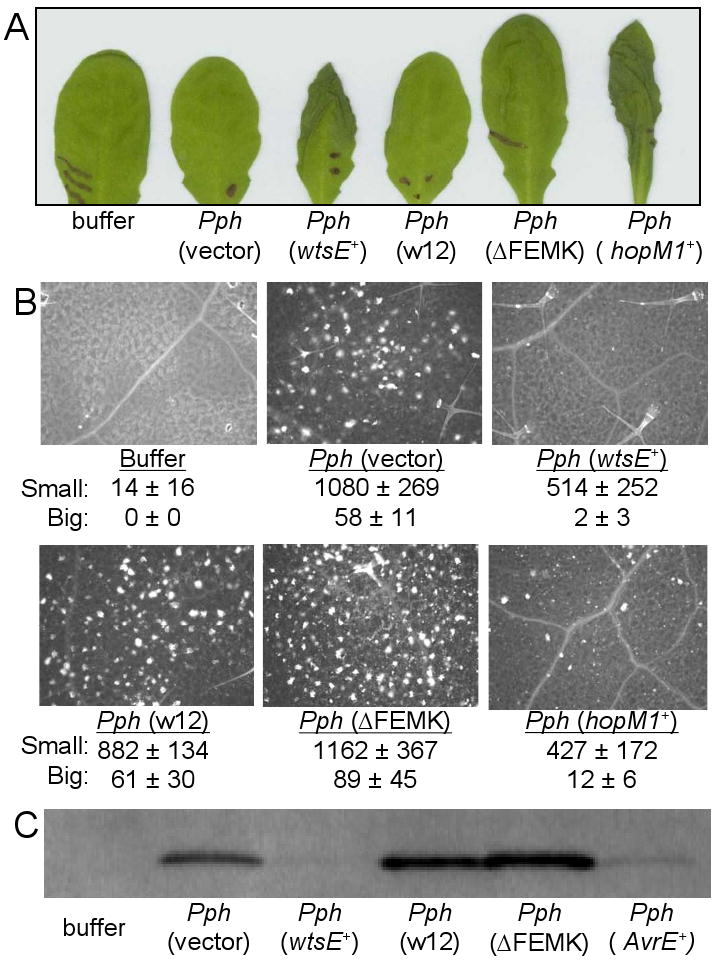
Effect of mutating the WxxxE and ERMRS motifs in WtsE on cell death-inducing and defense-suppressing activities in A. thaliana. Wild-type WtsE, derivatives of WtsE, HopM1 or AvrE were delivered by infiltration of P. syringae pv. phaseolicola (Pph) strain NPS3121 (at 1 × 108 CFU/ml) carrying the following expression plasmids: pJH082 wtsEF+, pJH083 wtsE-ΔFEMK, pJH084 wtsE-wxe12, pORF43 hopM1+, and pAVRE avrE1+. Bacteria carrying pAVRE also carried pAVRF. A, tissue collapse of A. thaliana leaves. The photograph was taken 24 h after infiltration. B, callose deposition visualized by fluorescence microscopy at 15 h after infiltration. Counts of small (<20 μ diameter) and large (>20 μ diameter) callose deposits are the means ± SD from 5 replications. C, PR-1 protein expression detected by Western blot analysis at 48 h after infiltration.
The WxxxE and ERMRS motifs are required for AvrE1-mediated virulence in Pto
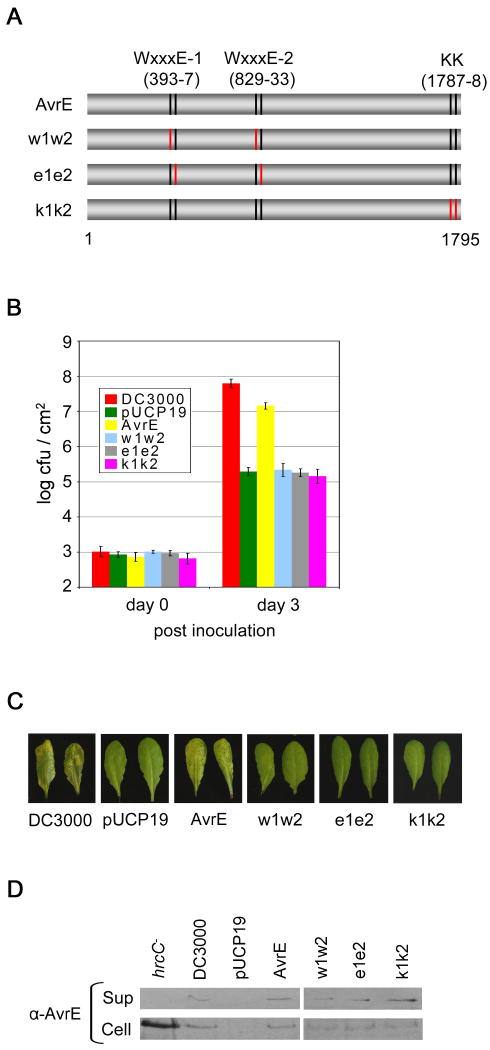
To determine whether the requirement for the WxxxE and predicted ERMRS motifs is unique to WtsE or a more general feature of AvrE-family effectors, we studied AvrE1, which is a major virulence factor of the A. thaliana and tomato pathogen Pto DC3000. We made two double mutations in the WxxxE motifs of AvrE1 [w1w2 (W393A/W829A) and e1e2 (E397A /E833A); Fig. 5A]. We also mutated the two K residues (KK1787-88AA) within the putative C-terminal ERMRS motif (Fig. 5A). All of the mutations were made in pAVRE. Together with pAVRF (encoding AvrF, the putative molecular chaperone for AvrE1 secretion), the wild-type or mutant pAVRE plasmid was transformed into the Pto ΔCEL mutant, which lacks the functionally redundant avrE and hopM1 genes (Alfano et al., 2000; DebRoy et al., 2004; Badel et al., 2006).
