Abstract
The transcription factor Nrf2 has emerged as a master regulator for the endogenous antioxidant response, which is critical in defending cells against environmental insults and in maintaining intracellular redox balance. However, whether Nrf2 has any role in neuronal cell differentiation is largely unknown. In this report, we have examined the effects of Nrf2 on cell differentiation using a neuroblastoma cell line, SH-SY5Y. Retinoic acid (RA) and 12-O-tetradecanoylphorbol-13-acetate (TPA), two well-studied inducers for neuronal differentiation, are able to induce Nrf2 and its target gene NAD(P)H quinone oxidoreductase 1 (NQO1) in a dose- and time- dependent manner. RA-induced Nrf2 up-regulation is accompanied by neurite outgrowth and an induction of two neuronal differentiation markers, neurofilament-M (NF-M) and microtubule-associated protein 2 (MAP-2). Overexpression of Nrf2 in SH-SY5Y cells promotes neuronal differentiation whereas inhibition of endogenous Nrf2 expression inhibited neuronal differentiation. More remarkably, the positive role of Nrf2 in neuronal differentiation was verified ex vivo in primary neuron culture. Primary neurons isolated from Nrf2-null mice showed a retarded progress in differentiation, compared to that from wild-type mice. Collectively, our data demonstrate a novel role for Nrf2 in promoting neuronal cell differentiation, which will open new perspectives for therapeutic uses of Nrf2 activators in patients with neurodegenerative diseases.
Keywords: Nrf2, Keap1, Oxidative Stress, Neuronal differentiation, SH-SY5Y, NQO1
Introduction
SH-SY5Y, a neuroblastoma cell line that is a well-established model system to study the initial phases of neuronal differentiation, was chosen to evaluate the possible role of Nrf2 in neuronal cell differentiation. This cell line was derived from a subclone of a neural crest tumor of a child [1, 2]. Several reagents including RA and TPA have been reported to induce differentiation of this cell line, as assessed by a neuron-like morphological change and increased expression of neuronal differentiation markers, including neurofilaments and microtubule-associated protein 2 (MAP-2) [3–6]. A robust increase in the length of neurites was readily observed and can be measured by phase-contrast microscopic images following 4 day-treatment with RA or TPA [3–6]. Although both RA and TPA are able to induce SH-SY5Y cell differentiation, it is believed that the mechanistic actions or pathways involved are different for each compond. For example, the length of neurite extension induced by TPA is generally shorter than that induced by RA [2, 4, 7, 8].
Neurite outgrowth requires the interplay of all three major types of protein filaments that form the cytoskeleton: actin filaments, microtubules and intermediate filaments. Precise regulation of cytoskeletal dynamics is important for neurite outgrowth through reorganization of cytoskeleton components. During neuronal differentiation, microtubules form a bundle while actin filaments reorganize to produce the growth cone, followed by elongation and maintenance of neurites through stabilization of microtubules. Neurofilament proteins, members of intermediate filaments specifically expressed in neuronal cells, play many roles in neurite outgrowth, including protein trafficking, cellular motility, and maintaining neurite structures [9–11]. Neurofilaments represent major cytoskeleton elements of neurons for both the central and the peripheral nervous systems. There are five different classes of neurofilaments that are heteropolymers composed of three different subunits named neurofilament-H (NF-H), neurofilament-M (NF-M), and neurofilament-L (NF-L) [12]. Expression of each class of neurofilaments changes during different developmental stages. For instance, NF-M and NF-L subunits are expressed early during embryonic neurogenesis while NF-H appears later during the postnatal period in rats [12, 13]. In cultured SH-SY5Y cells, dynamic regulation of neurofilament expression has also been reported. For instance, both NF-H and NF-M are expressed in naïve cells and their expression is significantly increased following RA treatment [6, 14]. In addition, increased phosphorylation of NF-H in the tail region was observed in SH-SY5Y cells following 5 day RA treatment [6]. According to in vitro studies, phosphorylation of neurofilaments, especially NF-H, decreased degradation of neurofilaments by calpain [15, 16]. In vivo, phosphorylated NF subunits are expressed primarily in maturing neurites within the axonodendritic structure of neurons. Furthermore, cytoskeleton proteins other than neurofilaments, are also known to be associated with the development of neurites during neuronal cell differentiation. Microtubule-associated proteins (MAPs) are one class of such proteins. Differentiation-inducing agents are able to regulate the dynamics of microtubules through increased expression and phosphorylation of MAPs [17, 18]. In primary neurons, MAP-2 preferentially associates with the somatodendritic domain.
The central role of Nrf2 in cell survival has been well established over a decade of research. Nrf2 is activated in response to oxidative stress to induce expression of an array of genes whose functions are to counteract redox imbalance and to eliminate harmful reactive species [19–24]. NAD(P)H quinone oxidoreductase-1 (NQO1), glutamate cysteine ligase (GCL), glutathione peroxidase (GPx), thioredoxin (Trx), thioredoxin reductase (TrxR), peroxiredoxin (Prx), heme oxygenase-1 (HMOX-1), glutathione S-transferase (GST), and multidrug resistance-associated proteins (MRPs) are a few examples of well-characterized Nrf2-target genes [25–33]. Antioxidant response elements (AREs) have been identified in the promoters of these genes and are required for activation of these genes by Nrf2 [34–36]. Recently, great progress has been made in understanding how Nrf2 is able to activate expression of the ARE-bearing genes in response to oxidative stress or chemopreventive compounds. Keap1, a negative regulator of Nrf2, is a substrate adaptor for the Cul3-E3 ubiquitin ligase that constantly targets Nrf2 for ubiquitin-mediated proteasomal degradation under the redox-balanced conditions to maintain low constitutive levels of Nrf2 [37–39]. In response to Nrf2 activators, the Keap1-dependent ubiquitination of Nrf2 is inhibited, resulting in stabilization of Nrf2 and its translocation to the nucleus where it forms a heterodimer with one of the small Maf proteins [37–39]. The Nrf2-Maf heterodimer then binds the ARE to activate transcription. This Keap1-dependent regulation has been considered the primary control mechanism of the Nrf2 signaling pathway, although multi-levels of regulation may exist. Meanwhile, functional studies with Nrf2 have generated a growing body of literature that supports the protective role of Nrf2 against environmental toxicants and carcinogens. Cell-based studies have demonstrated that sensitivity of cells to a vast majority of toxic chemicals is modulated by intracellular levels of Nrf2. Pretreatment of cells with small doses of Nrf2 activators enhance cell resistance to subsequent challenge with toxic chemicals [40, 41]. In vivo studies with Nrf2-null mice indicate that these mice are highly susceptible to toxicants and carcinogens, compared to wild-type mice [42–48]. These pioneering studies have provided strong evidence that Nrf2 is critical in the cellular defense system and cell survival. Furthermore, the protective role of Nrf2 against neurodegenerative diseases have been demonstrated in cultured cell models and in animal models [49–54]. However, whether Nrf2 has any role in neuronal differentiation has not been investigated. In the present study, using both overexpression and knockdown approaches and by assessing three neuronal differentiation indexes, such as neurite outgrowth, increased protein levels of NF-M, and enhanced MAP-2 immunostaining, we have provided the first evidence that Nrf2 promotes neuronal cell differentiation both in SH-SY5Y cells and in primary neurons isolated from wild-type and Nrf2-null mice.
Materials and Methods
Chemicals
Most chemicals, including RA, TPA, tBHQ, Hoechst 33258, Na2SO4, K2SO4, MgCl2, CaCl2, Hepes, cysteine, HCl, papain latex, and glucose were from Sigma Chemical Co. (St. Louis, MO).
Cell cultures and establishment of stable cell lines
SH-SY5Y cells (American Type Culture Collection, Manassas, VA) were grown in Minimum Essential Medium (MEM)/F12 medium (Invitrogen, Grand Island, NY) with 10% fetal bovine serum at 37°C in a humidified incubator containing 5% CO2. The hemagglutinin (HA)–Nrf2 cDNA was cloned into the cloning site of a lentiviral vector purchased from System Biosciences (Mountain View, CA) using the standard recombinant DNA technique. For establishment of stable cell lines harboring HA–Nrf2, the lentiviral vectors were transfected into 293TN cells alone with the packaging plasmids according to the manufacturer’s instruction. Viruses produced in the supernatant were collected and used to infect SH-SY5Y in the presence of 8 µg/ml polybrene. At 48 hr post-infection, cells were grown in media containing 1.5 µg/ml of puromycin for selection. Stable cell lines were established once all the cells in the negative control plate were killed. Stable cell lines were continuously grown in media containing 1.5 µg/ml of puromycin.
RA Treatment
All RA treatments were performed under dark condition. A final concentration of 10 µM RA was used to treat SH-SY5Y cells with or without differentiation. No toxicity was observed with the RA-differentiation treatments (Data not shown).
mRNA extraction and qRT-PCR
Total mRNA was extracted from cells using TRI reagent (Sigma, St. Louis, MO), and the equal amounts of RNA were reverse-transcripted into cDNA using the Transcriptor First Strand cDNA synthesis kit (Roche, Indianapolis, IN). The following Taqman probes from the universal probe library were used (Roche, Indianapolis, IN): hNrf2 (#70), hNQO1 (#87), hNF-M (#42), and hGAPDH (#25). The following primers were synthesized by Integrated DNA Technologies (Coralville, IA): hNrf2: forward (acacggtccacagctcatc) and reverse (tgtcaatcaaatccatgtcctg); hNQO1: forward (atgtatgacaaaggacccttcc) and reverse (tcccttgcagagagtacatgg); hNF-M: forward (acgacctcagcagctacca) and reverse (gagccatttcccactttgtg); and hGAPDH: forward (ctgacttcaacagcgacacc) and reverse (tgctgtagccaaattcgttgt). The real-time PCR condition was as follows: one cycle of initial denaturation (95°C for 10 min), 40 cycles of amplification (95°C for 10 sec and 60°C for 20 sec), and a cooling period (50°C for 5 sec). The data presented are relative mRNA levels normalized to GAPDH, and the value from the undifferentiated cells was set as 1. PCR assays were performed two times with duplicated samples were used to determine the mean ± SD.
