Abstract
Aims
A recent study found that variation in camptothecin pharmacodynamic genes (TOP1, PARP1, TDP1 and XRCC1) correlated with efficacy and risk of neutropenia in irinotecan-treated cancer patients (median dose: 180 mg/m2), which suggests that these genes might predict outcomes to irinotecan-based therapies. The present study was conducted to evaluate previous gene associations using an independent sample of patients receiving irinotecan.
Materials & methods
DNA was isolated from 85 advanced cancer patients treated with 300 or 350 mg/m2 irinotecan and genotyped for haplotype-tag polymorphisms across TOP1, PARP1, TDP1 and XRCC1. Associations between genotypes and haplotypes and log(absolute neutrophil count nadirs) were assessed by linear regression.
Results
No associations were observed.
Conclusion
Our findings suggest that the genes we tested do not influence toxicity of irinotecan when adminstered at 300-350 mg/m2.
Keywords: irinotecan, PARP1, pharmacogenetics, TDP1, TOP1, XRCC1
Irinotecan is a semi-synthetic derivative of camptothecin. It is approved for first-line treatment of metastatic colorectal cancer in combination with 5-fluorouracil/leucovorin and as salvage therapy in 5-fluorouracil refractory disease in the USA. Results of Phase III trials indicate the addition of irinotecan to 5-fluorouracil/leucovorin improves rate of response, time to progression and median survival over 5-fluorouracil/leucovorin alone [1,2]. Irinotecan is also active when combined with bevacuzimab or cetuximab [3,4]. The dose limiting toxicities of irinotecan-based therapies include myelosuppression and late-onset diarrhea, the latter of which can be life threatening if not managed appropriately [5].
The anticancer activity of irinotecan is dependent upon enzymatic conversion by carboxylesterases to its active metabolite, 7-ethyl-10-hydroxycamptothecin (SN-38). Detoxification of SN-38 is predominantly via glucuronidation to SN-38-glucuronide (SN-38G) by hepatic uridine diphosphate-glucuronosyltransferases, principally UGT1A1. Many pharmacogenetic investigations of irinotecan pharmacokinetics and toxicity have been conducted [5-7]. The most studied gene to date is UGT1A1, which is located on chromosome 2q37. A common tandem repeat polymorphism in the TATA box of the promoter of the gene (-53A(TA)nTAA) has been described and an inverse relationship between the number of TA repeats and the transcriptional activity of the gene has been established [8]. Patients homozygous for seven TA repeats (UGT1A1*28/*28) have a reduced SN-38 glucuronidation rate (SN-38G:SN-38 area under the curve ratio), elevated SN-38 area under the curve and an increased risk of severe toxicity, especially neutropenia, compared with patients heterozygous or homozygous for six TA repeats (UGT1A1*1/*28 or UGT1A1*1/*1) [9-14]. The association between UGT1A1*28/*28 genotype and toxicity appears to be dose dependent, with strong associations observed at doses of more than 250 mg/m2 [15].
Pharmacodynamic genes have not been well studied for irinotecan. Alterations in topoisomerase I (topo I) activity (TOP1) that result in reduced levels of the SN-38-topo I-DNA complex and the events downstream from the ternary complex, for example apoptosis, cell cycle regulation, checkpoints and DNA repair might affect cell sensitivity to the cytotoxic effects of irinotecan [16,17]. Elevated expression of PARP1, TDP1 and XRCC1 are reported to abate the cytotoxic action of camptothecins in yeast and mammalian cancer cells [18-21]. Genetic variation in TOP1 and the cellular downstream effectors that lead to DNA repair or cell death are potential sources of patient-to-patient variation in irinotecan-induced toxicity and clinical response [22]. This variation might allow improved prediction of patient outcomes in response to irinotecan-based therapies.
