Abstract
Stem cells have great potential as cell sources for regenerative medicine due to both their self-renewal and multi-lineage differentiation capacity. Despite advances in the field of stem cell biology, major challenges remain before stem cells can be widely used for therapeutic purposes. One challenge is to develop reproducible methods to control stem cell growth and differentiation. The niche in which stem cells reside is a complex, multi-factorial environment. In contrast to using cells alone, biomaterials can provide initial structural support, and allow cells to adhere, proliferate and differentiate in a three-dimensional environment. Researchers have incorporated signals into the biomaterials that can promote desired cell functions in a spatially and temporally controlled manner. Despite progress in biomaterial design and methods to modulate cellular behavior, many of the complex signal networks that regulate cell-material interactions remain unclear. Due to the vast numbers of material properties to be explored and the complexity of cell-surface interactions, it is often difficult to optimize stem cell microenvironments using conventional, iterative approaches. To address these challenges, high throughput screening of combinatorial libraries has emerged as a novel approach to achieve rapid screening with reduced materials and costs. In this review, we discuss recent research in the area of high throughput approaches for characterization and optimization of cellular interactions with their microenvironments. In contrast to conventional approaches, screening combinatorial libraries can result in the discovery of unexpected material solutions to these complex problems.
INTRODUCTION
Stem cells have great potential as cell sources for regenerative medicine due to both their self-renewal and multi-lineage differentiation capacity. In general, stem cells can be divided into three categories: adult stem cells, embryonic stem cells (ES) and embryonic germ cells (EG) [1-3]. Adult stem cells are multipotent, and have been identified in many tissue types including bone marrow [1], adipose tissue [4], nervous tissue [5, 6], muscle [7], and umbilical cord blood [8]. However, adult stem cells can only be expanded for limited passages. In contrast, ES or EG cells can self-renew without differentiation for much longer and possess the potential to differentiate into any of the three germ layers [2, 3]. This makes them attractive candidates as cell sources for tissue engineering, where the often large cell number can be a challenge.
Despite advances in the field, major challenges remain before stem cells can be widely used for therapeutic purposes. One challenge is to develop reproducible methods to control stem cell growth and differentiation. Over the past decade, extensive research has been dedicated towards elucidating the molecular mechanisms that govern stem cell differentiation [9, 10]. Increasing evidence has shown that the interplay between stem cells and their surrounding microenvironments is critical [11]. The niche in which stem cells reside is a complex, multi-factorial environment. Stem cell phenotype is determined by numerous cues including cell-cell interactions, cell-extracellular matrix interactions, small molecules, soluble and insoluble growth factors and mechanical forces [12].
One area of particular interest for stem cell use is tissue engineering, where a significant number of cells is typically required. In tissue engineering, biomaterials play a key role by providing initial structural support, and allowing cells to adhere, proliferate and in the case of stem cells, differentiate in a three-dimensional environment. Early work in this area used homogenous scaffolds to provide a biodegradable environment for cell growth. More recently, researchers have incorporated signals directly into the biomaterials that can promote desired cell functions in a spatially and temporally controlled manner [13, 14]. These signals have been shown to modulate a variety of cellular functions such as cell adhesion, migration and differentiation [15-19]. Despite progress in biomaterial design and methods to modulate cellular behavior, many of the complex signal networks that regulate cell-material interactions remain unclear. Conventional approaches evaluate a few interactions at a time, and often require large number of cells/materials. To address these limitations, high throughput screening of combinatorial libraries has emerged as a novel approach to achieve rapid screening with reduced materials and costs [20, 21]. In this review, we discuss recent research in the area of high throughput approaches for characterization and optimization of cellular interactions with their microenvironments.