Antibodies and immunoblot analysis
Antibodies for Nrf2, Keap1, and α-Tubulin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), NF-M from Covance (Emeryville, CA), MAP-2 from Calbiochem (San Diego, CA). Cells were lysed in sample buffer [50 mM Tris-HCl (pH 6.8), 2% SDS, 10% glycerol, 100 mM DTT, 0.1% bromophenol blue]. After sonication, cell lysates were electrophoresed through SDS-polyacrylamide gel and subjected to immunoblot analysis.
Protein half-life measurement
To measure the half-life of Nrf2, cells were either undifferentiated or RA-differentiated for 24 hr. 50 µM cycloheximide was added to block protein synthesis. Total cell lysates were collected at different time points and subjected to immunoblot analysis with an anti-Nrf2 antibody. The relative intensity of bands was quantified by the ChemiDoc CRS gel documentation system and Quantity One software from BioRad (Hercules, CA, USA). The pulse-chase method was also used to measure the rate of protein synthesis and half-life. SH-SY5Y cells were either undifferentiated or RA-differentiated for 24 hr. Cells were then incubated for 15 min in labeling MEM without methionine, cysteine and L-glutamine, complemented with 10% dialyzed FBS. Cells were cultured for 30 min with 0.3mCi/ml [35S]-methionine and [35S]-cysteine labeling media. Cells were washed once with complete media and incubated in complete media at 37 °C in a humidified incubator containing 5% CO2. Total cell lysates were collected at different time points. Immunoprecipitation was performed using an anti-Nrf2 antibody in RIPA buffer. Immunoprecipitated proteins were subjected to SDS-PAGE gel resolution and autoradiography.
Transient transfection of siRNA and lentiviral vector
Nrf2-siRNA and control siRNA were purchased from Ambion (Austin, TX). Transient transfection of siRNA was performed using siPORT NeoFX Transfection Reagent according to the manufacturer's protocol (Ambion, Austin, TX). Cells were then treated with 10µM RA for 24 hr and lysed in sample buffer for SDS-polyacrylamide gel and immune blot analysis. The Keap1–chitin-binding domain (CBD) or hemagglutinin (HA)–Nrf2 cDNA was cloned into the cloning sites of the lentivector purchased from System Biosciences (Mountain View, CA). The lentivectors were transfected into 293TN cells with the packaging plasmids according to the manufacturer’s instruction. Viruses produced in the supernatant were collected and used to infect cancer cell lines in the presence of 8 µg/ml polybrene. At 48 hr post-infection, cells were treated with 10 µM RA for 24 hr. Cells were lysed in sample buffer. After sonication, cell lysates were electrophoresed through an SDS-polyacrylamide gel and subjected to immunoblot analysis.
Immunofluorescence cellular staining
SH-SY5Y cells were grown on poly-D-lysine coated (Sigma, St.Lousi, MO) glass coverslips in 35-mm plates. Cells were fixed in pre-chilled methanol at −20 °C for one hour and incubated at room temperature for 50 min with primary antibodies diluted 1:100 in phosphate buffered saline (PBS) containing 10% fetal bovine serum. The glass coverslips were washed in PBS and incubated at room temperature for 50 min with secondary antibodies, Alexa Fluor 488-conjugated anti-mouse IgG or Alexa Fluor 593-conjugated anti-rabbit IgG (Invitrogen, Grand Island, NY), at 1:100 dilution in PBS containing 10% fetal bovine serum and 1% Hoechst. The glass coverslips were washed in PBS and mounted with ProLong Gold Antifade Reagent (Invitrogen, Grand Island, NY) on glass slides. Cells were scored and imaged using 3i Marianas Ziess Observer Z1 system and Slidebook (Intelligent Imaging Innovations, Denver, Co). The images were exported from Slidebook to tiff files. Adobe Photoshop was used to construct the figures. Minimal alterations were performed on the digital images.
ChIP assay
ChIP assay was done as previously reported [55]. SH-SY5Y cells (approximately 1 × 106) were cross-linked with formaldehyde, collected in PBS, resuspended in 200µl SDS lysis buffer with PMSF and a protease inhibitor cocktail, and sonicated on ice. The lysates were then diluted in chromatin immunoprecipitation (ChIP) dilution buffer to 2ml, precleared with protein A agarose, and then incubated with appropriate antibodies overnight. The immune complexes were collected with 50µl protein A agarose, washed with low-salt buffer, high-salt buffer, LiCl buffer, and TE buffer. The complexes were eluted in 500µl fresh elution buffer. The cross-links were reversed by heating at 65°C for 5 h after addition of 20µl of 5 M NaCl. Samples were treated with RNase and proteinase K. DNA was recovered by phenol-chloroform extraction and ethanol precipitation. Relative amounts of DNA in the complex were quantified by the real-time PCR method using the LightCycler 480 DNA SYBR green I kit (Roche, Indianapolis, IN). Primers used were as follows: human NQO1 ARE forward, 5'-GCAGTCACAGTGACTCAGC-3'; human NQO1 ARE reverse, 5'-TGTGCCCTGAGGTGCAA-3'; tubulin promoter forward, 5'-GTCGAGCCCTACAACTCTATC-3'; tubulin promoter reverse, 5'-CCGTCAAAGCGCAGAGAA-3'. PCR cycling was performed as follows: initial denaturation at 95°C for 5 min (1 cycle), 40 cycles of amplification of 95°C for 10 s, 60°C for 10 s, and 72°C for 20 s, with a single fluorescence acquisition. The amplification was followed by a melting curve program (65 to 95°C with a heating rate of 0.1°C per second and a continuous fluorescence measurement) and then a cooling program at 40°C for 30 s. The mean crossing-point values and standard deviations for NQO1 and tubulin were determined for the different samples. The crossing point is defined as the point at which the fluorescence rises appreciably above the background fluorescence. A nontemplate control was run for each primer pair to assess the overall specificity and to ensure that primer dimers were not interfering with amplification detection. Amplification specificity was checked using melting-curve and agarose gel electrophoresis. Melting-curve analysis showed a single sharp peak for all samples, and agarose gel electrophoresis showed a single band at the expected size. Data are presented as n-fold change. The real-time PCR assays were performed two times, each with duplicated samples.
DNA affinity precipitation assay
A 39 bp ARE sequence from the human NQO1 promoter was used as a probe. The sequence used was 5’-AATCGCAGTCACAGTGACTCAGCAGAATCTGAGCCTAGGG-3’. Two 5’-biotinylated ssDNA oligos with complimentary sequences were synthesized by Integrated DNA Technologies and annealed to form a dsDNA probe. The probe was incubated with cell lysates from transfected SH-SY5Y cells in RIPA buffer (0.1% SDS). Proteins were pulled down using streptavidin beads (PIERCE, ROCKFORD, IL) and were subjected to immunoblot analysis.
Evaluation of neuritogenesis
Percent of differentiated cells and the length of neuritis was done according to previous methods [7]. To determine if Nrf2 promotes the differentiation of SH-SY5Y cells induced by RA, we grew SH-SY5Y cells in poly D-lysine coated glass coverslips in the absence or presence of 10µM RA in the media. The cells were photographed 4 days post-treatment of RA on a phase contrast microscope (3i Marianas, Intelligent Imaging Innovations, Denver, Co). The percent of differentiated cells and the length of neurites were quantified using Slidebook (Intelligent Imaging Innovations, Denver, Co) to count 500 cells in 10 randomly chosen fields (100x magnification) in each treatment group. Cells were considered differentiated if they had one neurite longer than the control group mean neurite length. The length of the longest neurite was measured in at least 500 cells in 10 randomly chosen fields (100x magnification) for each treatment group.
Isolation of primary neuron
Coverslips were coated with poly D-lysine prior to experiments. Neocortex was harvested in 10ml dissociation media (DM, 82 mM Na2SO4, 60mM K2SO4 , 5.8mM MgCl2, 2.5uM CaCl2, 1mM Hepes, 10mM Glucose, adjust pH to 7.4), rinsed twice with 10ml DM, and once with DM/Ky.Mg (DM 45ml, 10xKy/Mg 5ml) (10xKy.Mg, 10 mM kynurenic acid, 100 mM MgCl2, 5mM Hepes adjust pH to 7.4). Tissues were minced and washed with DM/Ky.Mg. Activated enzyme solution (DM/Ky.Mg 9.5ml, Cysteine.HCl 0.45mg/ml, papain latex 100 units, adjust pH to 7.4) was added and tissues were incubated 30 min at 37°C. Tissues were then rinsed with DM/Ky.Mg and pipetted up and down until dispersed, and then washed with 10ml DMEM 3 times. Before plating, cells were counted with the hemocytometer and diluted in plating media. Cultures were incubated at 37°C in a humidified incubator containing 5% CO2 for 24 hr. Plating media was then removed and replaced with neuronal maintenance media.
Statistical analysis
All statistical results were expressed as means ±S.E.M. Statistical significance between different experiment groups were evaluated by one-way analysis of variance (ANOVA) followed by Tukey-test performed with the SPSS software(version 16.0; SPSS Chicago, IL) A P-value less than 0.05 was considered statistically significant.