Hoskins et al. previously tested the hypothesis that genetic variation in the pharmacodynamic genes, including TOP1, PARP1, TDP1, XRCC1, CDC45L and NFKB1, influence patient outcomes to irinotecan therapy in an exploratory study that utilized a haplotype-based study approach [23]. In the study they assessed the relationships between genetic variability in the pharmacodynamic genes and irinotecan treatment outcomes in a cohort of 107 white advanced colorectal cancer patients treated with a median irinotecan dose of 180 mg/m2 (range: 80-350 mg/m2), administered as a single-agent or in combination with 5-fluorouracil or raltitrexed [23]. By univariate analyses, genetic variation in TOP1 and PARP1 was associated with grade III/IV neutropenia while variation in XRCC1 and TDP1 was associated with clinical response in this cohort [23]
The goal of the present study was to evaluate the previous associations between TOP1, PARP1, TDP1 and XRCC1 and irinotecan-related toxicity of neutropenia using an independent sample of 85 cancer patients treated with single-agent 300-350 mg/m2 irinotecan using a haplotype marker (htSNP) approach [23]. The influence of UGT1A1*28 on neutropenia has previously been investigated in this patient cohort [10,13] and was also included in the analysis.
Materials & methods
Patients
A total of 85 patients with solid tumors or lymphoma of a type known to respond to irinotecan, or for which no proven therapy of benefit exists, received 90 min intravenous infusions of 300 mg/m2 (n = 20) or 350 mg/m2 irinotecan (n = 65) once every 21 days (National Cancer Institute [MD, USA] protocol #T98-0039, University of Chicago [IL, USA] protocol #9531) [10,13]. Patients gave written informed consent prior to being enrolled in the study. The present study was approved by Washington University Medical Center Human Studies Committee, Washington University School of Medicine, (MO, USA) (#05-1152) and The University of Chicago Institutional Review Board (#05-125). The initial objectives of the studies were to evaluate the influence of UGT1A1*28 genotype on irinotecan-related toxicities and the findings were published else-where [10,13]. Patient pathological characteristics are outlined in Table 1.
Table 1. Baseline characteristics of cancer patients and response to treatment.
| Characteristic | Value | No. of patients (%) |
|---|---|---|
| Patients | 85 | |
| Sex | Male | 48 (56) |
| Female | 37 (44) | |
| Age (years), median (range) | 59 (34-85) | |
| Ethnicity | European-American | 67 (79) |
| African-American | 11 (13) | |
| Hispanic | 4 (5) | |
| Asian-American | 2 (2) | |
| Pacific Islander | 1 (1) | |
| BSA (m2), median (range) | 1.82 (1.41-2.55) | |
| Irinotecan therapy (mg/m2) | 350 | 65 (76) |
| 300 | 20 (24) | |
| UGT1A1*28 genotype | *28/*28 | 11 (13) |
| *1/*28 | 34 (40) | |
| *1/*1 | 40 (47) | |
| Toxicities | Grade III/IV neutropenia | 12 (14) |
| Grade III/IV diarrhea | 5 (6) | |
DNA was collected for genetic analysis from all 85 patients. Absolute neutrophil counts (ANCs) were collected weekly. Presence and grade of neutropenia and diarrhea were recorded for all patients in accordance with the criteria of WHO. In the total cohort of 85 patients, 5 patients (6%) experienced grade III/IV diarrhea and 12 (14%) grade III/IV neutropenia.
Genotyping
Germline DNA samples from 85 patients were genotyped for six htSNPs (one insertion/deletion and five SNPs) across TOP1, PARP1, TDP1 and XRCC1 (see Table 2 for a list of polymorphisms). htSNPs were chosen as previously described from haplotype networks and distinguished between three major haplotypes in a European sample [23]. Genotyping was performed using PCR and Pyrosequencing® on the PyroMark MD (Qiagen, Hilden, Germany) as previously described [23].
Table 2. Allele and genotype frequencies for haplotype marker in the candidate genes in 85 cancer patients.