ASSESSING CELL-MATERIAL INTERACTIONS USING COMBINATORIAL POLYMER ARRAYS
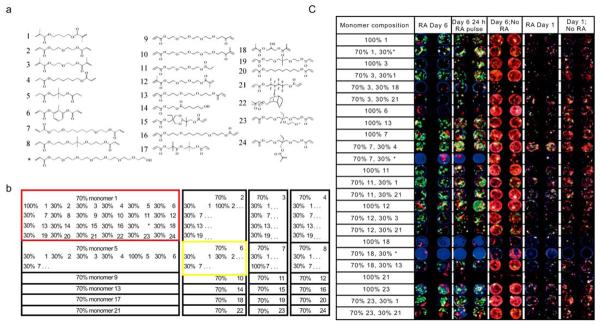
Optimizing biomaterial effects on cell behavior remains challenging. The surface properties of biomaterials play an important role in controlling cellular behaviors, and investigations of the interactions between stem cells and biomaterials have received significant attention [22]. However, due to the vast numbers of material properties to be explored and the complexity of cell-surface interactions, it is often difficult to optimize polymer surface chemistry using conventional, iterative approaches. To address these challenges, combinatorial methods have been developed to screen cell responses to material surfaces. Anderson et al. described a nanoliter-scale synthesis platform of biomaterial libraries in an array format by combining a robotic liquid handling system with photopolymerization technology [23] (Fig. 1). To generate a diverse biomaterial library, 24 photopolymerizable monomers that vary in chain length, polarity and branching were mixed in a multi-well format. Using this method, more than 1,700 cellular-materials interactions were characterized simultaneously on a conventional glass slide, and their effects on human embryonic stem cell growth and differentiations were examined using fluorescence microscopy. A number of unexpected cell-material interactions were identified, including materials that facilitate high levels of the differentiation of human embryonic stem cells into epithelial cells.
Fig. (1).
Biomaterial microarray design. (a) Monomers used for microarray synthesis. (b) Monomers were mixed at a 70:30 ratio pairwise in all possible combinations with the exception of monomer 17, which was substituted with * to increase polymer hydrophilicity. To facilitate analysis, all 24 polymers composed of 70% of a particular monomer were printed as a 6*4 group on the array, as highlighted by the red and yellow boxes. Three blocks of 576 polymers were printed on each side, with a center to center spacing of 740 μm. (c) “Hit” polymer effects on hES cell attachment, growth and proliferation. Four million hES cell embryoid body day-6 cells were grown on the “hit” polymer arrays in the presence of retinoic acid, the absence of retinoic acid and with a 24-h pulse of the retinoic acid for 1 or 6 d. Cells were then stained for cytokeratin 7 (green), vimentin (red) and DNA (blue). Representative images at each time point for each polymer are shown for all conditions. RA, retinoic acid. (Figure from reference [23] with permission).
In a later study, this technology was expanded to develop polymer microarrays containing 3456 individual polymer composites. A number of commonly used medical biomaterials such as polyesters were tested. In particular, different forms of poly (lactide-co-glycolide) (PLGA), a clinically approved biodegradable material were examined [24]. As proof of principle, the effects of the polymer properties on a variety of cell types including with human mesenchymal stem cells, neural stem cells, and primary articular chondrocytes were simultaneously characterized using these arrays.
COMBINATORIAL SCREENING OF CELL-MATERIAL INTERACTIONS USING GRADIENT LIBRARIES
Materials in a microarray are presented to cells as distinct material regions with boundaries. In contrast, gradient techniques produce surfaces with a position-dependent and gradually changing chemistry. Various gradient substrates have been generated to explore biological response to different physical, chemical cues, and they can provide quantitative information about a cell's precise point of transition along a gradually-changing surface [25-29]. For example, Mei and coworkers prepared gradient samples with a tenfold difference in graft density of poly(2-hydroxyethyl methacrylate) (pHEMA) along the surface, pre-exposed to bovine fibronectin, and induced a significant change of fibroblast cell spreading on the surface [27]. Maximal cell adhesion and cell spreading were found to occur at fibronectin surface densities of 50 ng/cm2 and 100 ng/cm2, respectively. These results demonstrate that the gradient methodology is a valuable tool for exploring biological response to materials systems in a single, well-controlled experiment. In a similar study, Bhat and coworkers generated 2D gradient substrates, in which both the molecular weight and graft density of pHEMA vary along two orthogonal directions on the specimen, to study the cell adhesion in a combinatorial fashion [25]. In addition to protein, the bioactive peptides such as RGD have also been incorporated into substrates in a gradient fashion [26, 28].
High throughput methods have also been applied to analyze cellular response to crystallinity in polymer materials [30]. Variations in crystallinity lead to changes in surface roughness on nanometer length scales. Gradients of polymer crystallinity were fabricated on films of poly(L-lactic acid). The use of these gradient libraries revealed that cell proliferation can be effected by nanometer-scale roughness. Zapata and coworkers expanded the combinatorial screening to measure quantitatively the osteoblast attachment, spreading, and proliferation on the polymer surface with various features [29]. Their findings demonstrated that cell attachment was favored on the more hydrophilic poly(D,L-lactide) domains while cell spreading was strongly influenced by phase-separated microstructures on the polymer surfaces.