Results
RA and TPA up-regulated expression of Nrf2 and its downstream genes
SH-SY5Y cells were treated with 10 µM of RA for different time periods and expression of Nrf2, NQO1, and NF-M was analyzed by immunoblot analysis. Induction of Nrf2 was observed as early as 2 hr and remained elevated up to 48 h, with the maximal induction at 24 hr (Fig. 1A). Elevated expression of NQO1 was observed at 24 hr and 48h. As expected, NF-M was also up-regulated 12 hr following RA treatment and remained elevated at 48 hr, whereas, the levels of control α-tubulin remained the same (Fig. 1A). These data indicate that there might be a positive correlation between Nrf2 activation and NF-M expression. Another parallel set of samples treated with 10µM of RA was also evaluated for mRNA expression of Nrf2, NQO1, and NF-M by real-time qPCR. While there was only a marginal induction of Nrf2 at the mRNA level, expression of NQO1 and NF-M mRNA showed a significant increase (Fig. 1B). Next, the 24 hr time point was chosen for induction of Nrf2, NQO1, and NF-M by different doses of RA. As shown in figure 1C, RA induced the protein levels of Nrf2 and NQO1 at all the doses used. Similarly, RA had no effect on Nrf2 transcription while NQO1 mRNA expression was up-regulated significantly (Fig. 1D). Induction of NF-M mRNA was observed with any treatment and reached peak with 10 µM RA treatment (Fig. 1D). In order to understand whether Nrf2 induction is due to the differentiation processes, or is specific to RA treatment, induction of Nrf2 by TPA, which induces neuronal differentiation through a different mechanism, was also determined. Similar to RA, TPA enhanced Nrf2 and NQO1 protein levels (Fig 1E). Furthermore, TPA-induced Nrf2 activation correlated with enhanced expression of NF-M (Fig 1E). Collectively, these data indicate that the Nrf2 signaling pathway is up-regulated during neuronal differentiation processes.
Fig. 1.
RA up-regulated expression of Nrf2 and its downstream genes. (A) Endogenous Nrf2 increased in a time-dependent manner following 10µM RA treatment. SH-SY5Y cells were treated with 10µM RA for the indicated time periods. Cells were lysed in sample buffer and total cell lysates were subjected to immunoblot analysis using anti-Nrf2, -NQO1, -NF-M, and –Tubulin (Tub) antibodies. (B) RA increased mRNA levels of Nrf2, NQO1, and NF-M in a time-dependent manner. Total mRNA was extracted from another parallel set of experiments and converted into cDNA. The same amount of cDNA was used for quantification by qRT-PCR. *p < 0.05 versus 0 hr of Treatment. **p < 0.01 versus 0 hr of Treatment (C) Endogenous Nrf2 increased in a dose-dependent manner following 24 hr treatment of RA. Cells were treated with different doses of RA for 24 hr and total cell lysates were subjected to immunoblot analysis using the indicated antibodies. (D) mRNA levels of Nrf2, NQO1 and NF-M increased with RA treatment. Another parallel set of samples was analyzed using qRT-PCR as described in (B). *p < 0.05 versus 0 µM of Treatment. (E) TPA up-regulated Nrf2. Cells were treated with several doses of TPA for 24 hr. Expression of Nrf2, NQO1, and NF-M was measured by immunoblot analysis.
RA up-regulated Nrf2 through enhancing the rate of Nrf2 protein synthesis
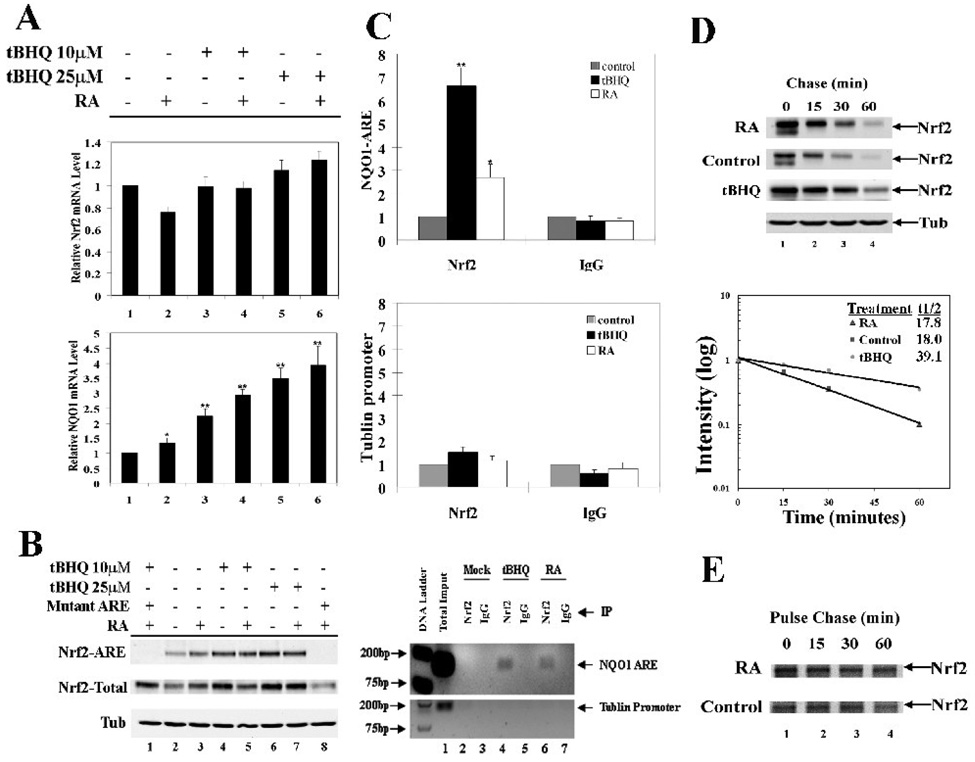
Next, the mechanism of RA-mediated up-regulation of Nrf2 was explored. SH-SY5Y cells were treated with two doses of tBHQ, RA, or in combination as indicated, and mRNA expression of Nrf2 and NQO1 was measured by qRT-PCR. Nrf2 mRNA expression was not affected by any treatment (Fig 2A, upper panel). However, the mRNA expression of NQO1 was enhanced slightly by RA treatment alone (Fig. 2A, lower panel, bar 2). Cotreatment of RA and tBHQ resulted in a higher induction of NQO1 (Fig. 2A, compare lane 2 to lane 4 or lane 6). To test any change in the amount of Nrf2 bound to the ARE, parallel-treated cells were subjected to the ARE affinity immunoprecipitation analysis. Nrf2 expression in the total lysate was shown (Fig. 2B) and the mutant ARE was included as a negative control (Fig. 2B, lanes 1 and 8). RA treatment alone increased Nrf2 binding to the ARE (Fig. 2B, compare lane 3 with lane 2) and combined treatment further enhanced binding of Nrf2 with the ARE (Fig. 2B, lane 4–7). To demonstrate that there is more Nrf2 bound to the ARE after RA treatment in vivo, we performed chromatin immunoprecipitation analysis (ChIP). tBHQ induced Nrf2 binding to the ARE significantly while RA enhanced Nrf2 binding to the ARE about 2 fold (Fig. 2C). These data indicate that RA is able to activate the Nrf2-mediated signaling pathway by increasing the total protein level of Nrf2 and, thus, facilitating more Nrf2 binding to the ARE. Previously, it has been well-demonstrated that most Nrf2-inducers, including tBHQ and sulforaphane, upregulate Nrf2 by suppressing Nrf2 degradation. Therefore, the half-life of Nrf2 in response to RA treatment was measured using the cycloheximide/immunoblot method. To our surprise, RA did not affect the half-life of Nrf2 and the half-life was approximately 18 min in the absence or presence of RA (Fig. 2D), whereas tBHQ increased the half-life of Nrf2 to 39 min. Next, protein synthesis and degradation were measured by the pulse chase method (Fig. 2E). The half-life of Nrf2 measured by the pulse chase method was consistent with that measured by the cycloheximide/immunoblot method. Interestingly, the newly synthesized proteins in the 30-min pulse period were higher in the RA-treated sample, compared to that in the non-treated control (Fig. 2E, lane 1, compare upper panel to lower panel). These data indicate that RA is able to upregulate the Nrf2-signaling pathway by enhancing the rate of Nrf2 protein synthesis.
Fig. 2.
RA up-regulated Nrf2 by enhancing the rate of Nrf2 protein synthesis. (A) RA potentiated the effect of tBHQ on mRNA levels of Nrf2 and NQO1 in SH-SY5Y cells. Cells were treated with tBHQ (10µM or 20µM) and/or RA (10µM) simultaneously for 24 hr. mRNA was extracted, followed by qRT-PCR. *p < 0.05 versus line 1. **p < 0.01 versus line 1. (B) RA and tBHQ up-regulated Nrf2 target genes by enhancing the Nrf2 protein level and thus binding to the ARE. Biotinylated double strand oligonucleotides containing 39 bp of the human NQO1 ARE sequence were incubated with cell lysates from SH-SY5Y that were simultaneously treated with tBHQ and/or RA. A DNA affinity precipitation assay was performed. A biotinylated mutant-NQO1 ARE sequence was used as a negative control. (C) RA increased Nrf2 binding to the ARE. ChIP analysis was performed in SH-SY5Y cells with the indicated antibodies after 24 hr treatment with tBHQ or RA. Genomic DNA fragments containing the NQO1 ARE region in the immunoprecipitates were quantified by real-time PCR and visualized on an agarose gel. *p < 0.05 versus control. **p < 0.01 versus control. (D) The half-life of Nrf2 was unaffected by RA treatment. SH-SY5Y cells were treated with cycloheximide for the indicated time periods. Cell lysate was collected and analyzed by immunoblot analysis. (E) RA enhanced the rate of Nrf2 protein synthesis. Pulse-chase assay was conducted with SH-SY5Y cells that were pulsed labeled with media containing [35S]-methionine and [35S]-cysteine for 30 min and chased with normal media for the indicated time points. Cell lysates were immunoprecipitated using an anti-Nrf2 antibody and the immunoprecipitates were resolved on a SDS-PAGE gel and visualized by autoradiography.