| Gene | Polymorphism | dbSNP ID | Allele 1 (frequency) | Allele 2 (frequency) | Genotype, no. of patients (frequency) | ||
|---|---|---|---|---|---|---|---|
| 1/1 | 1/2 | 2/2 | |||||
| PARP1 | c.852 (A284A) | rs1805414 | T (0.62) | C (0.38) | 35 (0.41) | 35 (0.41) | 15 (0.18) |
| PARP1 | IVS19-297 | rs3219142 | C (0.81) | T (0.19) | 59 (0.69) | 19 (0.22) | 7 (0.08) |
| TDP1 | IVS12+79 | rs2401863 | T (0.64) | G (0.36) | 37 (0.44) | 35 (0.41) | 13 (0.15) |
| TOP1 | IVS4+61 | rs6072262 | G (0.88) | A (0.12) | 67 (0.79) | 15 (0.18) | 3 (0.04) |
| XRCC1 | -1449delGGCC | rs3213239 | GGCC (0.58) | del (0.42) | 29 (0.34) | 41 (0.48) | 15 (0.18) |
| XRCC1 | c.1196 (R399Q) | rs25487 | G (0.74) | A (0.26) | 47 (0.55) | 32 (0.38) | 6 (0.07) |
Statistics
Polymorphism and Haplotype Analysis Suite (PHASE version 0.9) was used to estimate pair wise linkage disequilibrium (Dʑ) and haplotype identities and to assign diplotypes for PARP1 and XRCC1 [24]. The prevalence of diarrhea was too low to permit statistical analysis of this outcome. Our analyses concerned ANC and related grade III/IV neutropenia data collected during the first cycle of chemotherapy and were performed for the total cohort and European-American patients alone. Linear regression with the log(ANC nadir) as the outcome variable was used to test associations between htSNP genotypes and diplotypes and irinotecan-induced hematological toxicity. Analyses were adjusted for irinotecan dose (300 vs 350 mg/m2), sex, ethnicity (European-American vs African-American), age and baseline ANC, regardless of statistical significance. Grade III/IV neutropenia was compared between genotype and diplotype groups using the χ2 test. We constructed a model to fit with an interaction term to assess whether TOP1, PARP1, TDP1 and XRCC1 htSNPs and haplotypes modulate the association between UGT1A1*28 genotype and ANC. A p-value of 0.05 was set for statistical significance. No p-value adjustments were made for multiple comparisons. The population size was limited by the availability of samples and data. In a post hoc calculation of power, based on the known sample size (81 patients), number of grade III/IV neutropenia events (12 patients), a two-sided type I error of 5%, and assuming a minor allele frequency of 20%, we had 80% power to detect an odds ratio of 7.2 or greater using the Fisher’s exact test (two-by-two table) and an odds ratio of 5.9 using an uncorrected χ2 test (two-by-two table).
Results
TOP1, PARP1, TDP1 & XRCC1 genotypes
Patient DNA samples were typed for six htSNPs across the candidate genes. Allele and genotype frequencies of the six htSNPs are presented in Table 2. For the patients of European descent (which is the majority of patients, n = 67), all observed genotype frequencies were in agreement with the Hardy-Weinberg equation (p > 0.05).
PARP1 & XRCC1 haplotype identities
Haplotypes were estimated for PARP1 and XRCC1. For each gene, some individuals were homozygous at both loci and others heterozygous at one locus, which provided unambiguous information about the specific combinations of alleles. For both genes, some individuals were homozygous for all loci and others were heterozygous at a single site, which provided unambiguous information about the specific combinations of alleles. Other diplotypes were inferred by PHASE. The estimated haplotypes for PARP1 were T-C, C-C and T-T from c.852T>C and IVS19-297C>T with frequencies of 0.42, 0.38 and 0.19, respectively. For XRCC1, del-G (frequency = 0.42), GGCC-A (0.26) and GGCC-G (0.32) were estimated from alleles -1449delGGCC and c.1196G>A. The numbers of patients with (and frequencies of) each diplotype are presented in Tables 3 & 4.
Table 3. The number and frequency of PARP1 diplotypes observed.
| Diplotype | c.852 (A284A) | IVS19-297 | No. of patients | Frequency |
|---|---|---|---|---|
| T-C/T-C | T/T | C/C | 15 | 0.18 |
| T-C/T-T | T/T | C/T | 13 | 0.15 |
| T-T/T-T | T/T | T/T | 7 | 0.08 |
| T-C/C-C | C/T | C/C | 29 | 0.34 |
| C-C/T-T | C/T | C/T | 6 | 0.07 |
| C-C/C-C | C/C | C/C | 15 | 0.18 |
Table 4. The number and frequency of XRCC1 diplotypes observed.