Fig. 5.
The WxxxE and ERMRS motifs are required for the virulence activity of P. syringae pv. tomato (Pto) AvrE1 in A. thaliana. A, a diagram of the AvrE1 protein, indicating the locations of the motifs and various mutations introduced. Numbers indicate the amino acid positions. B, bacterial populations in A. thaliana Col-0 leaves at 3 days after inoculation. Leaves were inoculated by infiltration with 1 × 106 CFU/ml of Pto ΔCEL bacteria carrying wild-type or mutant pAVRE and pAVRF. C, disease symptoms at 3 days after inoculation. D, immunoblot analysis of the production and secretion of wild-type and mutant AvrE1 proteins in Pto ΔCEL. Bacteria were grown in hrp-inducing liquid medium. Cultures were separated into supernatant (Sup) and cell (Cell) fractions by centrifugation. AvrE1 proteins in total lysates were detected by immunoblotting using an AvrE1 antibody.
We then examined the ability of the avrE1 mutants to complement the virulence defect of the ΔCEL mutant. As reported previously (DebRoy et al., 2004), the ΔCEL mutant was severely reduced in multiplication (wild-type DC3000 reached >100 fold higher populations at 3 days after inoculation) and caused little or no disease symptoms in A. thaliana (Figs. 5B and 5C). pAVRE (along with pAVRF) restored the in planta growth of the ΔCEL mutant and production of disease symptoms to near the level of wild-type DC3000 (Figs. 5B and 5C). All mutant derivatives of AvrE1 were defective in the virulence function(s) assessed by bacterial growth and disease symptoms (Figs. 5B and 5C). As a control, we verified that none of the mutations significantly impaired AvrE1 production or secretion in the ΔCEL mutant (Fig. 5D).
Discussion
AvrE-family proteins are conserved in several important genera of Gram-negative plant pathogenic bacteria and there is clear genetic evidence that they are crucial to their virulence. Since the avrE-family genes are part of a conserved TTSS PAI, it is likely that these effectors have played a major role in the evolution of plant pathogenic bacteria from commensals. For technical reasons, it has been historically difficult to study the virulence mechanisms of AvrE-family effectors. Not only are these effectors extremely large proteins, but they also induce cell death when expressed in plants or yeast (Meng et al., 2006; Ham et al., 2008). While induction of cell death in both plants and yeast suggest that AvrE-family proteins may target a cellular process that is fundamental to eukaryotic cells, it has precluded the use of several experimental approaches (e.g., transgenic expression and yeast two-hybrid assays with full-length effector proteins) that have proven to be useful in the study of other effectors. The lack of overall sequence similarities to proteins of known functions has also hindered hypothesis-driven experiments. In this context, the identification of WxxxE motifs putatively involved in G-protein-mimicry and the genetic demonstration of their importance in virulence mark a significant advance in our understanding of how these important effectors contribute to bacterial pathogenesis in plants. Moreover, our results suggest that manipulating host pathways by mimicking host G-proteins could be a widespread virulence mechanism employed not only by animal pathogens, but also by plant pathogens. At this point, we are not in a position to demonstrate biochemically that WtsE or AvrE1 have the same effects on plant cells as a known activated G-protein because the targets of these effectors and their effects on plant cells are unknown. However, the likelihood that changing only two amino acids at different positions in two such large proteins with so little aa identity to each other would have similar dramatic affects on their function seems rather remote. Furthermore, mutating these residues is unlikely to have a non-specific effect, such as disrupting protein structure, since mutating each residue alone in WtsE has little to no effect on its function.
Although the presence of WxxxE motifs indicates that G-protein mimicry may be a common mode of action among most AvrE-family members, the activity of individual AvrE-family effectors may be host-adapted. For example, unlike native AvrE1, WtsE is unable to promote the growth of Pto ΔCEL in A. thaliana and P. agglomerans pv. gypsophiliae DspE is unable to complement a Pnss wtsE mutant in maize (data not shown). Furthermore, AvrE-family members are likely to have additional virulence activities independent of G-protein mimicry. This notion is supported by the large size of the AvrE-family proteins and our finding that Pnss strains carrying the wtsE-wxe12 and wtsE-ΔFEMK mutations still produce a few small lesions and grow in corn significantly better than a nonpathogenic wtsE null mutant and nearly as well as the wild-type parent. Such putative additional virulence activities may explain why P. atrosepticum retains an AvrE effector gene that lacks a WxxxE motif. Alternatively, variant motifs, such as WxxxD, may also function in G-protein mimicry, in which case all AvrE-family members might engage in this activity.
As discussed above, AvrE-family effectors are very interesting because they both promote host cell death and suppress basal defenses (DebRoy et al., 2004; Boureau et al., 2006; Ham et al., 2008). Prior to this study, however, it was unknown whether these dual functions were mediated by distinct activities or resulted from interconnected effects of a single activity. The large size of the AvrE-family effectors would be consistent with their being multifunctional. For example, in susceptible hosts these effectors could use one domain (activity) to disrupt membrane permeability by inducing HR-like cell death while employing another domain to control the damage, thus avoiding a full-blown HR and instead causing the water-soaking symptom common to many bacterial diseases. However, we provide evidence here that disease-associated cell death, HR elicitation and defense suppression all require the same WxxxE and putative ERMRS motifs in WtsE and AvrE1. Therefore, to carry out these activities, AvrE-family effectors may target a single pathway or, alternatively, mimic more than one G-protein to target multiple host cellular processes. In the case of the nonhost HR, this common target(s) could be guarded by an R-protein. Interestingly, a function of WtsE that is independent of its WxxxE and putative ERMRS motifs also contributes to the growth of Pnss in corn. The nature of this additional activity is unknown.