RA-induced neuronal outgrowth was accompanied by enhanced expression and nuclear translocation of Nrf2
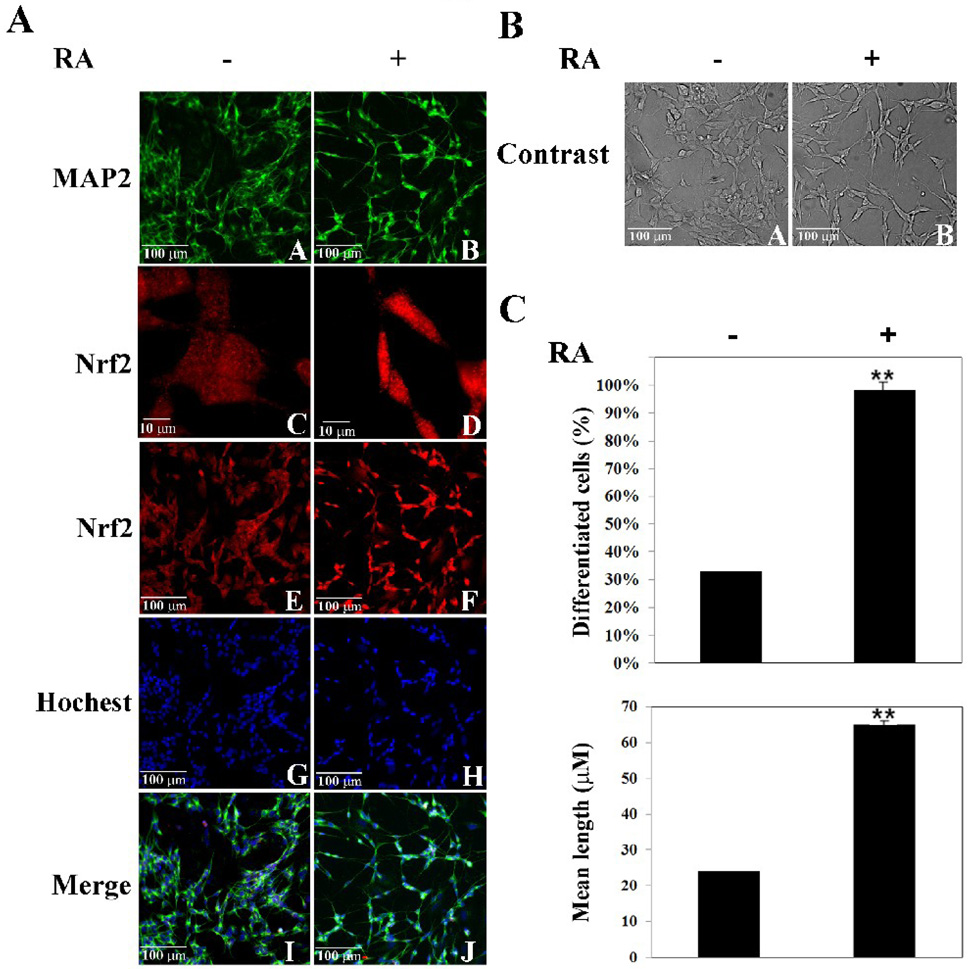
To test the coordinated up-regulation of Nrf2 with a neuron-like morphological change in response to RA treatment, an indirect double immunofluorescence stain with anti-Nrf2 and anti-MAP-2 was performed in SH-SY5Y cells in the absence or presence of RA. In the undifferentiated cells, MAP-2 expression was low but increased upon treatment with RA (Fig. 3A, compare panel A with B). The cell density was low in the RA-treated sample, compared to the non-treated sample, even though the same amount of cells were seeded for the two groups (Fig. 3A, compare panel A with B). This is due to a reduced growth rate after RA stimulation. In addition, neurite extension was clearly seen following RA treatment (Fig. 3A, compare panel A with B). Cellular localization of Nrf2 was switched from the whole cell to the nucleus in response to RA treatment, (Fig. 3A, compare panel C with D). Furthermore, the intensity of Nrf2 was enhanced markedly, indicating an increase in the Nrf2 protein level in response to RA treatment (Fig. 3A, compare panel C with D; panel E with F). Figure 3B show a phase-contrast image of the same set of samples taken prior to the immunofluorescence staining. The percentage of differentiated cells was counted and the mean neurite length was measured using phase-contrast images. Both parameters were significantly increased in the RA-treated samples (Fig. 3C). Clearly, RA induced both Nrf2 and neuronal cell differentiation.
Fig. 3.
RA-induced neuronal outgrowth was accompanied by enhanced expression and nuclear translocation of Nrf2. (A) RA induced morphological changes and up-regulated MAP-2 and Nrf2. Cells were treated with 10µM RA for 4 days, fixed in methanol, and immunostained with anti-MAP-2 and anti-Nrf2 antibodies. (B) and (C) RA caused neurite extension. Phase-contrast images were taken before cells were fixed for immunostaining. 500 cells in 10 randomly chosen fields for each group were counted to calculate the percentage of differentiated cells and the mean length of neurites. **p < 0.01 versus cells control.
Stable-overexpression of Nrf2 promoted neuronal differentiation
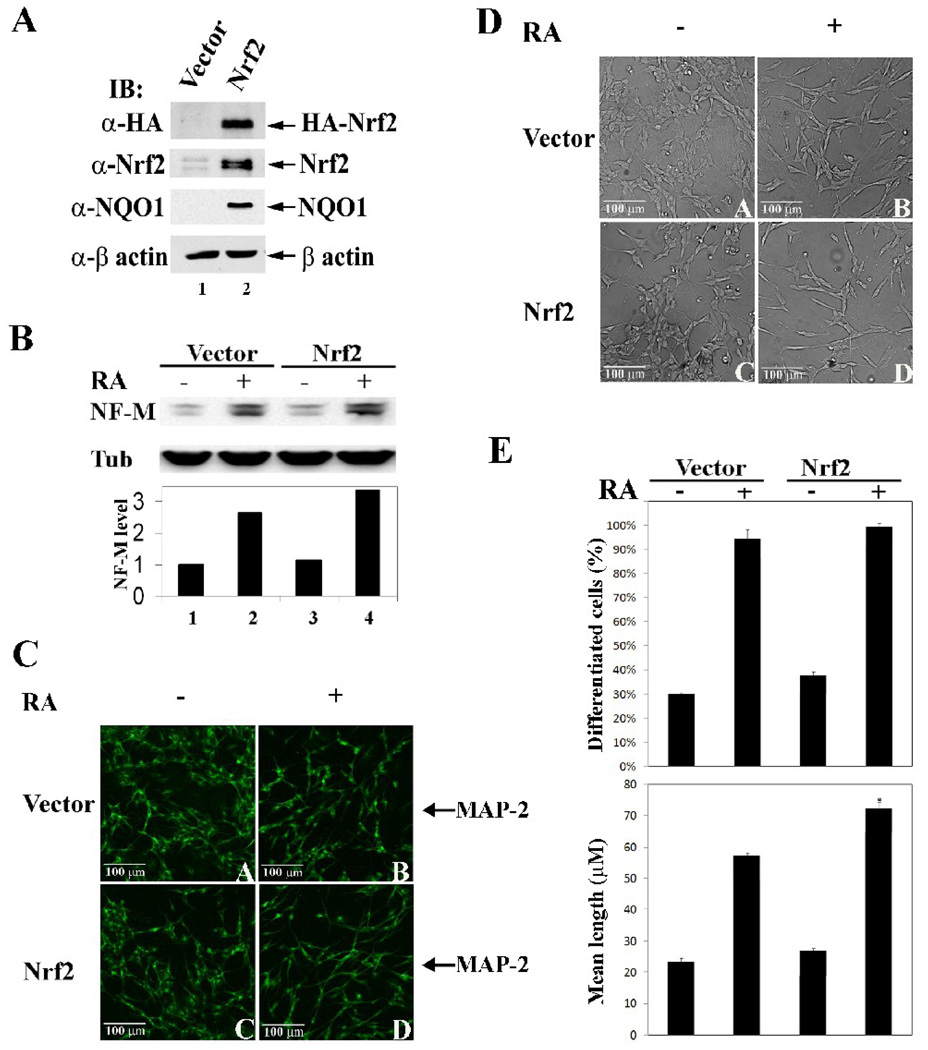
Next, the possible correlation between Nrf2 up-regulation and neuronal cell differentiation was tested by stable overexpression of Nrf2. A SH-SY5Y-derived stable cell line expressing HA-Nrf2 was established using a lentivirus delivery and puromycin selection system. As shown in figure 4A, HA-Nrf2 was expressed as detected by immunoblot analysis with anti-HA antibodies (Fig. 4A). Similarly, overall expression of Nrf2 was enhanced in this stable cell line as detected with anti-Nrf2 antibodies (Fig. 4A). Overexpression of Nrf2 resulted in enhanced expression of NQO1, indicating up-regulation of Nrf2 and its downstream events (Fig. 4A). In this cell line, overexpression of Nrf2 promoted RA-induced up-regulation of NF-M although it had no effect on the levels of NF-M in the absence of RA (Fig. 4B, compare lane 1 with lane 3, lane 2 with 4), indicating that upregualtion of the Nrf2 signal pathway itself is insufficient to initiate neuronal differentiation processes. Morphological differences in the vector-transfected and HA-Nrf2 transfected cells in the absence or presence of RA were also determined. Overexpression of HA-Nrf2 had no effect on neurite outgrowth in the absence of RA, but potentiated neuronal differentiation in RA-treated samples as determined by the indirect immunofluorescence stain with MAP-2 (Fig. 4C, compare panel A with C; panel B with D). Similarly, neurite outgrowth promoted by Nrf2 overexpression was observed in phase-contrast images (Fig 4D, compare panel B with panel D). Moreover, the conclusion that Nrf2 positively regulates neuronal differentiation was confirmed by quantifying the percentage of differentiated cells and the mean neurite length (Fig. 4E).