| Diplotype | -1449delGGCC | c.1196 (R399Q) | No. of patients | Frequency |
|---|---|---|---|---|
| del-G/del-G | del/del | G/G | 15 | 0.18 |
| del-G/GGCC-A | GGCC/del | A/G | 21 | 0.25 |
| del-G/GGCC-G | GGCC/del | G/G | 20 | 0.24 |
| GGCC-A/GGCC-A | GGCC/GGCC | A/A | 6 | 0.07 |
| GGCC-A/GGCC-G | GGCC/GGCC | A/G | 11 | 0.13 |
| GGCC-G/GGCC-G | GGCC/GGCC | G/G | 12 | 0.14 |
Associations between irinotecan-induced hematological toxicities & pharmacodynamic genes
TOP1, PARP1, TDP1 and XRCC1 htSNPs were not associated with first-cycle log(ANC nadir) in the total cohort (Figure 1A & 1B) nor in the European-American patients analyzed alone. In addition, PARP1 and XRCC1 haplotypes (Figure 1C) were not associated with first-cycle log(ANC nadir). Moreover, genotypes and diplotypes were not associated with the incidence of first-cycle grade III/IV neutropenia in the total cohort or European-American patients tested alone (p > 0.30).
Figure 1.
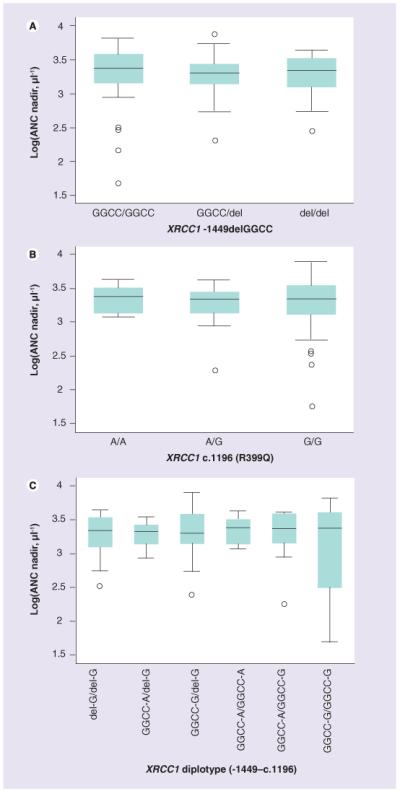
Relationships between first-cycle log(ANC nadir) and (A) XRCC1 -1449delGGCC and (B) XRCC1 c.1196 genotypes and (C) XRCC1 (-1449delGGCC-c.1196) diplotypes (p > 0.10). ANC: Absolute netrophil count.
Gene-gene interactions between pharmacodynamic genes & UGT1A1*28 UGT1A1*28
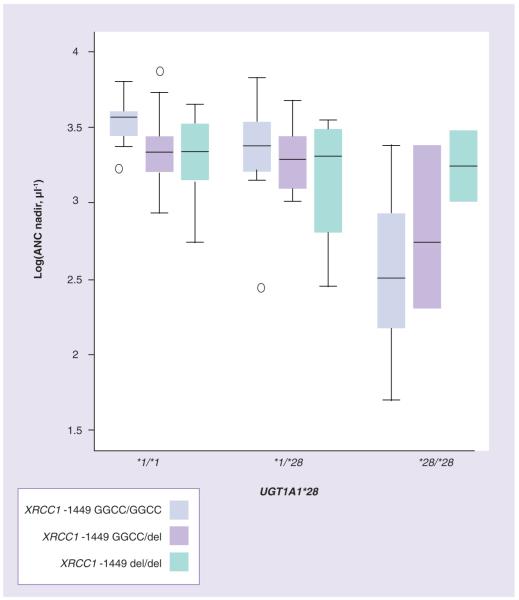
genotype was associated with first-cycle log(ANC nadir), with a significant trend towards lower ANC nadir from *1/*1 to *1/*28 to *28/*28 (p > 0.0001), as previously reported [10,13]. Patients homozygous for UGT1A1*28 and XRCC1 -1449 GGCC had significantly lower nadirs than the other patients (p = 0.002, test for interaction) (Figure 2). We did not find significant interactions between UGT1A1*28/*28 and the other genotypes with respect to first-cycle ANCs in the total cohort and European-American patients alone (p > 0.1).
Figure 2.
Interactions between UGT1A1*28 and XRCC1 -1449delGGCC genotypes and first-cycle log(ANC nadir) (p < 0.0001). ANC: Absolute netrophil count.