Small G-proteins are central regulators of cytoskeleton dynamics and intracellular vesicle trafficking in eukaryotes, including higher plants (Vernoud et al., 2003; Samaj et al., 2006). Future research will be aimed at determining the mechanisms by which AvrE-family effectors manipulate host G-protein pathways to perturb the cytoskeleton and/or vesicular trafficking. Notably, P. syringae HopM1 interacts with and promotes degradation of several host targets, including AtMIN7, a guanine exchange factor that is predicted to activate small G-proteins (Nomura et al., 2006). Though sequence unrelated, HopM1 is functionally redundant with AvrE1 for the virulence of Pto. Thus, AvrE-family effectors may suppress defenses by perturbing G-protein regulated processes including export of antimicrobial cargo, such as phytoalexins and/or PR-proteins, and production of cell-wall defenses, such as callose deposition. Meng et al. (2006) have shown that DspA interacts with apple receptor-like kinases that are putatively involved in host defense. G-protein mimicry could also suppress activation associated internalization of is class of proteins (Robatzek et al., 2006). In addition, sustained perturbation of normal vesicle trafficking or cytoskeleton dynamics may lead to host cell death. Thus, G-protein-mimicry may explain the dual function of AvrE-family effectors in suppression of host basal defenses and induction of disease symptoms.
Materials and Methods
Bacterial strains and their growth conditions
Bacterial strains and plasmids used in this study are described in Table 1. E. coli, Pnss, and A. tumefaciens strains were routinely grown and maintained in Luria-Bertani (LB) media (Sambrook and Russel, 2001) at 28°C to 30°C. P. syringae pvs. phaseolicola and tomato strains were grown in King's B agar at 30°C (Schaad et al., 2001) or on LB without NaCl (for Pto). Appropriate antibiotics were added at the following concentrations (μg/ml): ampicillin (Ap), 200; kanamycin (Km), 100 for P. syringae and 50 for Pnss and E. coli; nalidixic acid (Nal), 20; spectinomycin (Sp), 50; rifampicin (Rif), 100; and tetracycline (Tc), 25.
Table 1.
Plasmids and bacterial strains used in this study
| Description | Source/Reference | |
|---|---|---|
| Agrobacterium tumefaciens | ||
| C58C1 | Vir-, prototrophic strain, Rifr | Van Larebeke et al., 1974 |
| Escherichia coli | ||
| DH5α | mcrBC+ supE44 ΔlacU169 (f80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Invitrogen |
| MC4100 | araD139 Δ(argF-lac)205 λ− flbB5301 ptsF25 relA1 rpsL150 deoC1 | CGSCa Casadaban, 1976 |
| Pantoea stewartii subsp. Stewartii | ||
| DC283 | A Nalr derivative of the wild-type strain SS104 | Coplin et al., 1986 |
| DM711 | DC283 hrpJ79∷Tn3HoHoI, Apr | Frederick et al., 2001 |
| DM5101 | A wtsE∷miniTn5gus derivative of DC283, Kmr | Ham et al., 2006 |
| AA005 | A wtsE∷aphT derivative of DC283, Kmr | Ham et al., 2006 |
| JH030 | A derivative of DC283 expressing WtsE-W840A | This study |
| JH033 | A derivative of DC283 expressing WtsE Δaa1831-1834 | This study |
| JH037 | A derivative of DC283 expressing WtsE-W694A | This study |
| JH039 | A derivative of DC283 expressing WtsE-W694A/W840A | This study |
| JH042 | A derivative of DC283 expressing WtsE-W694A/W840A and WtsE-Δaa1-236 | This study |
| Pseudomonas syringae pv. Phaseolicola | ||
| NPS3121 | A wild-type strain, Rifr | Lindgren et al., 1986 |
| Pseudomonas syringae pv. Tomato | ||
| DC3000 | Wild-type, Rifr | Cuppels, 1986 |
| DC3000 ΔCEL | DC3000, ΔCEL∷ΩSp/Sm, Rifr | Alfano et al., 2000 |
| DC3000 hrcC | DC3000, hrcC∷miniTn5Cm, Rifr Cmr | Yuan and He, 1996 |
| Plasmids | ||
| pAA007 | pLAFR3 with 8.9-kb EcoRI/BamHI fragment carrying wtsE and wtsF, Tcr | Ham et al., 2006 |
| pAVRE | pUCP19 containing avrE1 and its own promoter, Apr | Nomura et al., 2006 |
| pAVRF | pDSK519 containing avrF with its own promoter, Kmr | Nomura et al., 2006 |
| pAVRE-w1w2 | Derivative of pAVRE expressing AvrE1-W393A/W829A, Apr | This study |
| pAVRE-e1e2 | Derivative of pAVRE expressing AvrE1-E397A/E833A, Apr | This study |
| pAVRE-k1k2 | Derivative of pAVRE expressing AvrE1-K1787A/K1788A, Apr | This study |
| pBluescript KS and SK (+) | ColE1 α-lacZ, Apr | Stratagene |
| pCR®4-TOPO® | A TOPO® vector used for cloning PCR products, Apr, Kmr. | Invitrogen |
| pCPP2156 | A cosmid clone carrying the D. dadantiii hrp cluster, Spr | Ham et al., 1998 |
| pGDG | Binary vector for agroinfiltration, derivative of pCAMBIA with 35S promoter and GFP, Kmr | Goodin et al., 2002 |
| pJH021 | pBluescriptSK(+) clone of the 6.4-Kb EcoRI/BamHI fragment from pJH001 carrying the wtsEF operon with its native promoter, Apr | Ham et al., 2008 |
| pJH023 | Derivative of pJH021 with WtsE-Δaa1831-1834, Apr | Ham et al., 2008 |
| pJH050 | Derivative of pJH021 with WtsE-W694A, Apr | This study |
| pJH052 | Derivative of pJH021 with WtsE-W840A, Apr | This study |
| pJH055 | Derivative of pJH021 with WtsE-W694A/W840A, Apr | This study |
| pJH059 | Derivative of pJHI02G with ΔFEMK in WtsE, Kmr | This study |
| pJH063 | Derivative of pJH102G with W694A/W840A in WtsE, Kmr | This study |
| pJH101G | pGDG∷wtsE, Kmr | This study |
| pJH102G | pGDG∷wtsE-Δaa1-236, Kmr | This study |
| pJH103G | pGDG∷wtsE-Δaa1-492, Kmr | This study |
| pLD55 | A suicide vector with R6K ori, αlacZ, tetAR, Apr | Metcalf et al., 1996 |
| pORF43 | pUCP19 carrying shcM-hopPtoM with the native shcM gene promoter, Apr | Badel et al., 2003 |
| pRF205 | pVK100 with a 1.7-kb HindIII fragment containing Plac-hrpS+, Tcr | Frederick et al., 1993 |
The Coli Genetic Stock Center at Yale University.