Fig. 4.
Stable-overexpression of Nrf2 promoted neuronal differentiation. (A) A SH-SY5Y-derived stable cell line has enhanced expression of Nrf2 and its downstream gene, NQO1. SH-SY5Y cells were infected using a lentivirus delivery system to express HA-Nrf2 and were selected using puromycin. Cells were lysed under denaturing conditions and subjected to immunoblot analysis with anti-HA, -Nrf2, -NQO1, and –β-actin antibodies. (B) Nrf2 overexpression up-regulated NF-M in the presence of RA. The vector control and HA-Nrf2 SH-SY5Y stable cell lines were undifferentiated or RA-differentiated for 24 hr. Cell lysates were subjected to immunoblot analysis with anti-NF-M and –Tub antibodies. (C) Overexpressed Nrf2 promotes neurite outgrowth. Cells were treated with 10 µM RA for 4 days before fixation for indirect immunofluorescence staining using an antibody for MAP-2. (D) and (E) Stable overexpression of Nrf2 promoted neuronal cell differentiation. Phase-contrast images were taken before cells were fixed. Quantification of neuronal differentiation of the stable cell lines were conducted as previously described in Figure 3. *p < 0.05 versus vector with treatment.
Up-regulation of endogenous Nrf2 potentiated differentiation
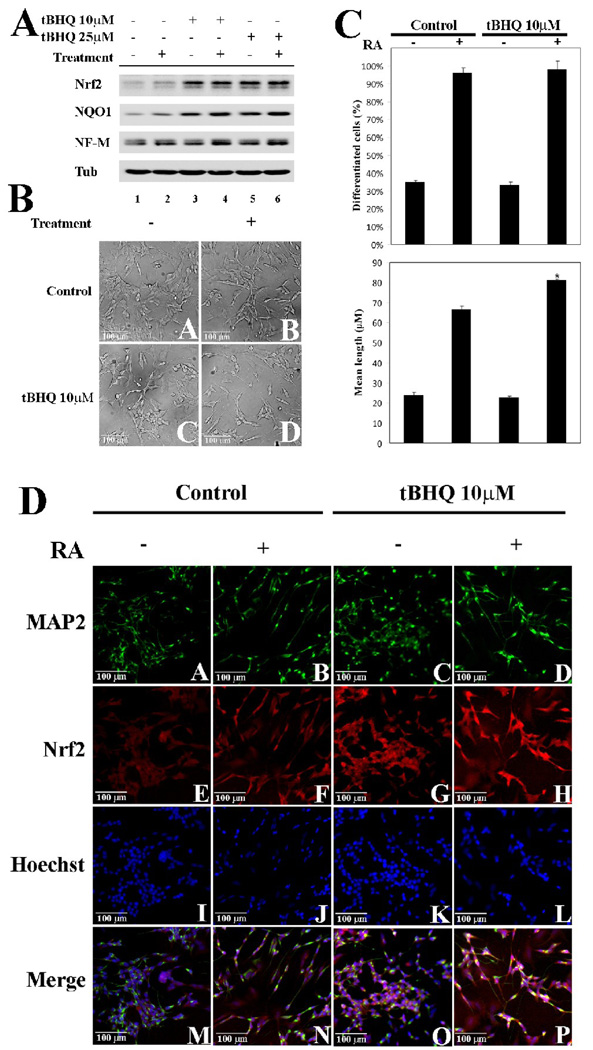
Next, we tested whether up-regulation of endogenous Nrf2 by tBHQ is able to modulate neuronal differentiation. SH-SY5Y cells were treated with tBHQ, RA, or in combination as indicated and cell lysates were subjected to immunoblot analysis. Although tBHQ up-regulated Nrf2 and NQO1 (Fig. 5A lanes 3–6), tBHQ alone had no effect on NF-M expression (Fig. 5A, lanes 1, 3, and 5). Significantly, tBHQ enhanced the RA-mediated upregualtion of NF-M protein levels (Fig. 5A, compare lanes 4 and 6 with lane 2), demonstrating the positive role of Nrf2 in potentiating neuronal differentiation processes. Phase contrast images were taken before cells were lysed (Fig. 5B), and these images were used for quantification of the percentage of differentiated cells and the mean neurite length (Fig. 5C). Co-treatment of tBHQ with RA increased the percentage of differentiated cells slightly and significantly increased the mean neurite length, compared with RA-only treatment (Fig. 5C). Furthermore, tBHQ had no effect in the absence of RA (Fig 5C). tBHQ treatment enhanced Nrf2 (Fig. 5D, compare panels G and H with panels E and F) and MAP-2 staining (Fig. 5D, panel D). Together, these data demonstrate that induction of endogenous Nrf2 facilitates RA-induced neuronal differentiation, although Nrf2 activation itself is insufficient to induce neuronal differentiation.
Fig. 5.
Up-regulation of endogenous Nrf2 potentiated differentiation. (A) tBHQ enhanced RA-mediated up-regulation of NF-M. SH-SY5Y cells undifferentiated or RA-differentiated and/or the indicated concentration of tBHQ were lysed and subjected to immunoblot analysis using the indicated antibodies. (B) and (C) Up-regulation of endogenous Nrf2 by tBHQ promoted neuronal differentiation. Phase-contrast images and quantification of neuronal differentiation were conducted as previously described in Figure 3. *p < 0.05 versus control with treatment. (D) Nrf2 activation by tBHQ potentiated the RA-mediated neurite outgrowth. SH-SY5Y cells were untreated or treated with tBHQ and/or undifferentiated or RA-differentiated with RA for 24 hr. Cells were fixed in methanol and subjected to immunofluorescence staining with MAP-2 and Nrf2 antibodies.
Down-regulation of Nrf2 compromised neuronal differentiation
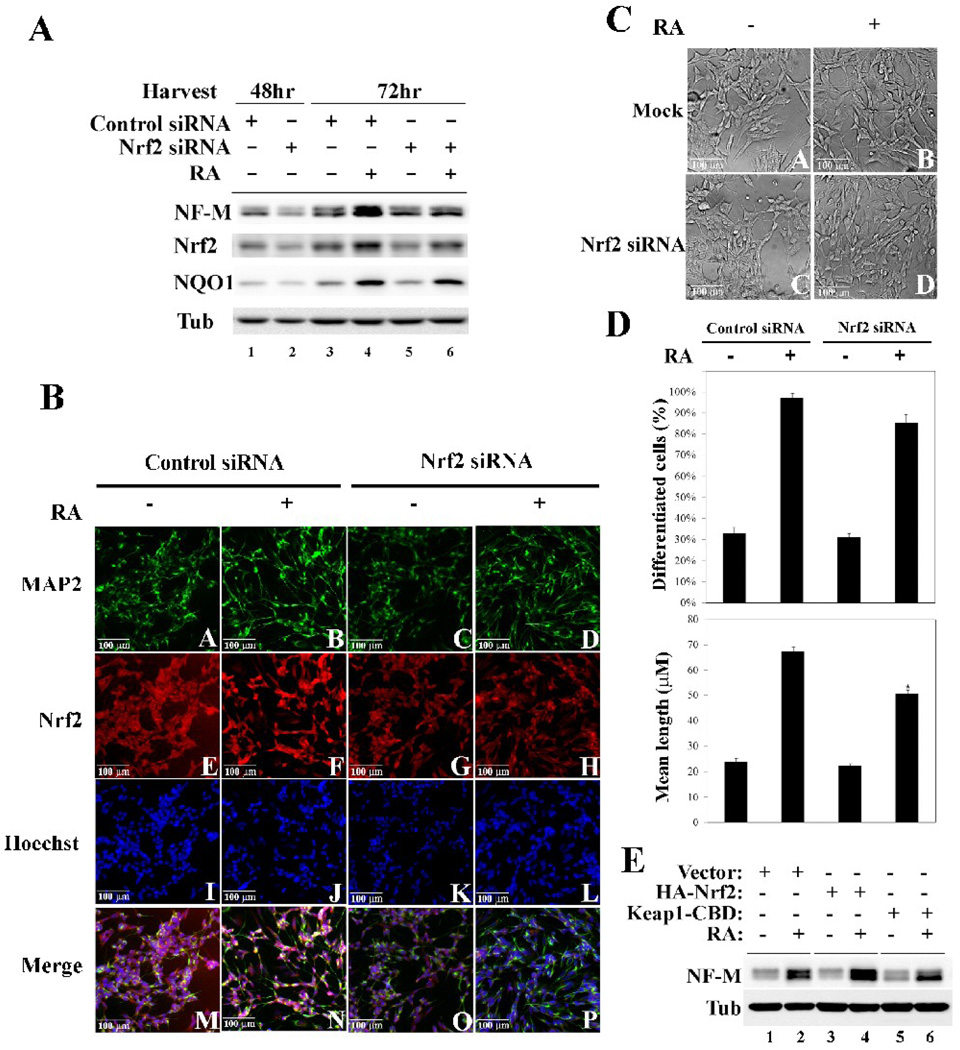
To further verify the importance of Nrf2 in neuronal differentiation, the possible requirement of Nrf2 in neuronal differentiation was tested. Two approaches were used to knockdown the expression of endogenous Nrf2. In one set of experiments, Nrf2-siRNA or control siRNA were transfected into SH-SY5Y cells and levels of Nrf2 were measured. Transfection of Nrf2-siRNA for 48 hr decreased levels of Nrf2 to 50% (Fig. 6, compare lane 1 and 2, Nrf2 panel). At 48 hr post-transfection, cells were further treated with RA for an additional 24 hr and NF-M expression was determined by immunoblot analysis. Knockdown of Nrf2 expression itself had no effect on NF-M levels (Fig. 6A, compare lane 1 with 2; 3 and 5; NF-M panel). However, Nrf2-siRNA blocked induction of NF-M in response to RA (Fig. 6A, compare lane 4 with lane 6). Morphological analysis showed retarded neurite outgrowth in cells transfected with Nrf2 siRNA, but not with the control siRNA, in response to RA treatment (6B, compare panel B with panel D; panel A with panel B). In addition, knockdown of Nrf2 expression was clearly shown by the reduced Nrf2-fluorescence staining (Fig. 6B, compare panels G and H with panels E and F). The phase contrast images of these cells were taken before cells were fixed for immunofluorescence staining (Fig. 6C). Changes in the percentage of differentiated cells and the mean neurite length by RA treatment were diminished in Nrf2-siRNA transfected cells (Fig. 6D). The fact that inhibition of Nrf2 by Nrf2-siRNA was unable to completely block the RA-mediated neurite outgrowth can be due to two reasons: (i) Nrf2 is not absolutely required for the neuronal differentiation process; (ii) complete inhibition of Nrf2 is not achieved and Nrf2-siRNA only reduced Nrf2 expression by 50%. Another approach was also used to inhibit expression of Nrf2 by overexpression of Keap1, the substrate adapter for E3 ubiquitin ligase responsible for Nrf2 degradation. SH-SY5Y cells were infected with lentivirus carrying HA-Nrf2 and Keap1-CBD. Transient overexpression of Nrf2 and Keap1 was confirmed by immunoblot analysis with anti-HA and anti-CBD antibodies (data not shown). In response to RA treatment, overexpression of Nrf2 resulted in further up-regulation of NF-M, whereas knockdown of Nrf2 by Keap1 overexpression suppressed upregualtion of NF-M by RA (Fig. 6E, compare lanes 4 and 6 with lane 2). Together, these data demonstrate that inhibition of Nrf2 expression compromised neuronal differentiation. Noticeably, up- or down-regulation of Nrf2 had no effect on the levels of NF-M in the absence of RA treatment, indicating that Nrf2 itself neither can initiate neuronal differentiation nor is absolutely required for neuronal differentiation.