Discussion
Camptothecins, including SN-38, exert their cytotoxic effects by reversibly stabilizing the topo I-DNA covalent complex, trapping the complex and preventing religation of nicked DNA. The camptothecin-topo I-DNA complex collides with advancing replicating forks, which convert the single-strand DNA breaks to double-strand breaks that can not be easily resealed. This induces irreversible arrest of the replication fork and may lead to cell death. Genetic variation in the drug target of SN-38, TOP1 and cellular downstream effectors that lead to DNA repair or cell death might be sources of patient-to-patient variation in irinotecan-induced toxicity and clinical response [22]. In the present study, we investigated whether genetic variation in TOP1, and genes encoding other camptothecin pharmacodynamic factors, including PARP1, TDP1 and XRCC1, influences first-cycle ANC nadirs and related first-cycle grade III/IV neutropenia in 85 cancer patients with solid tumors or lymphoma treated with moderately high-dose, single-agent irinotecan (300 or 350 mg/m2 every 21 days). Although a previous study showed that genotypes and diplotypes of TOP1, PARP1, TDP1 and XRCC1 influenced outcomes to irinotecan-based therapies [23], these results could not be replicated in an independent cohort. Our analysis suggests genetic variation in TOP1, PARP1, TDP1 and XRCC1 does not explain severe neutropenia in this irinotecan treated cohort.
It is plausible that associations between polymorphisms in pharmacodynamic genes and toxicity are irinotecan dose-dependent, such that differences in irinotecan doses that were administered among the studies explains the inconsistent genetic associations [23]. In the present study, patients received moderately high doses of irinotecan (300 or 350 mg/m2, every 21 days) and UGT1A1*28/*28 genotype was significantly associated with grade III/IV neutro penia (seven of 12 UGT1A1*28/*28 patients experienced neutropenia [58%], p = 0.0002) [10,13]. In the previous study, where patients were treated with irinotecan monotherapy or in combination with 5-fluorouracil or raltitrexed at a median dose of 180 mg/m2 irinotecan every 14 days, with doses ranging from 80-350 mg/m2 irinotecan (administered every 7, 14 or 21 days), UGT1A1*28 genotype was not associated with grade III/IV neutropenia (p = 0.5) [23]. These genotype-toxicity associations are consistent with the findings of a recent meta-analysis of 821 cancer patients treated with irinotecan [15]. Specifically, a large effect size (odds ratio [OR]: 27.8; p = 0.005) for the association of the UGT1A1*28/*28 genotype with neutropenia was observed at higher doses of irinotecan (>250 mg/m2), a moderate effect size (OR: 3.2; p = 0.008) at medium doses (150-250 mg/m2), and a weak, nonsignificant effect size (OR: 1.8; p = 0.41) at lower doses of irinotecan (<150 mg/m2) [15]. These findings suggest UGT1A1*28/*28 genotype is a good determinant of risk of neutropenia when patients receive higher doses of irinotecan but is not predictive when they receive lower doses (<150 mg/m2). The larger effect size of the UGT1A1*28 on irinotecan-related neutro penia at higher irinotecan doses may have overshadowed the smaller effect sizes of the pharmacodynamic gene variants on neutropenia, thereby reducing the power of the present study to detect the latter genetic associations.
The present study was undertaken to replicate the findings of Hoskins et al. [23]. In that study, patients of European ancestry were genotyped for htSNPs that distinguished between major haplotypes in a European-American sample. The htSNPs are not considered functional or known to be in strong linkage with alleles of known functional importance [23]. In the present study we evaluated whether the same set of htSNPs were associated with irinotecan-related neutropenia in an independent cohort of patients. Some polymorphisms in the pharmacodynamic genes have been shown to alter the function of these proteins in vitro and to be risk factors for various cancers [25-28]. However, patients in the present study were not genotyped for these functional variants because they were not associated with outcomes to irinotecan therapy in a prior study [23]. The present cohort was of mixed ethnicity, with the majority of patients being European-American (79%). The distribution of haplotypes for the pharmacodynamic genes varies among ethnic groups [23] and so too might the degree of linkage disequilibrium between htSNPs and irinotecan response altering alleles in the genes [23]. This may have limited our power to detect associations in the present study. However, associations between the htSNPs and irinotecan-related neutropenia were not observed for the European-American patients when analyzed alone suggesting that this does not explain the inconsistent genetic associations among the studies [23].