Arabidopsis Biological Resource Center at The Ohio State University.
DNA manipulation, transformation and sequencing
Standard protocols described by Sambrook and Russel (2001) were used for gene cloning procedures, such as DNA isolation, digestion with restriction enzymes, and ligation. Appropriate Qiagen (Valencia, CA, U.S.A.) kits were utilized for isolation of DNA fragments from agarose gels or PCR reactions. Cloning of PCR products was conducted with Platinum® Taq DNA polymerase High Fidelity and TOPO® vectors (Invitrogen, Carlsbad, CA, U.S.A.) following the manufacturer's protocols and all the clones obtained were confirmed by DNA sequencing with an ABI 3700 capillary electrophoresis sequencer (Applied Biosystems, Foster City, CA, U.S.A.) at The Ohio State University Plant-Microbe Genomic Facility. Oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, IA, U.S.A.) Bacterial transformation was conducted by either electroporation using a GenePulser™ (Bio-Rad Laboratories, Hercules, CA, U.S.A.) apparatus set to 200 Ω and 1.5 kv or triparental mating as previously described (Merighi et al., 2003).
Plant assays
Sweet corn seedlings (Zea mays cv. Seneca Horizon), tobacco (Nicotiana tabacum cv. Bottom Special), N. benthamiana, and beet (Beta vulgaris cv. Red Ace Hybrid) were grown and maintained in growth chambers set to 30°C, 90% humidity, and 18/6 h day/night cycle. A. thaliana (ecotype Col-0) plants were grown in a growth room at 23°C for 8 h of light and 16°C for 16 h of darkness for WtsE studies or at 20°C with a 12 h light/dark cycle for AvrE1 studies. Pnss and E. coli suspensions for the cell death assays in host and non-hosts were prepared as previously described (Ham et al., 2006). For the cell death assay in sweet corn, 5-day-old seedlings in 5-inch-pots were inverted, submerged in beakers containing the bacterial suspensions (109 CFU/ml), and vacuum infiltrated (Ham et al., 2006). For the cell death assays in non-host plants, the bacterial suspensions were infiltrated into the abaxial side of the leaves with needleless syringes. Conductivity assays to measure electrolyte leakage from corn leaf disks were done as previously described (Ham et al., 2007).
Procedures for virulence assays of Pnss strains in sweet corn seedlings are described in our previous studies (Frederick et al., 2001; Ham et al., 2006). Briefly, bacterial suspensions containing 1 × 107 CFU/ml in buffered 0.2% Tween 40 were added to the whorls of 7-day-old seedlings without wounding and disease severity was rated on a 0 to 3 scale (0 = no symptoms, 1 = a few lesions but no ooze, 2 = many lesions and some ooze, and 3 = confluent lesions and ooze). For in planta growth assays, individual seedlings were homogenized with a Powergen 700 homogenizer (Fisher Scientific, Pittsburg, PA, U.S.A.) in 5 ml of 10 mM potassium phosphate buffer (pH 7.0) and dilution-plated on LB agar supplemented with Nal. Bacterial growth and disease assays in A. thaliana with P. syringae strains were performed following previously described methods (Hauck et al., 2003).
Bioinformatic techniques
Amino acid sequences of AvrE-family proteins were obtained from the National Center for Biotechnology Information (NCBI) database (www.ncbi.nlm.nih.gov) and the putative ERMRS motifs of the AvrE-family proteins were identified by the PSORT II program (Nakai and Kanehisa, 1992); (psort.nibb.ac.jp). Alignment of AvrE-family proteins and creation of a neighbor-joining tree were conducted by using the software MEGA 4.1 (Tamura et al., 2007).
Callose staining and PR-1 detection
For callose staining, the infiltrated leaves were bleached with lactophenol solution, then stained with 0.01% aniline blue in 150 mM K2HPO4, and observed under a Nikon Eclipse 80i epifluorescence microscope (Nikon, Melville, NY, U.S.A.) as previously described (Kim et al., 2005). PR-1 proteins produced in A. thaliana were detected by Western blot analysis (Kim et al., 2005) using ECL-plus reagents and a Storm phosphorimager (GE Healthcare, Piscataway, NJ, U.S.A.).