Most high throughput biomaterials work conducted to date has been limited to the assessment of the interactions between cells and material surfaces in a two-dimensional environment. However, cells in vivo reside in three-dimensional structures composed of extracellular matrix proteins, and cells behavior in a three-dimensional scaffold can vary significantly from that in two-dimensions [31-33]. In a recent study, Simon and coworkers generated 3D PDLLA scaffold libraries with syringe pumps and a static mixer [34]. The presence of the gradients in the libraries was confirmed with the continuously color change from the Sudan IV dye along the scaffold. This work demonstrates the potential of performing screening with combinatorial three-dimensional scaffold libraries.
COMBINATORIAL EXTRACELLULAR MATRIX MICROARRAYS FOR EVALUATING CELL-ECM INTERACTIONS
In addition to synthetic materials, proteins represent another source of signaling materials that the cells would frequently encounter in vivo. MacBeath and coworkers developed high throughput synthesis and screening of proteins microarrays [35]. A high-precision contact printing robot, originally designed to manufacture DNA microarrays, was used to spot nanoliter volumes of proteins onto chemically modified glass slides, yielding spots about 150 to 200 μm in diameter, with 1600 spots per square centimeter. Using this technology, the proteins attached covalently to the slide surface yet retained their ability to specifically interact with other proteins or small molecules.
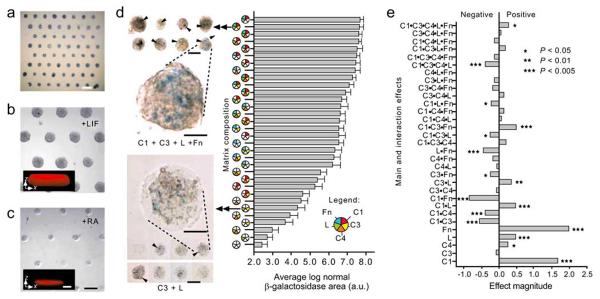
Differentiation of stem cells in vivo is regulated by a variety of cues including insoluble ECM proteins and soluble growth factors. Recently, Flaim and coworkers developed a robotic method to generate the ECM protein microarrays [36] (Fig. 2). In this study, 32 different combinations of natural ECM proteins such as collagen, laminin and fibronectin were deposited on a hydrogel slide, and their effects on mouse embryonic stem cells and hepatocytes were studied. Interestingly, the optimized combinations of ECM proteins that can impact both hepatocyte function and ES cell differentiation were identified. The roles of each ECM protein in the combinations were also elucidated. For example, collagen IV was shown to increase the level of intracellular albumin, a marker of liver-specific function, while collagen III and laminin resulted in decreased albumin levels.
Fig. (2).
Mouse ES cells differentiate on ECM microarrays. (a) Bright-field alkaline phosphatase staining of day 1 ES cultures on ECM microarrays in 15% serum medium (scale bar, 1 mm). (b, c) Phase-contrast images of day 3 arrays cultured with LIF (b) and with RA (c). Cells cultured with LIF showed three-dimensional features (in b, inset x-z confocal section, ~77 μm thickness). In contrast, RA-induced cells grew as a relatively thin sheet (in c, inset x-z section, ~25 μm thickness). Scale bars, 250 μm (insert scale bars, 50 μm). (d) Bright-field micro-graph of selected X-gal stained ECM microarray conditions after 3 d of culture in RA. C1 + C3 + L +Fn (top left images) induced higher Ankrd17 reporter activity (arrowheads) than was seen in cells cultured on C3+L (bottom left images). Scale bars, 250 μm. Magnified views of reporter activity: scale bars, 50 μm. Bar graph: hierarchical depiction of ‘blue’ image area (pooled data from four microarrays) for each of the matrix mixtures. Error bars, s.e.m. (n=32). The C1 + C3 + L + Fn culture condition induced ~27-fold more β-galactosidase image area than the C3 + L cultures. (e) Results of 25 full factorial analysis on β-galactosidase positive ‘blue’ image area (four microarray data sets). The relative magnitude of main effects as well as 2-, 3-, 4- and 5-factor interactions are shown. C1, collagen I; C3, collagen III; C4, collagen IV; L, Laminin; Fn, fibronectin. (Figure from reference [36] with permission).