We may not have detected any associations between pharmacodynamic genes and neutrophil nadir in the present study because there were none to find (i.e., true negatives). There may be additional variants that were not evaluated in this study. Certainly other genes may also contribute to variation in irinotecan effect. Alternatively, the observations of the earlier exploratory study may have been false positives due to limitations in sample size [23]. Genetic variations in TOP1 and PARP1 were found to predict grade III/IV neutropenia by univariate statistical tests but not by multiple logistic regression, suggesting the associations detected by univariate analyses may have been false positives [23]. However, it is possible the associations detected in the earlier study by univariate analyses were true positives but the study may have lacked adequate power to detect these associations by multivariate analysis because the numbers of homozygous variants were low and the size of effects were small-to-moderate and the study was powered to detect large effect sizes.
Patients homozygous for UGT1A1*28 and XRCC1 -1449 GGCC appeared to have lower first-cycle ANC nadirs than the other patients (p < 0.0001). However, the limited sample size, the small number of individuals with the combined genotype (n = 5), the lack of influence of the XRCC1 insertion/deletion on first-cycle ANCs (by univariate statistical test), and that the relationship between XRCC1 -1449 genotypes and ANCs for patients with the UGT1A1*1/*1 and *1/*28 genotypes appears to be in the opposite direction, suggest that this may not be a true association and caution should be exercised when interpreting this gene-gene interaction observation.
Conclusion
In conclusion, the present study represents an attempt to confirm observations from a previous pharmacogenetic study in which Hoskins et al. demonstrated germline variations in genes that play a role in camptothecin pharmacodynamics influenced outcomes to irinotecan-based therapies in a separate patient population [23]. Using a sample of cancer patients treated with moderately high-dose single-agent irinotecan, we failed to confirm the findings of the initial study suggesting that in the case of moderately high-dose irinotecan, TOP1, PARP1, TDP1 and XRCC1 play minor or negligible roles in patient-to-patient variation in irinotecan treatment outcomes.
Executive summary.
-
■
Patients with the UGT1A1*28/*28 genotype have reduced detoxification of the active metabolite of irinotecan, SN-38 and an elevated risk of severe irinotecan-related neutropenia. However this genetic variant does not explain all of the patient-to-patient variation in irinotecan-related toxicity or efficacy.
-
■
A recent study found that variation in camptothecin pharmacodynamic genes, TOP1, PARP1, TDP1 and XRCC1, correlated with efficacy and risk of neutropenia in irinotecan-treated cancer patients (median dose: 180 mg/m2).
-
■
These genetic variants may influence outcomes to irinotecan-based therapies.
Materials & methods
-
■
A total of five SNPs and an insertion/deletion polymorphism across TOP1, PARP1, TDP1 and XRCC1 were determined in a total of 85 patients with cancer treated with moderately high-dose single-agent irinotecan (300 or 350 mg/m2) as part of a Phase I trial.
-
■
Genotyping was performed using PCR and Pyrosequencing® on the PyroMark MD (Qiagen, Hilden, Germany).
Results
-
■
TOP1, PARP1, TDP1 and XRCC1 genotypes, and PARP1 and XRCC1 diplotypes were not associated with first-cycle log(absolute neutrophil counts nadir) or incidence of first-cycle grade III/IV neutropenia in the total cohort and the European-Americans analyzed alone.
-
■
Patients homozygous for UGT1A1*28 and XRCC1 -1449 GGCC had significantly lower nadirs than the other patients (p = 0.002, test for interaction). Gene-gene interactions between UGT1A1*28/*28 and the other pharmacodynamic variants with respect to first-cycle ANCs or grade III/IV neutropenia were not observed.
Discussion
-
■
TOP1, PARP1, TDP1 and XRCC1 do not appear to be influencing factors of response to moderately high-dose single-agent irinotecan therapy in cancer patients.
-
■
In addition to UGT1A1*28, factors (genetic or nongenetic) other than the genetic variants studied in the current study may be determinants of response to irinotecan at higher doses of irinotecan.
Acknowledgements
The authors would like to thank Jacqueline Ramirez for her help writing the IRB proposal and providing tabulated patient data.
Footnotes
Financial & competing interests disclosure
This work was supported by the NIH Pharmacogenetics Research Network Comprehensive Research on Expressed Alleles in Therapeutic Evaluation (CREATE) (NIH/NIGMS grant U01 GM63340) and Pharmacogenetics of Anticancer Agents Research (PAAR) groups (http://pharmacogenetics.org) (NIH/NIGMS grant U01GM61393). Data will be deposited into PharmGKB (supported by NIH/NIGMS U01GM61374, http://pharmgkb.org/). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investi gations involving human subjects, informed consent has been obtained from the participants involved.