WtsE and AvrE1 constructs
The GeneTailor™ Site-Directed Mutagenesis System (Invitrogen) was used to create the W694A, W840A and Δaa1831-1834 mutations in wtsE. The procedure for generating WtsE-Δaa1831-1834 was described in our previous study (Ham et al., 2008). For W694A and W840A mutations, the 3.8-kb BamHI/NcoI-cut fragment of pAA008 containing the 5′-half portion of wtsE (Ham et al., 2006) was initially cloned into pET41b(+) (Novagen, San Diego, CA, U.S.A.). In turn, the 2.4-kb ClaI/NcoI region of wtsE, which contains both WxxxE1 and WxxxE2 motifs, was subcloned into pBSKS(+) using the ClaI site in wtsE and the KpnI site in pET41. The plasmid DNA of this subclone was then used as a template for PCR with overlapping primers. For the W694A mutation, the template plasmid DNA was initially methylated with SssI DNA methylase (New England Biolabs, Inc., Ipswich, MA, U.S.A.) and PCR was carried out with an overlapping primer set consisting of forward primer 5′-TTGATCAGCTTACCAGGGGCGCCACAGAAGCAGAG-3′ and reverse primer 5′-TGGTAAGCTGATCAAAATAGTGCACTTTGCCATCC-3′. The PCR products were subsequently introduced into E. coli DH5α, which degrades the methylated parental DNA. Plasmids with the W694A mutation were initially screened for the KasI site introduced by the forward primer (underlined), and then confirmed by DNA sequencing. The same procedure was used to create the W840A mutation, except that the overlapping primer set 5′-TTTCGCCAGGCACGTGAGCATGCGCAGCAGGGTGA-3′ and 5′-ACGTGCCTGGCGAAATATCTCTCCTTCGCGGGTCA-3′ was used and mutant plasmids were screened for the SphI site introduced by the forward primer (underlined). These were also confirmed by DNA sequencing. For the W694A/W840A double mutation, the same procedure was carried out using the plasmid with the W694A mutation as the template and the primer set for the W840A mutation. Then the 854-bp PstI-NcoI fragments carrying each mutation were substituted for the corresponding region of pJH001, which contains the entire wtsEF operon with its own promoter in pCR®4-TOPO® (Ham et al., 2008). These inserts were finally excised from pJH001, as 6.4-kb EcoRI/BamHI fragments, and recloned into pBSSK(+), generating pJH050, pJH052, and pJH055 for the W694A, W840A, and W694A/W840A derivatives of WtsE, respectively. In addition, the same inserts were also cloned in pDSK519 (Keen et al., 1988) to produce pJH082, pJH083, pJH084, and pJH085, which express WtsE derivatives for wild-type, Δaa1831-1834, W694A/W840A, and W694A/W840A/Δaa1831-1834 respectively.
For Agrobacterium-mediated transient expression, pJH101G wtsE+, pJH102G wtsE-Δaa1-236 and pJH103G wtsE-Δaa1-492 were constructed as described by Ham et al. (2008) for pJH101, pJH102 and pJH103, respectively. However, the vector pGDG (Goodin et al., 2002), which creates N-terminal GFP protein fusions, was used instead of pGD. For pJH059, which is a derivative of pJH102G withwtsE-ΔFEMK, the 5.5 kb XhoI/BamHI fragment from pJH023 was ligated into XhoI/BamHI-cut pGDG. For pJH063, which is a derivative of pJH102G with wtsE-wxe12, the 5.5 kb XhoI/BamHI fragment from pJH055 was ligated into XhoI/BamHI-cut pGDG.
For recombination into the chromosome of Pnss, the 2.4-kb SalI-KpnI fragments of wtsE with each W694A/W840 mutation and the 2.4-kb KpnI-BamHI fragment of pJH023, which carries the Δaa1831-1834 mutation, were cloned into suicide vector pLD55 and introduced into wild-type Pnss strain DC283 followed by screening the recombinant strains as described by Merighi et al. (2006). The resulting chromosomal wtsE mutants were JH030, JH033, JH037, JH039, and JH042, which carry the W840A, Δaa1831-1834, W694A, W694A/W840A, and W694A/W840A/Δaa1831-1834 mutations, respectively. Each Pnss mutant was confirmed by PCR and subsequent diagnostic restriction enzyme digestions of the incorporated restriction sites mentioned above and in our previous study (Ham et al., 2008).
Site-directed mutagenesis of avrE1 in pAVRE was performed using the Quikchange™ XL site-directed mutagenesis kit (Stratagene, La Jolla, CA, U.S.A.) with sequence-specific primers for each mutation (Supplementary Table 1). The constructs were transformed into ΔCEL mutant bacteria carrying pAVRF. All mutations were verified by sequencing.
Protein expression and secretion assays
For AvrE1 from Pto, bacteria were grown on low-salt LB plates for 24 h at 28°C and then suspended in sterile water. Bacteria were washed two times with sterile water, resuspended in hrp-inducing fructose minimal medium (Jin and He, 2001) and then incubated with shaking at 18°C for 15 h. Cultures were separated into cell and supernatant fractions by centrifugation at 12,000 × g. The cell and supernatant fractions were concentrated 5 and 50 times, respectively. Proteins were separated on SDS–PAGE gels and transferred to Immobilon-P membrane (Millipore Corp., Billerica, MA, U.S.A.). Immunoblot analyses were performed using rabbit polyclonal AvrE1 antibodies produced by Cocalico Biologicals, Inc., Reamstown, PA, U.S.A.). For expression and secretion of WtsE from Pnss, cells were grown in hrp-inducing medium, culture supernatants were concentrated 350-fold, and assays by Western blots, using rabbit DspEEa antibodies, were done as described previously (Ham et al., 2006).
Supplementary Material
Supplementary Fig. 1. AvrE family members contain seven conserved tryptophans (W). A, stick diagrams showing the aligned N-terminal halves of the proteins and their extreme C-termini. The locations of conserved tryptophans (w1 to w7) are indicated at top. Below are shown the residues at position +4 relative to W (or, where indicated by an asterisk, relative to a conserved tyrosine (Y) or phenylalanine (F) in the comparable position). Glutamates (E, which form prototype WxxxE motifs) are shown in red, conserved aspartates (D) are shown in green, positively charged amino acids are shown in blue, and other amino acids are shown in black. After breaks in the stick diagrams, the C-termini with predicted ERMRS motifs are indicated by magenta ERs. B, pile-ups of the sequences within AvrE-family effector proteins. In the columns w1 to w7, W is shaded dark gray and conserved Y or F substitutions are light gray. E is red, D is green, lysine (K), arginine (R), or asparagine (N) are blue. The underlined WxxxE1 and WxxxE2 motifs from Pnss and Pto are those that were mutated in this study. At the right, are those sequences predicted by PSORTII to function as an ERMRS. Contrary to PSORTII, the sequence of AvrE from Pvi-AS appears quite similar to the other predicted ERMRSs while the sequence of RopE from Pfl appears quite divergent. Species abbreviations are given in Fig. 1.