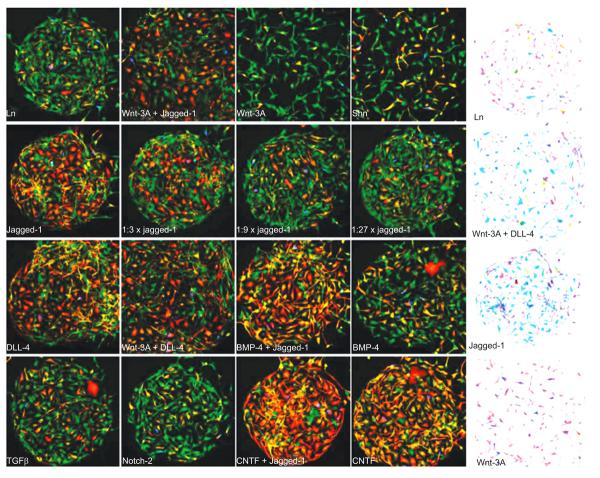
High throughput approaches also allow the analysis of responses to complex signals. For example, Soen and coworkers generated libraries of the ECM components and growth factors to screen their combinatorial effects on the stem cells differentiations [37] (Fig. 3). Using a piezoelectric arrayer, the pre-mixed combinations of signaling molecules including growth factors, ECM components, cell adhesion molecules, and morphogens were printed onto arrays. Primary bipotent human neural precursor cells were subsequently cultured on the arrays. Proliferation and differentiation of the neural progenitor cells was examined with immunostaining, followed by a quantitative image analysis. Co-stimulation by Wnt and Notch was shown to maintain the cells in an undifferentiated-like state, whereas bone morpho-genetic protein 4 induced an indeterminate differentiation phenotype, characterized by simultaneous expression of glial and neuronal markers.
Fig. (3).
Microenvironment-dependent differentiation and morphology. Human neural precursors were captured and cultured on a printed Ln/ligand array for 70 h under differentiation-promoting conditions. Following the differentiation period, the cells were fixed and counter-stained with GFAP (red), BrdU (blue), TUJ1 (green), and DAPI (not shown). (A) A small portion of the array with 16 different microenvironments each containing a few hundred cells. The balance between TUJ1 and GFAP staining on the reference Ln spot (top left) was skewed toward preferential expression of the neuronal marker TUJ1. This balance was shifted in a spot-dependent manner by some of the signal-containing spots. In particular, spots containing CNTF (bottom right) and Notch ligands (right panels on the 2nd and 3rd rows) led to a dramatic shift toward increased GFAP proportions, suggesting a gliogenic response to Notch stiumulation. Dilution series of Jagged-1 (2nd row panels) revealed dose-dependent response to Notch stimulation. Combination of some gliogenic signals (e.g. Jagged-1 and CNTF) led to further increase in the gliogenic response. A smaller shift toward increased neuronal proportions was observed on Wnt-3A spots. (B) Color inverted images demonstrating spot-dependent morphological differences. Cells that were exposed to a combination of Wnt-3A and a Notch ligand (second spot from the top) exhibited stronger and more elaborated processes compared to Ln alone (top). Typical spot diameter was 400 μm. Fields of view in all panels are identical in size. Wnt-3A-containing spots consistently larger (Figure from reference [37] with permission).
In an attempt to design biomaterials that can promote the expansion or differentiation of neural stem cells, Nakajima and coworkers employed an alkanethiol self-assembled monolayer (SAM) to screen extracellular matrix components and growth factors [38]. Using photo-assisted patterning technology, a variety of growth factors known to have neurotrophic activities were arrayed with natural and synthetic matrices in a combinatorial manner. The ECM components that can support the neural stem cell attachment and growth factors that can promote the directed differentiations to specific lineage were identified. Notably, their results showed that the effects of growth factor signaling were influenced by the incorporated matrix proteins, suggesting the crosstalk between ECM molecules and growth factor receptors.
CELL PATTERNING CHIP FOR CONTROLLING CELL-CELL INTERACTIONS
Cell-cell signaling is an important component of the stem cell microenvironment, affecting both differentiation and self-renewal. To facilitate microenvironment optimization, Rosenthal et al. created the Bio Flip Chip (BFC), a microfabricated polymer chip containing thousands of microwells, each sized to trap down to a single stem cell [39]. This technology also allowed the patterning of small groups of cells, with and without cell-cell contact, allowing incremental and independent control of contact-mediated signaling. Using this platform, cell-cell contact was shown to play an important role in depressing mESC colony formation, and Ecadherin was found to be involved in this negative regulatory pathway.