Bibliography
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
- 1.Douillard JY, Cunningham D, Roth AD, et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial. Lancet. 2000;355:1041–1047. doi: 10.1016/s0140-6736(00)02034-1. [DOI] [PubMed] [Google Scholar]
- 2.Saltz LB, Cox JV, Blanke C, et al. Irinotecan Study Group: Irinotecan plus fluorouracil and leucovorin for metastatic colorectal cancer. N. Engl. J. Med. 2000;343:905–914. doi: 10.1056/NEJM200009283431302. [DOI] [PubMed] [Google Scholar]
- 3.Hurwitz H, Fehrenbacher L, Novotny W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N. Engl. J. Med. 2004;350:2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 4.Vincenzi B, Santini D, Rabitti C, et al. Cetuximab and irinotecan as third-line therapy in advanced colorectal cancer patients: a single centre Phase II trial. Br. J. Cancer. 2006;94:792–797. doi: 10.1038/sj.bjc.6603018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Innocenti F, Ratain MJ. Pharmacogenetics of irinotecan: clinical perspectives on the utility of genotyping. Pharmacogenomics. 2006;7:1211–1221. doi: 10.2217/14622416.7.8.1211. [DOI] [PubMed] [Google Scholar]
- 6.Marsh S, McLeod HL. Pharmacogenetics of irinotecan toxicity. Pharmacogenomics. 2004;5:835–843. doi: 10.1517/14622416.5.7.835. [DOI] [PubMed] [Google Scholar]
- 7.Smith NF, Figg WD, Sparreboom A. Pharmacogenetics of irinotecan metabolism and transport: an update. Toxicol. In Vitro. 2006;20:163–175. doi: 10.1016/j.tiv.2005.06.045. [DOI] [PubMed] [Google Scholar]
- 8.Beutler E, Gelbart T, Demina A. Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: a balanced polymorphism for regulation of bilirubin metabolism? Proc. Natl Acad. Sci. USA. 1998;95:8170–8174. doi: 10.1073/pnas.95.14.8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ando Y, Saka H, Ando M, et al. Polymorphisms of UDP-glucuronosyltransferase gene and irinotecan toxicity: a pharmacogenetic analysis. Cancer Res. 2000;60:6921–6926. [PubMed] [Google Scholar]
- 10.Innocenti F, Undevia SD, Iyer L, et al. Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan. J. Clin. Oncol. 2004;22:1382–1388. doi: 10.1200/JCO.2004.07.173.■■ Phase I trial of advanced cancer patients treated with moderately high-dose single-agent irinotecan (350 mg/m2 every 21 days) showed that patients with the UGT1A1*28/*28 genotype had an elevated risk of severe neutropenia in cycle one.
- 11.Rouits E, Boisdron-Celle M, Dumont A, Guerin O, Morel A, Gamelin E. Relevance of different UGT1A1 polymorphisms in irinotecan-induced toxicity: a molecular and clinical study of 75 patients. Clin. Cancer Res. 2004;10:5151–5159. doi: 10.1158/1078-0432.CCR-03-0548. [DOI] [PubMed] [Google Scholar]
- 12.Marcuello E, Altes A, Menoyo A, Del Rio E, Gomez-Pardo M, Baiget M. UGT1A1 gene variations and irinotecan treatment in patients with metastatic colorectal cancer. Br. J. Cancer. 2004;91:678–682. doi: 10.1038/sj.bjc.6602042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iyer L, Das S, Janisch L, et al. UGT1A1*28 polymorphism as a determinant of irinotecan disposition and toxicity. Pharmacogenomics J. 2002;2:43–47. doi: 10.1038/sj.tpj.6500072. [DOI] [PubMed] [Google Scholar]
- 14.Paoluzzi L, Singh AS, Price DK, et al. Influence of genetic variants in UGT1A1 and UGT1A9 on the in vivo glucuronidation of SN-38. J. Clin. Pharmacol. 2004;44:854–860. doi: 10.1177/0091270004267159. [DOI] [PubMed] [Google Scholar]
- 15.Hoskins JM, Goldberg RM, Qu P, Ibrahim JG, McLeod HL. UGT1A1*28 genotype and irinotecan-induced neutropenia: dose matters. J. Natl Cancer Inst. 2007;99:1290–1295. doi: 10.1093/jnci/djm115.■■ Meta-analysis of irinotecan-UGT1A1*28 pharmacogenetic studies showed that the risk of hematological toxicity was higher among patients with a UGT1A1*28/*28 genotype than among other patients at both medium and high doses of irinotecan. However, risk was similar at lower doses.