Supplementary Fig. 2. The HR in tobacco following delivery of WtsE or the indicated derivatives from E. coli. The photograph was taken 16 h after infiltration. Results are representative of three independent experiments.
Supplementary Fig. 3. Cell death phenotypes of WtsE derivatives transiently expressed from A. tumefaciens or delivered from Pnss into N. benthamiana. A, Agrobacterium-mediated transient expression. A derivative of WtsE missing the first 236 aa is slightly more active in this system (Ham et al., 2008), so mutations were introduced into this truncated protein. A. tumefaciens C58C1 strains containing the following plasmids were infiltrated: 1) pGDG (empty vector control), 2) pJH101G full length wtsE, 3) pJH103G inactive wtsE-Δaa1-492, 4) pJH103G wtsE-Δaa1-236, 5) pJH063 wtsE-Δaa1-236-wxe12, 6) pJH059 wtsE-Δaa1-236-ΔFEMK. The leaf was photographed at 72 h after infiltration with a suspension of A. tumefaciens at A600 = 0.6. Note the weak residual cell death caused by expression of the wtsE-Δaa1-236-wxe12 mutant. B, cell death induced by wild-type and wtsE mutant Pnss strains. We have shown previously that Pnss induces HR-like cell death in N. benthamiana that is dependent on WtsE (Ham et al., 2008). Numbers correspond to infiltration of the following strains: 1) DC283 (wild-type), 2) DM711 hrpJ (Hrp-), 3) AA005 wtsE∷aphT, 4) buffer control, 5) DM5101 wtsE∷miniTn5gus, 6) JH039 wtsE-wxe12, and 7) JH033 wtsE-ΔFEMK. The leaf was photographed at 24 h after infiltration with a suspension of bacteria at 109 CFU/ml. Note the weak residual cell death caused by the Pnss wxe12 and ΔFEMK mutants. C, WxxxE and ERMRS mutations do not affect secretion of WtsE from Pnss. Western blots of concentrated culture supernatants using anti-DspEEa serum to detect WtsE. The following strains were tested; DM711 hrpJ (Hrp-), DC283 (wild-type), JH039 wtsE-wxe12, JH033 wtsE-ΔFEMK, JH042 wtsE-wxe12/ΔFEMK and AA005 wtsE∷aphT. Similar results were obtained with the pellet fractions and the experiment was done twice.
Acknowledgments
We thank S. V. Beer for the anti-DspEEa antibodies. This project was supported by grants from the National Research Initiative of the United States Department of Agriculture Cooperative State Research, Education, and Extension Service (grant number 2005-35319-15328) and the Ohio Agricultural Research and Development Center to DLC and DM, from the National Science Foundation (MCB-0718882) to DM, and from National Institute of Health and Department of Energy to SYH.
Footnotes
*The e-Xtra logo stands for “electronic extra” and indicates that three supplementary figures and one supplementary table are published online and that Figures 2, 4 and 5 are published in color online.
Literature Cited
- Alfano JR, Charkowski AO, Deng WL, Badel JL, Petnicki-Ocwieja T, van Dijk K, Collmer A. The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc Natl Acad Sci USA. 2000;97:4856–4861. doi: 10.1073/pnas.97.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alto NM, Shao F, Lazar CS, Brost RL, Chua G, Mattoo S, McMahon SA, Ghosh P, Hughes TR, Boone C, Dixon JE. Identification of a bacterial type III effector family with G protein mimicry functions. Cell. 2006;124:133–145. doi: 10.1016/j.cell.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Araki H, Innan H, Kreitman M, Bergelson J. Molecular evolution of pathogenicity-island genes in Pseudomonas viridiflava. Genetics. 2007;177:1031–1041. doi: 10.1534/genetics.107.077925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeloa A, Bulgin RR, MacKenzie G, Shaw RK, Pallen MJ, Crepin VF, Berger CN, Frankel G. Subversion of actin dynamics by EspM effectors of attaching and effacing bacterial pathogens. Cell Microbiol. 2008;10:1429–1441. doi: 10.1111/j.1462-5822.2008.01136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badel JL, Shimizu R, Oh HS, Collmer A. A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol Plant-Microbe Interact. 2006;19:99–111. doi: 10.1094/MPMI-19-0099. [DOI] [PubMed] [Google Scholar]
- Badel JL, Nomura K, Bandyopadhyay S, Shimizu R, Collmer A, He SY. Pseudomonas syringae pv. tomato DC3000 HopPtoM (CEL ORF3) is important for lesion formation but not growth in tomato and is secreted and translocated by the Hrp type III secretion system in a chaperone-dependent manner. Mol Microbiol. 2003;49:1239–1251. doi: 10.1046/j.1365-2958.2003.03647.x. [DOI] [PubMed] [Google Scholar]
- Bogdanove A, Kim JF, Wei Z, Kolchinsky P, Charkowski AO, Conklin AK, Collmer A, Beer SV. Homology and functional similarity of an hrp-linked pathogenicity locus, dspEF, of Erwinia amylovora and the avirulence locus avrE of Pseudomonas syringae pathovar tomato. Proc Natl Acad Sci USA. 1998;95:1325–1330. doi: 10.1073/pnas.95.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boureau T, ElMaarouf-Bouteau H, Garnier A, Brisset MN, Perino C, Pucheu I, Barny MA. DspA/E, a type III effector essential for Erwinia amylovora pathogenicity and growth in planta, induces cell death in host apple and nonhost tobacco plants. Mol Plant-Microbe Interact. 2006;19:16–24. doi: 10.1094/MPMI-19-0016. [DOI] [PubMed] [Google Scholar]
- Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Coplin DL, Frederick RD, Majerczak DR, Haas ES. Molecular cloning of virulence genes from Erwinia stewartii. J Bacteriol. 1986;168:619–623. doi: 10.1128/jb.168.2.619-623.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuppels DA. Generation and Characterization of Tn5 Insertion Mutations in Pseudomonas syringae pv. tomato. Appl Environ Microbiol. 1986;51:323–327. doi: 10.1128/aem.51.2.323-327.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cunha L, Sreerekha MV, Mackey D. Defense suppression by virulence effectors of bacterial phytopathogens. Curr Opin Plant Biol. 2007;10:349–357. doi: 10.1016/j.pbi.2007.04.018. [DOI] [PubMed] [Google Scholar]
- DebRoy S, Thilmony R, Kwack YB, Nomura K, He SY. A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci USA. 2004;101:9927–9932. doi: 10.1073/pnas.0401601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick RD, Majerczak DR, Coplin DL. Erwinia stewartii WtsA, a positive regulator of pathogenicity gene expression, is similar to Pseudomonas syringae pv. phaseolicola HrpS. Mol Microbiol. 1993;9:477–485. doi: 10.1111/j.1365-2958.1993.tb01709.x. [DOI] [PubMed] [Google Scholar]
- Frederick RD, Ahmad M, Majerczak DR, Arroyo-Rodriguez AS, Manulis S, Coplin DL. Genetic organization of the Pantoea stewartii subsp. stewartii hrp gene cluster and sequence analysis of the hrpA, hrpC, hrpN and wtsE operons. Mol Plant-Microbe Interact. 2001;14:1213–1222. doi: 10.1094/MPMI.2001.14.10.1213. [DOI] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 2002;31:375–383. doi: 10.1046/j.1365-313x.2002.01360.x. [DOI] [PubMed] [Google Scholar]
- Ham JH, Bauer DW, Fouts DE, Collmer A. A cloned Erwinia chrysanthemi Hrp (type III protein secretion) system functions in Escherichia coli to deliver Pseudomonas syringae Avr signals to plant cells and to secrete Avr proteins in culture. Proc Natl Acad Sci USA. 1998;95:10206–10211. doi: 10.1073/pnas.95.17.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham JH, Kim MG, Lee SY, Mackey D. Layered basal defenses underlie non-host resistance of Arabidopsis to Pseudomonas syringae pv. phaseolicola. Plant J. 2007;51:604–616. doi: 10.1111/j.1365-313X.2007.03165.x. [DOI] [PubMed] [Google Scholar]
- Ham JH, Majerczak DR, Arroyo-Rodriguez AS, Mackey D, Coplin DL. WtsE, an AvrE-family effector protein from Pantoea stewartii subsp. stewartii, causes disease-associated cell death in corn and requires a chaperone protein for stability. Mol Plant-Microbe Interact. 2006;19:1092–1102. doi: 10.1094/MPMI-19-1092. [DOI] [PubMed] [Google Scholar]
- Ham JH, Majerczak DR, Ewert S, Mysore-Venkatara S, Mackey D, Coplin DL. WtsE, an AvrE-family type III effector protein of Pantoea stewartii subsp. stewartii, causes cell death in nonhost plants. Mol Plant Pathol. 2008;9:633–643. doi: 10.1111/j.1364-3703.2008.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck P, Thilmony R, He SY. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci USA. 2003;100:8577–8582. doi: 10.1073/pnas.1431173100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He SY, Nomura K, Whittam TS. Type III protein secretion mechanism in mammalian and plant pathogens. Biochim Biophys Acta. 2004;1694:181–206. doi: 10.1016/j.bbamcr.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Holeva MC, Bell KS, Hyman LJ, Avrova AO, Whisson SC, Birch PRJ, Toth IK. Use of a pooled transposon mutation grid to demonstrate roles in disease development for Erwinia carotovora subsp. atroseptica putative type III secreted effector (DspE/A) and helper (HrpN) proteins. Mol Plant-Microbe Interact. 2004;17:943–950. doi: 10.1094/MPMI.2004.17.9.943. [DOI] [PubMed] [Google Scholar]
- Jin Q, He SY. Role of the Hrp pilus in type III protein secretion in Pseudomonas syringae. Science. 2001;294:2556–2558. doi: 10.1126/science.1066397. [DOI] [PubMed] [Google Scholar]
- Keen NT, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- Kim MG, da Cunha L, McFall AJ, Belkhadir Y, DebRoy S, Dangl JL, Mackey D. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005;121:749–759. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Lindgren PB, Peet RC, Panapoulos NJ. Gene cluster of Pseudomonas syringae pv. phaseolicola controls pathogenicity on bean plants and hypersensitivity on nonhost plants. J Bacteriol. 1986;168:512–522. doi: 10.1128/jb.168.2.512-522.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang JM, Keen NT. Characterization of avrE from Pseudomonas syringae pv. tomato: a hrp-linked avirulence locus consisting of at least two transcriptional units. Mol Plant-Microbe Interact. 1995;8:49–57. doi: 10.1094/mpmi-8-0049. [DOI] [PubMed] [Google Scholar]
- Meng X, Bonasera JM, Kim JF, Nissinen RM, Beer SV. Apple proteins that interact with DspA/E, a pathogenicity effector of Erwinia amylovora, the fire blight pathogen. Mol Plant-Microbe Interact. 2006;19:53–61. doi: 10.1094/MPMI-19-0053. [DOI] [PubMed] [Google Scholar]
- Merighi M, Majerczak DR, Stover EH, Coplin DL. The HrpX/HrpY two-component system activates hrpS expression, the first step in the regulatory cascade controlling the Hrp regulon in Pantoea stewartii subsp. stewartii. Mol Plant-Microbe Interact. 2003;16:238–248. doi: 10.1094/MPMI.2003.16.3.238. [DOI] [PubMed] [Google Scholar]