HIGH THROUGHPUT SCREENING OF CELLULAR RESPONSES TO A SMALL MOLECULE LIBRARY
Small molecules play an important role in regulating cell signaling pathways, and screening stem cell responses to small molecules library offers a powerful tool to better understand and control the processes that regulate stem cell fate. Ding et al. developed a high throughput phenotypic cell-based screen of kinase-directed combinatorial libraries and identified TWS119, a 4,6-disubstituted pyrrolopyrimidine that can induce neurogenesis in murine ESCs [40]. In another related study, small molecules were identified that can differentiate ES cells down cardiomyocyte lineages [41]. Using similar approach, chen et al. discovered molecules that can induce the dedifferentiation of myogenic lineage-committed cells back to multipotent mesenchymal progenitor cells that can re-differentiate into bone and fat cells [42]. This approach is likely to provide insights into the molecular mechanisms that control stem cell fate, and may ultimately be useful to in vivo stem cell biology and therapy.
In addition to the microtiter-based screening approach as mentioned above, small molecule libraries can also be presented in a microarray format, which is compatible with image-based screens and requires only small cell number [43]. Bailey et al. developed a method where small molecules can be impregnated in a microarray of of poly-(D),(L)-lactide/glycolide polymer and compounds would diffuse out to affect cells seeded on the scaffolds. This study represents an early attempt in applying microfabrication approach for small molecule screening.
Among the small molecules, RNA interference (RNAi) has become one of the most important research tools in functional genomics analysis since the discovery of the phenomenon. Large-scale screens of RNAi libraries have been constructed and initial screening efforts have demonstrated that these libraries and methods are practical and powerful tools for high throughput lentivirus RNAi screens [44]. Bailey and coworkers develop a lentivirus-infected cell microarray (LICM), whose features include clusters of mammalian cells, each transduced with a single type of lentivirus [45]. These microarrays enabled robust and localized infection of mammalian cells with lentiviruses containing overexpression cassettes or RNAi cassettes. This technology provides a platform for loss-of-function and overexpression screens in a broad range of mammalian cell types. Such screening is very useful for establishing the functional links between gene and phenotypes, thus offers valuable insights on dissecting the biology of cellular pathways. For a comprehensive discussion on available RNAi library resources and application of these strategic tools, see review by Chen et al [46].
PARALLEL SYNTHESIS AND SCREENING OF LARGE POLYMER LIBRARIES FOR GENE DELIVERY
Biomaterials can not only influence cell fate and function through extracellular interactions, but can also directly effect cell behavior through genetic regulation. For various tissue engineering applications, a target gene can be transferred to specific cell types, such as stem cells, to promote the desired cell differentiation and tissue formation [47]. There are two general techniques for delivering genetic materials into mammalian cells: viral and non-viral. The viral approach employs the naturally evolved mechanism for viral self-replication, and is highly efficient. However, viruses are inherently immunogenic and potentially pathogenic, and safety concerns have always been a major issue for the clinical applications of viral-based gene delivery. Furthermore, viral vectors do not facilitate the design of target cell specificity and are associated with relatively high manufacturing costs.