Fig. 6.
Down-regulation of Nrf2 compromised neuronal differentiation. (A) Nrf2 siRNA effectively decreased Nrf2 protein levels and blocked induction of NF-M in response to RA. SH-SY5Y cells were transiently transfected for 48 hr with either control siRNA or Nrf2 siRNA (lane 1 and 2). Another set of samples was transfected with siRNA for 48 hr and treated with RA for an additional 24 hr (lanes 3–6). Cells were lysed and subjected to immunoblot analysis with antibodies for NF-M, Nrf2, NQO1, and Tubulin. (B) Knockdown of Nrf2 affects cell morphology and inhibits neurite outgrowth when treated with RA. Cells were transfected with control siRNA or Nrf2 siRNA for 48 hr followed by 10µM RA treatment for 4 days. Cells were then fixed in methanol and immunofluorescence analysis was conducted using antibodies for MAP-2 and Nrf2. (C) and (D) Knockdown of Nrf2 by siRNA compromised neuronal differentiation. Cells transfected with control siRNA or Nrf2 siRNA, and undifferentiated or RA-differentiated with 10µM RA for 4 days were subjected to phase-contrast microscopy. Calculation of the percentage of differentiated cells and mean length was conducted as previously described in Fig 3. *p < 0.05 versus control with siRNA. (E) Overexpression of Nrf2 or Keap1 in conjunction with RA treatment caused an increase or decrease in NF-M, respectively. Cells were transiently infected by lentiviruses containing either HA-Nrf2 or Keap1-CBD. Total cell lysates were subjected to immunoblot analysis using the indicated antibodies.
Primary neurons isolated from Nrf2-null mice were retarded in neurite outgrowth
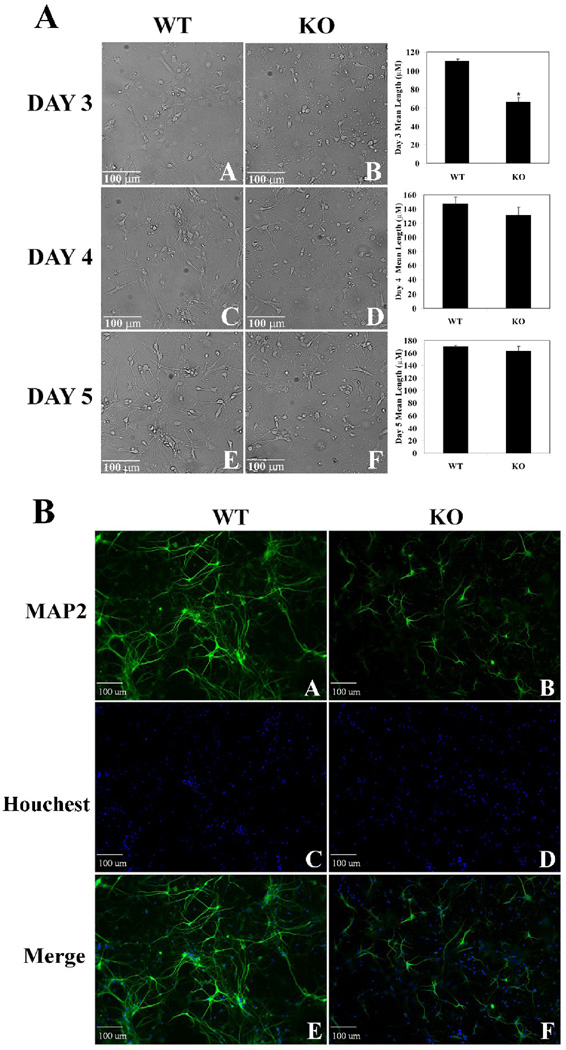
To further verify the important function of Nrf2 in regulating neuronal cell differentiation, primary neurons were isolated from wild-type and Nrf2-null mice. The primary neurons were allowed to grow in culture and morphological changes were observed by taking phase contrast images every 24 hr for 5 consecutive days (Fig 7A). The neuron mean length at day 3, 4 and 5 was measured (Fig. 7A, bar graphs). The mean neurite length was significantly reduced in primary neurons from Nrf2-null mice, especially at day 3, demonstrating a slow differentiation process in neurons that are deficient in Nrf2 expression. Primary neurons that had been growing in culture for three days were subjected to immunofluorescence staining with anti-MAP-2 (Fig. 7B). Expression of MAP-2 was lower in neurons from Nrf2-null mice as shown by a reduced intensity of MAP-2 staining (Fig. 7B, compare panel A with panel B). In addition, primary neurons isolated from Nrf2-null mice had less neurite outgrowth (Fig. 7B, compare panel A with panel B). Similar amounts of cells were seeded in these two groups as shown by nuclear staining (Fig. 7B, panel C and D), indicating that the difference in neurite outgrowth is not due to a difference in cell confluency, but rather, it is due to the status of Nrf2. These results clearly demonstrate the functional role of Nrf2 in potentiating neuronal differentiation.
Fig. 7.
Primary neurons isolated from Nrf2-null mice were retarded in neurite outgrowth. (A) Cell morphology and neurite lengths were compromised in Nrf2-null mice compared to wild-type. Primary neurons were isolated from wild-type and Nrf2-null mice and grown in cell culture for 3–5 days. *p < 0.05 versus wild-type. (B) Neurite outgrowth is delayed in primary neurons from Nrf2-null mice. Immunofluorescence staining was conducted on primary neurons isolated on day 3 from wild-type and Nrf2-null mice using a MAP-2 antibody.