- Merighi M, Majerczak DR, Zianni M, Tessanne K, Coplin DL. Molecular characterization of Pantoea stewartii subsp. stewartii HrpY, a conserved response regulator of the Hrp type III secretion system, and its interaction with the hrpS promoter. J Bacteriol. 2006;188:5089–5100. doi: 10.1128/JB.01929-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf WW, Jiang W, Daniels LL, Kim SK, Haldimann A, Wanner BL. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K, Debroy S, Lee YH, Pumplin N, Jones J, He SY. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, Boller T. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes & Develop. 2006;20:537–542. doi: 10.1101/gad.366506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaj J, Muller J, Beck M, Bohm N, Menzel D. Vesicular trafficking, cytoskeleton and signaling in root hairs and pollen tubes. Trends Plant Sci. 2006;11:594–600. doi: 10.1016/j.tplants.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Schaad NW, Jones JB, Chun W. Laboratory Guide for Identification of Plant Pathogenic Bacteria. APS Press; St. Paul: 2001. [Google Scholar]
- Speth EB, Lee YN, He SY. Pathogen virulence factors as molecular probes of basic plant cellular functions. Curr Opin Plant Biol. 2007;10:580–586. doi: 10.1016/j.pbi.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Van Larebeke N, Engler G, Holsters M, Van den Elsacker S, Zaenen I, Schilperoort RA, Schell J. Large plasmid in Agrobacterium tumefaciens essential for crown gall-inducing ability. Nature. 1974;252:169–170. doi: 10.1038/252169a0. [DOI] [PubMed] [Google Scholar]
- Vernoud V, Horton AC, Yang Z, Nielsen E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 2003;131:1191–1208. doi: 10.1104/pp.013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, He SY. The Pseudomonas syringae Hrp regulation and secretion system controls the production and secretion of multiple extracellular proteins. J Bacteriol. 1996;178:6399–6402. doi: 10.1128/jb.178.21.6399-6402.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. AvrE family members contain seven conserved tryptophans (W). A, stick diagrams showing the aligned N-terminal halves of the proteins and their extreme C-termini. The locations of conserved tryptophans (w1 to w7) are indicated at top. Below are shown the residues at position +4 relative to W (or, where indicated by an asterisk, relative to a conserved tyrosine (Y) or phenylalanine (F) in the comparable position). Glutamates (E, which form prototype WxxxE motifs) are shown in red, conserved aspartates (D) are shown in green, positively charged amino acids are shown in blue, and other amino acids are shown in black. After breaks in the stick diagrams, the C-termini with predicted ERMRS motifs are indicated by magenta ERs. B, pile-ups of the sequences within AvrE-family effector proteins. In the columns w1 to w7, W is shaded dark gray and conserved Y or F substitutions are light gray. E is red, D is green, lysine (K), arginine (R), or asparagine (N) are blue. The underlined WxxxE1 and WxxxE2 motifs from Pnss and Pto are those that were mutated in this study. At the right, are those sequences predicted by PSORTII to function as an ERMRS. Contrary to PSORTII, the sequence of AvrE from Pvi-AS appears quite similar to the other predicted ERMRSs while the sequence of RopE from Pfl appears quite divergent. Species abbreviations are given in Fig. 1.
Supplementary Fig. 2. The HR in tobacco following delivery of WtsE or the indicated derivatives from E. coli. The photograph was taken 16 h after infiltration. Results are representative of three independent experiments.
Supplementary Fig. 3. Cell death phenotypes of WtsE derivatives transiently expressed from A. tumefaciens or delivered from Pnss into N. benthamiana. A, Agrobacterium-mediated transient expression. A derivative of WtsE missing the first 236 aa is slightly more active in this system (Ham et al., 2008), so mutations were introduced into this truncated protein. A. tumefaciens C58C1 strains containing the following plasmids were infiltrated: 1) pGDG (empty vector control), 2) pJH101G full length wtsE, 3) pJH103G inactive wtsE-Δaa1-492, 4) pJH103G wtsE-Δaa1-236, 5) pJH063 wtsE-Δaa1-236-wxe12, 6) pJH059 wtsE-Δaa1-236-ΔFEMK. The leaf was photographed at 72 h after infiltration with a suspension of A. tumefaciens at A600 = 0.6. Note the weak residual cell death caused by expression of the wtsE-Δaa1-236-wxe12 mutant. B, cell death induced by wild-type and wtsE mutant Pnss strains. We have shown previously that Pnss induces HR-like cell death in N. benthamiana that is dependent on WtsE (Ham et al., 2008). Numbers correspond to infiltration of the following strains: 1) DC283 (wild-type), 2) DM711 hrpJ (Hrp-), 3) AA005 wtsE∷aphT, 4) buffer control, 5) DM5101 wtsE∷miniTn5gus, 6) JH039 wtsE-wxe12, and 7) JH033 wtsE-ΔFEMK. The leaf was photographed at 24 h after infiltration with a suspension of bacteria at 109 CFU/ml. Note the weak residual cell death caused by the Pnss wxe12 and ΔFEMK mutants. C, WxxxE and ERMRS mutations do not affect secretion of WtsE from Pnss. Western blots of concentrated culture supernatants using anti-DspEEa serum to detect WtsE. The following strains were tested; DM711 hrpJ (Hrp-), DC283 (wild-type), JH039 wtsE-wxe12, JH033 wtsE-ΔFEMK, JH042 wtsE-wxe12/ΔFEMK and AA005 wtsE∷aphT. Similar results were obtained with the pellet fractions and the experiment was done twice.