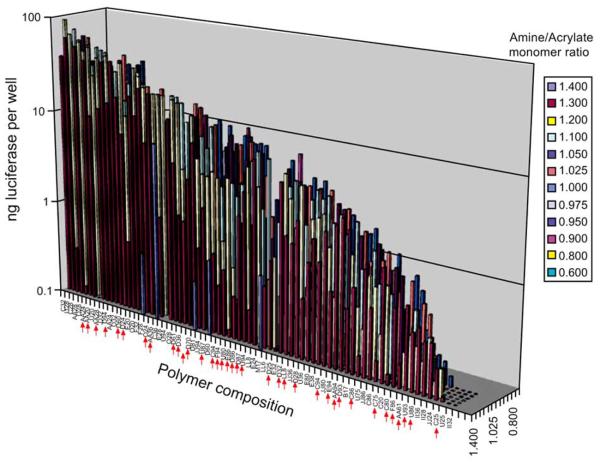
Non-viral synthetic vectors, on the other hand, overcome the problems associated with a viral-based approach and can be non-immunogenic. Many synthetic vectors are cationic materials that can electrostatically bind to DNA or RNA to form condensed nanoparticles (polyplexes or lipoplexes). Biomaterials that have been explored include cationic polymers, cationic lipids, liposomes, chitosans, dendrimers and inorganic nanoparticles nanoparticles [48-50]. These synthetic vectors also enable greater flexibility in structure design and integrating a targeting moiety, as well as relatively easy synthesis and lower manufacturing costs. However, non-viral based vectors have suffered from low transfection efficiency and occasional toxicity, and most synthetic vectors are unstable in the presence of serum, thus severely hindering their applications in vivo. High throughput synthesis and screening of a large polymer library using combinatorial methods enables faster discovery of potential polymer vectors, better understanding of the structure/property relationship, and rational design of novel polymers for gene delivery. Anderson et al. reported a semi-automated, solution-phase parallel synthesis and screening of a large library of 2350 structurally diverse, degradable poly(beta-amino esters) using commercially available monomers [51]. High throughput screening discovered 47 polymers that demonstrate better transfection efficiency than PEI, the best commercially available polymer transfection reagent [51]. Further structure/property analyses demonstrated structural similarity in the top-performing polymers, which are all formed from amino alcohols. Such structural convergence offers valuable insight on rational design of polymer vectors for gene delivery [52] (Fig. 4). The polymer with the highest transfection efficiency, C32, is an aminopentanol-terminated polymer with a molecular weight around 18kd relative to polystyrene standards. When injected intratumorally in vivo in mice, the C32 polymer demonstrates high biocompatibility and significantly reduces tumor size, a property that is attributable to cell apoptosis [53, 54]. The top-performing poly(β-amino esters) also showed great efficacy and low cytotoxicity in transfecting primary human vascular endothelial cells (HUVEC) in the presence of serum, which have been a great challenge [55].
Fig. (4).
Transfection proficiency of the polymer library. COS-7 cells were transfected with polymer/DNA complexes at six polymer/DNA ratios in quadruplicates: 10:1, 20:1, 30:1, 40:1, 60:1 and 100:1. The average transfection level in nanograms of luciferase per well of the optimal polymer/DNA ratio is presented. Polymers are organized from highest transfecting (C32) to lowest transfecting (II32) and amine/acrylate ratio from high (front) to low (back). Red arrows indicate polymers made with DMSO as described under experimental procedures (Figure from reference [52] with permission).
Amine-terminated poly(β-amino esters) have been shown to be generally more efficient in transfection efficiency. To examine further the effect of the type of amine group at the end chain on gene delivery, a generalized method has been presented to modify poly(β-amino esters) without the need for purification [56]. This system enables the rapid synthesis and screening of many structural variations at the polymer chain terminus. End modification of C32 significantly enhances its in vitro transfection efficiency. Most notably, the end-modification strategy has led to the discovery of many effective polymers that work very well in the presence of serum, which overcomes a great obstacle in using non-viral vectors for gene delivery. In vivo, intraperitoneal (IP) gene delivery using end-modified C32 polymers leads to expression levels over one order of magnitude higher than the levels attained by using unmodified C32.
CONCLUSIONS
Over the past decade, there has been remarkable progress in our understanding of some factors and signaling pathways involved in regulating stem cell behavior. Despite these advances, developing methods of controlling stem cell behavior has proven challenging. As discussed in this review, high throughput approaches have been developed to study cellular responses to complex, multi-factorial environments. In contrast to conventional approaches, screening combinatorial libraries can result in the discovery of unexpected material solutions to these complex problems. Advances in microfabrication technology have been a driving force behind the platform development for high throughput screening, and we believe further advances in the micro/nanofabrication technology will continue to push the capabilities of high throughput biomaterial screening. Finally, most high throughput studies on cell-material interactions to date have been performed on two-dimensional environments. We expect future studies using three-dimensional microenvironments will lead to new discoveries, and will further develop our understanding of the how the stem cell microenvironment can control cell behavior.