Discussion
Signaling mechanism controlling neuronal differentiation processes has not been well studied and is likely a complex process, requiring interplay of many signaling events. It is conceivable that multiple pathways converge to alter expression or phosphorylation states of cytoskeletal proteins, including MAPs and neurofilaments, to accommodate neurite outgrowth [5, 6, 14]. Many signaling pathways have been reported to regulate neuronal differentiation processes when the SH-SY5Y cell line is used as a model. For example, the integrin signaling pathway has been shown to be up-regulated during RA-induced differentiation processes, resulting in activation of cyclin-dependent kinase 5 that is required for enhanced expression and phosphorylation of NF-H as well as neurite outgrowth [6, 14]. During the early stages of neuronal differentiation following RA- or TPA-treatment, IκBβ is quickly degraded, resulting in activation of NFκB, which is required for neuronal differentiation [4]. Similar to what was observed with Nrf2 in our study, DAN, the founding member of the DAN family of secreted cytokines, was up-regulated in response to RA treatment [3]. Furthermore, stable-overexpression of DAN was shown to facilitate RA-induced neuronal differentiation [3]. As expected, activation of a variety of kinases has been implicated in neuronal differentiation processes. In one report, RA-induced neuronal differentiation was shown to be involved in the phosphatidylinositol 3-kinase (PI3K)/Akt survival signaling pathway, providing a link between cell survival and differentiation [8]. A tyrosine kinase, AATYK, is able to promote differentiation induced by RA, TPA, and insulin-like growth factor-1 [7]. More significantly, overexpression of AATYK induced differentiation without stimulation from other inducing agents [7]. A stress-associated kinase, JNK, was found to be required for RA-induced differentiation [56]. Interestingly, protein kinase C (PKC) and extracellular regulated kinase-1/2 (ERK-1/2) were demonstrated to mediate the activation of the adenosine receptor A1 and A2A, leading to neuritogenesis and differentiation in SH-SY5Y cells and primary neurons isolated from rats [5]. It is worth to mention that both adenosine receptor A1 and A2A have been demonstrated to play a neuroprotective role on adenosine during cerebral ischemia. Similarly, the neuroprotective role of Nrf2 has been increasingly reported [49–54]. Presently, there is data supporting our findings that RA activates Nrf2 and induces Nrf2-ARE binding in an in vivo mouse model and a liver cell model [57]. However, one paper suggests that toxic levels of RA reduced the binding of Nrf2 to the ARE and enhanced the formation of a complex with the retinoic acid receptor α (RARα), which inhibited the activity of Nrf2[58]. Although speculative at this time, it is plausible that the neuroprotective effects of the Nrf2-dependent pathway, activation of the adenosine receptors, the PI3K/Akt pathway, and many other signaling pathways, may have crosstalk because of their functions in promoting neuronal differentiation. The precise interplay between neuronal survival, differentiation, and apoptosis due to activation of the Nrf2 signaling pathway remains to be answered. Thus, revealing the novel role of Nrf2 in neuronal differentiation will open new perspectives for the therapeutic use of Nrf2 activators in patients with neurodegenerative diseases.
In this report, we have examined the effects of Nrf2 on cell differentiation using a neuroblastoma cell line, SH-SY5Y. RA and TPA, two well-studied inducers for neuronal differentiation, are able to induce Nrf2 and NQO1 in a dose- and time- dependent manner. Mechanistically, RA up-regulated Nrf2 by enhancing the rate of Nrf2 protein synthesis. RA-induced Nrf2 up-regulation is accompanied by neurite outgrowth and an induction of neuronal two differentiation markers, NF-M and MAP-2. Overexpression of Nrf2 in SH-SY5Y cells, by stable incorporation of Nrf2 cDNA, by transient infection of lentivirus containing Nrf2, or by chemical induction of endogenous Nrf2, promotes neuronal differentiation as measured by up-regulation of NF-M and MAP-2, as well as morphological changes. On the other hand, inhibition of endogenous Nrf2 expression, by transfection of Nrf2-siRNA or by transient infection of lentivirus containing Keap1, a negative regulator of Nrf2, inhibited neuronal differentiation. More remarkably, the positive role of Nrf2 in neuronal differentiation was verified ex vivo in primary neuron culture. Primary neurons isolated from Nrf2-null mice showed a retarded progress in differentiation, compared to that from wild-type mice. The different growing rate of neurons isolated from wild-type and Nrf2-null mice was prominently observed at day 3. However, the neuronal differentiation process most likely reached completion on day 4, since neurite outgrowth in the Nrf2-null neurons was similar to that in wild-type mice at day 4 and day 5. Collectively, our data demonstrate a novel role for Nrf2 in promoting neuronal cell differentiation, which will open new perspectives for therapeutic uses of Nrf2 activators in patients with neurodegenerative diseases. Nevertheless, further detailed evaluation of the possible morphologic differences between wild-type and Nrf2-null mice during the course of brain development will be valuable in verifying the important role of Nrf2 in promoting neuronal differentiation. Although it seems that there is no apparent developmental defect with Nrf2-null mice, there are no studies addressing the possible delay in neuronal differentiation. Furthermore, it is possible that such experiments should be performed under stress conditions to substantiate phenotypic differences between wild-type and Nrf2-null mice, because we live in an environment in which stress is inevitable.
Acknowledgments
We would like to thank Dr. Qiongman Kong for her advice in isolating primary neurons from animals. We thank Nicole Villeneuve for critical reading of the manuscript. This work was supported by research grants from NIEHS (ES015010) awarded to D. D. Zhang, American Cancer Society (RSG-07-154-CNE) awarded to D. D. Zhang, and Southwest Environmental Health Sciences Center (ES006694).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Fei Zhao, Department of Pharmacology and Toxicology, University of Arizona, Tucson, AZ, USA.
Tongde Wu, Department of Pharmacology and Toxicology, University of Arizona, Tucson, AZ, USA.
Alexandria Lau, Department of Pharmacology and Toxicology, University of Arizona, Tucson, AZ, USA.
Tao Jiang, Department of Pharmacology and Toxicology, University of Arizona, Tucson, AZ, USA.
Zheping Huang, Department of Pharmacology and Toxicology, University of Arizona, Tucson, AZ, USA.
Xiao-Jun Wang, Department of Pharmacology and Toxicology, University of Arizona, Tucson, AZ, USA.
Weimin Chen, Department of Pharmacology and Toxicology, University of Arizona, Tucson, AZ, USA.
Pak Kin Wong, Department of Aerospace and Mechanical Engineering, University of Arizona, Tucson, AZ, USA.
Donna D. Zhang, Department of Pharmacology and Toxicology, University of Arizona, Tucson, AZ, USA.
References
- 1.Encinas M, Iglesias M, Liu Y, Wang H, Muhaisen A, Cena V, et al. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J Neurochem. 2000;75:991–1003. doi: 10.1046/j.1471-4159.2000.0750991.x. [DOI] [PubMed] [Google Scholar]
- 2.Pahlman S, Hoehner JC, Nanberg E, Hedborg F, Fagerstrom S, Gestblom C, et al. Differentiation and survival influences of growth factors in human neuroblastoma. Eur J Cancer. 1995;31A:453–458. doi: 10.1016/0959-8049(95)00033-f. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura Y, Ozaki T, Ichimiya S, Nakagawara A, Sakiyama S. Ectopic expression of DAN enhances the retinoic acid-induced neuronal differentiation in human neuroblastoma cell lines. Biochem Biophys Res Commun. 1998;243:722–726. doi: 10.1006/bbrc.1998.8112. [DOI] [PubMed] [Google Scholar]
- 4.Feng Z, Porter AG. NF-kappaB/Rel proteins are required for neuronal differentiation of SH-SY5Y neuroblastoma cells. J Biol Chem. 1999;274:30341–30344. doi: 10.1074/jbc.274.43.30341. [DOI] [PubMed] [Google Scholar]
- 5.Canals M, Angulo E, Casado V, Canela EI, Mallol J, Vinals F, et al. Molecular mechanisms involved in the adenosine A and A receptor-induced neuronal differentiation in neuroblastoma cells and striatal primary cultures. J Neurochem. 2005;92:337–348. doi: 10.1111/j.1471-4159.2004.02856.x. [DOI] [PubMed] [Google Scholar]
- 6.Li BS, Zhang L, Gu J, Amin ND, Pant HC. Integrin alpha(1) beta(1)-mediated activation of cyclin-dependent kinase 5 activity is involved in neurite outgrowth and human neurofilament protein H Lys-Ser-Pro tail domain phosphorylation. J Neurosci. 2000;20:6055–6062. doi: 10.1523/JNEUROSCI.20-16-06055.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raghunath M, Patti R, Bannerman P, Lee CM, Baker S, Sutton LN, et al. A novel kinase, AATYK induces and promotes neuronal differentiation in a human neuroblastoma (SH-SY5Y) cell line. Brain Res Mol Brain Res. 2000;77:151–162. doi: 10.1016/s0169-328x(00)00048-6. [DOI] [PubMed] [Google Scholar]
- 8.Lopez-Carballo G, Moreno L, Masia S, Perez P, Barettino D. Activation of the phosphatidylinositol 3-kinase/Akt signaling pathway by retinoic acid is required for neural differentiation of SH-SY5Y human neuroblastoma cells. J Biol Chem. 2002;277:25297–25304. doi: 10.1074/jbc.M201869200. [DOI] [PubMed] [Google Scholar]
- 9.Muley PD, McNeill EM, Marzinke MA, Knobel KM, Barr MM, Clagett-Dame M. The atRA-responsive gene neuron navigator 2 functions in neurite outgrowth and axonal elongation. Dev Neurobiol. 2008;68:1441–1453. doi: 10.1002/dneu.20670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.da Silva JS, Dotti CG. Breaking the neuronal sphere: regulation of the actin cytoskeleton in neuritogenesis. Nat Rev Neurosci. 2002;3:694–704. doi: 10.1038/nrn918. [DOI] [PubMed] [Google Scholar]