- 16.Pommier Y, Pourquier P, Urasaki Y, Wu J, Laco GS. Topoisomerase inhibitors I: selectivity and cellular resistance. Drug Resist. Updat. 1999;2:307–318. doi: 10.1054/drup.1999.0102. [DOI] [PubMed] [Google Scholar]
- 17.Rasheed ZA, Rubin EH. Mechanisms of resistance to topoisomerase I-targeting drugs. Oncogene. 2003;22:7296–7304. doi: 10.1038/sj.onc.1206935. [DOI] [PubMed] [Google Scholar]
- 18.Barrows LR, Holden JA, Anderson M, D’Arpa P. The CHO XRCC1 mutant, EM9, deficient in DNA ligase III activity, exhibits hypersensitivity to camptothecin independent of DNA replication. Mutat. Res. 1998;408:103–110. doi: 10.1016/s0921-8777(98)00022-6. [DOI] [PubMed] [Google Scholar]
- 19.Park JY, Lee SY, Jeon HS, et al. Polymorphism of the DNA repair gene XRCC1 and risk of primary lung cancer. Cancer Epidemiol. Biomarkers Prev. 2002;11:23–27. [PubMed] [Google Scholar]
- 20.Malanga M, Althaus FR. Poly(ADP-ribose) reactivates stalled DNA topoisomerase I and induces DNA strand break resealing. J. Biol. Chem. 2004;279:5244–5248. doi: 10.1074/jbc.C300437200. [DOI] [PubMed] [Google Scholar]
- 21.Barthelmes HU, Habermeyer M, Christensen MO, et al. TDP1 overexpression in human cells counteracts DNA damage mediated by topoisomerases I and II. J. Biol. Chem. 2004;279:55618–55625. doi: 10.1074/jbc.M405042200. [DOI] [PubMed] [Google Scholar]
- 22.Pommier Y. Camptothecins and topoisomerase I: a foot in the door. Targeting the genome beyond topoisomerase I with camptothecins and novel anticancer drugs: importance of DNA replication, repair and cell cycle checkpoints. Curr. Med. Chem. Anticancer Agents. 2004;4:429–434. doi: 10.2174/1568011043352777. [DOI] [PubMed] [Google Scholar]
- 23.Hoskins JM, Marcuello E, Altes A, et al. Irinotecan pharmacogenetics: influence of pharmacodynamic genes. Clin. Cancer Res. 2008;14:1788–1796. doi: 10.1158/1078-0432.CCR-07-1472.■ Retrospective study that used the same patient cohort as [12], found by multivariate analyses that XRCC1 diplotype was an independent predictor of objective response in advanced colorectal cancer patients receiving irinotecan-containing regimens.
- 24.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lockett KL, Hall MC, Xu J, et al. The ADPRT V762A genetic variant contributes to prostate cancer susceptibility and deficient enzyme function. Cancer Res. 2004;64:6344–6348. doi: 10.1158/0008-5472.CAN-04-0338. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Miao X, Liang G, et al. Polymorphisms in DNA base excision repair genes ADPRT and XRCC1 and risk of lung cancer. Cancer Res. 2005;65:722–726. [PubMed] [Google Scholar]
- 27.Hu Z, Ma H, Chen F, Wei Q, Shen H. XRCC1 polymorphisms and cancer risk: a meta-analysis of 38 case-control studies. Cancer Epidemiol. Biomarkers Prev. 2005;14:1810–1818. doi: 10.1158/1055-9965.EPI-04-0793. [DOI] [PubMed] [Google Scholar]
- 28.Hu Z, Ma H, Lu D, et al. A promoter polymorphism (-77T>C) of DNA repair gene XRCC1 is associated with risk of lung cancer in relation to tobacco smoking. Pharmacogenet. Genomics. 2005;15:457–463. doi: 10.1097/01.fpc.0000167329.85163.0d. [DOI] [PubMed] [Google Scholar]