REFERENCES
- 1.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 2.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 3.Shamblott MJ, Axelman J, Wang S, Bugg EM, Littlefield JW, Donovan PJ, Blumenthal PD, Huggins GR, Gearhart JD. Derivation of pluripotent stem cells from cultured human primordial germ cells. Proc. Natl. Acad. Sci. USA. 1998;95(23):13726–31. doi: 10.1073/pnas.95.23.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, Benhaim P, Lorenz HP, Hedrick MH. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7(2):211–28. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 5.Song HJ, Stevens CF, Gage FH. Neural stem cells from adult hippocampus develop essential properties of functional CNS neurons. Nat. Neurosci. 2002;5(5):438–45. doi: 10.1038/nn844. [DOI] [PubMed] [Google Scholar]
- 6.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255(5052):1707–10. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 7.Jankowski RJ, Deasy BM, Huard J. Muscle-derived stem cells. Gene Ther. 2002;9(10):642–7. doi: 10.1038/sj.gt.3301719. [DOI] [PubMed] [Google Scholar]
- 8.Broxmeyer HE, Douglas GW, Hangoc G, Cooper S, Bard J, English D, Arny M, Thomas L, Boyse EA. Human umbilical cord blood as a potential source of transplantable hematopoietic stem/progenitor cells. Proc. Natl. Acad. Sci. USA. 1989;86(10):3828–32. doi: 10.1073/pnas.86.10.3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyer LA, Mathur D, Jaenisch R. Molecular control of pluripotency. Curr. Opin. Genet. Dev. 2006;16(5):455–62. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Eckfeldt CE, Mendenhall EM, Verfaillie CM. The molecular repertoire of the ‘almighty’ stem cell. Nat. Rev. Mol. Cell. Biol. 2005;6(9):726–37. doi: 10.1038/nrm1713. [DOI] [PubMed] [Google Scholar]
- 11.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441(7097):1075–9. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 12.Metallo CM, Mohr JC, Detzel CJ, Pablo JJ, Wie BJ, Palecek SP. Engineering the stem cell microenvironment. Biotechnol Prog. 2007;23(1):18–23. doi: 10.1021/bp060350a. [DOI] [PubMed] [Google Scholar]
- 13.Matsumoto T, Mooney DJ. Cell instructive polymers. Adv. Biochem. Eng. Biotechnol. 2006;102:113–37. doi: 10.1007/b137207. [DOI] [PubMed] [Google Scholar]
- 14.Chen RR, Silva EA, Yuen WW, Mooney DJ. Spatio-temporal VEGF and PDGF delivery patterns blood vessel formation and maturation. Pharm. Res. 2007;24(2):258–64. doi: 10.1007/s11095-006-9173-4. [DOI] [PubMed] [Google Scholar]
- 15.Pratt AB, Weber FE, Schmoekel HG, Müller R, Hubbell JA. Synthetic extracellular matrices for in situ tissue engineering. Biotechnol. Bioeng. 2004;86(1):27–36. doi: 10.1002/bit.10897. [DOI] [PubMed] [Google Scholar]
- 16.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat. Biotechnol. 2005;23(1):47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 17.Wang DA, Williams CG, Yang F, Cher N, Lee H, Elisseeff JH. Bioresponsive phosphoester hydrogels for bone tissue engineering. Tissue Eng. 2005;11(12):201–13. doi: 10.1089/ten.2005.11.201. [DOI] [PubMed] [Google Scholar]
- 18.Yang F, Williams CG, Wang DA, Lee H, Manson PN, Elisseeff J. The effect of incorporating RGD adhesive peptide in polyethylene glycol diacrylate hydrogel on osteogenesis of bone marrow stromal cells. Biomaterials. 2005;26(30):5991–8. doi: 10.1016/j.biomaterials.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 19.Elisseeff J, Ferran A, Hwang S, Varghese S, Zhang Z. The role of biomaterials in stem cell differentiation: applications in the musculoskeletal system. Stem. Cells. Dev. 2006;15(3):295–303. doi: 10.1089/scd.2006.15.295. [DOI] [PubMed] [Google Scholar]
- 20.Kohn J. New approaches to biomaterials design. Nat. Mater. 2004;3(11):745–747. doi: 10.1038/nmat1249. [DOI] [PubMed] [Google Scholar]
- 21.Anderson DG, Burdick JA, Langer R. Materials science. Smart biomaterials. Science. 2004;305(5692):1923–4. doi: 10.1126/science.1099987. [DOI] [PubMed] [Google Scholar]
- 22.Spradling A, Drummond-Barbosa D, Kai T. Stem cells find their niche. Nature. 2001;414(6859):98–104. doi: 10.1038/35102160. [DOI] [PubMed] [Google Scholar]