- 11.Helfand BT, Mendez MG, Pugh J, Delsert C, Goldman RD. A role for intermediate filaments in determining and maintaining the shape of nerve cells. Mol Biol Cell. 2003;14:5069–5081. doi: 10.1091/mbc.E03-06-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lazarides E. Intermediate filaments: a chemically heterogeneous, developmentally regulated class of proteins. Annu Rev Biochem. 1982;51:219–250. doi: 10.1146/annurev.bi.51.070182.001251. [DOI] [PubMed] [Google Scholar]
- 13.Carden MJ, Trojanowski JQ, Schlaepfer WW, Lee VM. Two-stage expression of neurofilament polypeptides during rat neurogenesis with early establishment of adult phosphorylation patterns. J Neurosci. 1987;7:3489–3504. doi: 10.1523/JNEUROSCI.07-11-03489.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma M, Sharma P, Pant HC. CDK-5-mediated neurofilament phosphorylation in SHSY5Y human neuroblastoma cells. J Neurochem. 1999;73:79–86. doi: 10.1046/j.1471-4159.1999.0730079.x. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein ME, Sternberger NH, Sternberger LA. Phosphorylation protects neurofilaments against proteolysis. J Neuroimmunol. 1987;14:149–160. doi: 10.1016/0165-5728(87)90049-x. [DOI] [PubMed] [Google Scholar]
- 16.Pant HC. Dephosphorylation of neurofilament proteins enhances their susceptibility to degradation by calpain. Biochem J. 1988;256:665–668. doi: 10.1042/bj2560665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Markus A, Patel TD, Snider WD. Neurotrophic factors and axonal growth. Curr Opin Neurobiol. 2002;12:523–531. doi: 10.1016/s0959-4388(02)00372-0. [DOI] [PubMed] [Google Scholar]
- 18.Heraud C, Hilairet S, Muller JM, Leterrier JF, Chadeneau C. Neuritogenesis induced by vasoactive intestinal peptide, pituitary adenylate cyclase-activating polypeptide, and peptide histidine methionine in SH-SY5y cells is associated with regulated expression of cytoskeleton mRNAs and proteins. J Neurosci Res. 2004;75:320–329. doi: 10.1002/jnr.10866. [DOI] [PubMed] [Google Scholar]
- 19.Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev. 2006;38:769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi M, Yamamoto M. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid Redox Signal. 2005;7:385–394. doi: 10.1089/ars.2005.7.385. [DOI] [PubMed] [Google Scholar]
- 21.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 22.Osburn WO, Kensler TW. Nrf2 signaling: an adaptive response pathway for protection against environmental toxic insults. Mutat Res. 2008;659:31–39. doi: 10.1016/j.mrrev.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Surh YJ, Kundu JK, Na HK. Nrf2 as a master redox switch in turning on the cellular signaling involved in the induction of cytoprotective genes by some chemopreventive phytochemicals. Planta Med. 2008;74:1526–1539. doi: 10.1055/s-0028-1088302. [DOI] [PubMed] [Google Scholar]
- 24.Owuor ED, Kong AN. Antioxidants and oxidants regulated signal transduction pathways. Biochem Pharmacol. 2002;64:765–770. doi: 10.1016/s0006-2952(02)01137-1. [DOI] [PubMed] [Google Scholar]
- 25.Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD. Dual roles of Nrf2 in cancer. Pharmacol Res. 2008;58:262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishii T, Itoh K, Takahashi S, Sato H, Yanagawa T, Katoh Y, et al. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J Biol Chem. 2000;275:16023–16029. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- 27.Moinova HR, Mulcahy RT. Up-regulation of the human gamma-glutamylcysteine synthetase regulatory subunit gene involves binding of Nrf-2 to an electrophile responsive element. Biochem Biophys Res Commun. 1999;261:661–668. doi: 10.1006/bbrc.1999.1109. [DOI] [PubMed] [Google Scholar]
- 28.Banning A, Deubel S, Kluth D, Zhou Z, Brigelius-Flohe R. The GI-GPx gene is a target for Nrf2. Mol Cell Biol. 2005;25:4914–4923. doi: 10.1128/MCB.25.12.4914-4923.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim YC, Masutani H, Yamaguchi Y, Itoh K, Yamamoto M, Yodoi J. Hemin-induced activation of the thioredoxin gene by Nrf2. A differential regulation of the antioxidant responsive element by a switch of its binding factors. J Biol Chem. 2001;276:18399–18406. doi: 10.1074/jbc.M100103200. [DOI] [PubMed] [Google Scholar]
- 30.Sakurai A, Nishimoto M, Himeno S, Imura N, Tsujimoto M, Kunimoto M, et al. Transcriptional regulation of thioredoxin reductase 1 expression by cadmium in vascular endothelial cells: role of NF-E2-related factor-2. J Cell Physiol. 2005;203:529–537. doi: 10.1002/jcp.20246. [DOI] [PubMed] [Google Scholar]
- 31.Vollrath V, Wielandt AM, Iruretagoyena M, Chianale J. Role of Nrf2 in the regulation of the Mrp2 (ABCC2) gene. Biochem J. 2006;395:599–609. doi: 10.1042/BJ20051518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maher JM, Cheng X, Slitt AL, Dieter MZ, Klaassen CD. Induction of the multidrug resistance-associated protein family of transporters by chemical activators of receptor-mediated pathways in mouse liver. Drug Metab Dispos. 2005;33:956–962. doi: 10.1124/dmd.105.003798. [DOI] [PubMed] [Google Scholar]
- 33.Ishii T, Yanagawa T. Stress-induced peroxiredoxins. Subcell Biochem. 2007;44:375–384. doi: 10.1007/978-1-4020-6051-9_18. [DOI] [PubMed] [Google Scholar]
- 34.Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 35.Wasserman WW, Fahl WE. Functional antioxidant responsive elements. Proc Natl Acad Sci U S A. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem J. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a "tethering" mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem. 2006;281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- 39.Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, Yokoyama S, et al. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol. 2007;27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang XJ, Sun Z, Chen W, Eblin KE, Gandolfi JA, Zhang DD. Nrf2 protects human bladder urothelial cells from arsenite and monomethylarsonous acid toxicity. Toxicol Appl Pharmacol. 2007;225:206–213. doi: 10.1016/j.taap.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W, Kong AN. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol Carcinog. 2009;48:91–104. doi: 10.1002/mc.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aoki Y, Sato H, Nishimura N, Takahashi S, Itoh K, Yamamoto M. Accelerated DNA adduct formation in the lung of the Nrf2 knockout mouse exposed to diesel exhaust. Toxicol Appl Pharmacol. 2001;173:154–160. doi: 10.1006/taap.2001.9176. [DOI] [PubMed] [Google Scholar]
- 44.Iida K, Itoh K, Kumagai Y, Oyasu R, Hattori K, Kawai K, et al. Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis. Cancer Res. 2004;64:6424–6431. doi: 10.1158/0008-5472.CAN-04-1906. [DOI] [PubMed] [Google Scholar]
- 45.Chan K, Han XD, Kan YW. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc Natl Acad Sci U S A. 2001;98:4611–4616. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho HY, Reddy SP, Yamamoto M, Kleeberger SR. The transcription factor NRF2 protects against pulmonary fibrosis. Faseb J. 2004;18:1258–1260. doi: 10.1096/fj.03-1127fje. [DOI] [PubMed] [Google Scholar]
- 47.Chan K, Kan YW. Nrf2 is essential for protection against acute pulmonary injury in mice. Proc Natl Acad Sci U S A. 1999;96:12731–12736. doi: 10.1073/pnas.96.22.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, Kensler TW, et al. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J Clin Invest. 2004;114:1248–1259. doi: 10.1172/JCI21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siebert A, Desai V, Chandrasekaran K, Fiskum G, Jafri MS. Nrf2 activators provide neuroprotection against 6-hydroxydopamine toxicity in rat organotypic nigrostriatal cocultures. J Neurosci Res. 2009;87:1659–1669. doi: 10.1002/jnr.21975. [DOI] [PubMed] [Google Scholar]
- 50.Johnson JA, Johnson DA, Kraft AD, Calkins MJ, Jakel RJ, Vargas MR, et al. The Nrf2-ARE pathway: an indicator and modulator of oxidative stress in neurodegeneration. Ann N Y Acad Sci. 2008;1147:61–69. doi: 10.1196/annals.1427.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clark J, Simon DK. Transcribe to Survive: transcriptional control of antioxidant defense programs for neuroprotection in Parkinson's disease. Antioxid Redox Signal. 2009;11:509–528. doi: 10.1089/ars.2008.2241. [DOI] [PubMed] [Google Scholar]
- 52.Calkins MJ, Johnson DA, Townsend JA, Vargas MR, Dowell JA, Williamson TP, et al. The Nrf2/ARE pathway as a potential therapeutic target in neurodegenerative disease. Antioxid Redox Signal. 2009;11:497–508. doi: 10.1089/ars.2008.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Satoh T, Okamoto SI, Cui J, Watanabe Y, Furuta K, Suzuki M, et al. Activation of the Keap1/Nrf2 pathway for neuroprotection by electrophilic [correction of electrophillic] phase II inducers. Proc Natl Acad Sci U S A. 2006;103:768–773. doi: 10.1073/pnas.0505723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shih AY, Imbeault S, Barakauskas V, Erb H, Jiang L, Li P, et al. Induction of the Nrf2-driven antioxidant response confers neuroprotection during mitochondrial stress in vivo. J Biol Chem. 2005;280:22925–22936. doi: 10.1074/jbc.M414635200. [DOI] [PubMed] [Google Scholar]
- 55.Sun Z, Zhang S, Chan JY, Zhang DD. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol Cell Biol. 2007;27:6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu YM, Han PL, Lee JK. JNK pathway is required for retinoic acid-induced neurite outgrowth of human neuroblastoma, SH-SY5Y. Neuroreport. 2003;14:941–945. doi: 10.1097/01.wnr.0000074341.81633.b8. [DOI] [PubMed] [Google Scholar]
- 57.Tan KP, Kosuge K, Yang M, Ito S. NRF2 as a determinant of cellular resistance in retinoic acid cytotoxicity. Free Radic Biol Med. 2008;45:1663–1673. doi: 10.1016/j.freeradbiomed.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 58.Wang XJ, Hayes JD, Henderson CJ, Wolf CR. Identification of retinoic acid as an inhibitor of transcription factor Nrf2 through activation of retinoic acid receptor alpha. Proc Natl Acad Sci U S A. 2007;104:19589–19594. doi: 10.1073/pnas.0709483104. [DOI] [PMC free article] [PubMed] [Google Scholar]