- 23.Anderson DG, Levenberg S, Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat. Biotechnol. 2004;22(7):863–866. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 24.Anderson DG, Putnam D, Lavik EB, Mahmood TA, Langer R. Biomaterial microarrays: rapid, microscale screening of polymer-cell interaction. Biomaterials. 2005;26(23):4892–4897. doi: 10.1016/j.biomaterials.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 25.Bhat RR, Chaney BN, Rowley J, Liebmann-Vinson A, Genzer J. Tailoring cell adhesion using surface-grafted polymer gradient assemblies. Adv. Mater. 2005;17(23):2802–2807. [Google Scholar]
- 26.Harris BP, Kutty JK, Fritz EW, Webb CK, Burg KJ, Metters AT. Photopatterned polymer brushes promoting cell adhesion gradients. Langmuir. 2006;22(10):4467–71. doi: 10.1021/la053417x. [DOI] [PubMed] [Google Scholar]
- 27.Mei Y, Elliott JT, Smith JR, Langenbach KJ, Wu T, Xu C, Beers KL, Amis EJ, Henderson L. Gradient substrate assembly for quantifying cellular response to biomaterials. J. Biomed. Mater. Res A. 2006;79A(4):974–988. doi: 10.1002/jbm.a.30883. [DOI] [PubMed] [Google Scholar]
- 28.Gallant ND, Lavery KA, Amis EJ, Becker ML. Universal gradient substrates for “click” biofunctionalization. Adv. Mater. 2007;19(7):965. [Google Scholar]
- 29.Zapata P, Su J, Garcia AJ, Meredith JC. Quantitative high-throughput screening of osteoblast attachment, spreading, and proliferation on demixed polymer blend micropatterns. Biomacro-molecules. 2007;8(6):1907–17. doi: 10.1021/bm061134t. [DOI] [PubMed] [Google Scholar]
- 30.Washburn NR, Yamada KM, Simon CG, Kennedy SB, Amis EJ. High-throughput investigation of osteoblast response to polymer crystallinity: influence of nanometer-scale roughness on proliferation. Biomaterials. 2004;25(78):1215–1224. doi: 10.1016/j.biomaterials.2003.08.043. [DOI] [PubMed] [Google Scholar]
- 31.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294(5547):1708–12. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 32.Bissell MJ, Radisky D. Putting tumours in context. Nat. Rev. Cancer. 2001;1(1):46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chun TH, Hotary KB, Sabeh F, Saltiel AR, Allen ED, Weiss SJ. A pericellular collagenase directs the 3-dimensional development of white adipose tissue. Cell. 2006;125(3):577–91. doi: 10.1016/j.cell.2006.02.050. [DOI] [PubMed] [Google Scholar]
- 34.Simon CG, Stephens JS, Dorsey SM, Becker ML. Fabrication of combinatorial polymer scaffold libraries. Rev. Sci. Instrum. 2007;78(7):072207. doi: 10.1063/1.2755761. [DOI] [PubMed] [Google Scholar]
- 35.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289(5485):1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 36.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat. Methods. 2005;2(2):119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 37.Soen Y, Mori A, Palmer TD, Brown PO. Exploring the regulation of human neural precursor cell differentiation using arrays of signaling microenvironments. Mol. Syst. Biol. 2006;2:37. doi: 10.1038/msb4100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakajima M, Ishimuro T, Kato K, Ko I-K, Hirata I, Arima Y, Iwata H. Combinatorial protein display for the cell-based screening of biomaterials that direct neural stem cell differentiation. Biomaterials. 2007;28(6):1048–60. doi: 10.1016/j.biomaterials.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Rosenthal A, Macdonald A, Voldman J. Cell patterning chip for controlling the stem cell microenvironment. Biomaterials. 2007;28(21):3208–16. doi: 10.1016/j.biomaterials.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding S, Wu TY, Brinker A, Peters EC, Hur W, Gray NS, Schultz PG. Synthetic small molecules that control stem cell fate. Proc. Natl. Acad. Sci. USA. 2003;100(13):7632–7. doi: 10.1073/pnas.0732087100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu X, Ding S, Ding Q, Gray NS, Schultz PG. Small molecules that induce cardiomyogenesis in embryonic stem cells. J. Am. Chem. Soc. 2004;126(6):1590–1. doi: 10.1021/ja038950i. [DOI] [PubMed] [Google Scholar]
- 42.Chen S, Zhang Q, Wu X, Schultz PG, Ding S. Dedifferentiation of lineage-committed cells by a small molecule. J. Am. Chem. Soc. 2004;126(2):410–1. doi: 10.1021/ja037390k. [DOI] [PubMed] [Google Scholar]
- 43.Bailey SN, Sabatini DM, Stockwell BR. Microarrays of small molecules embedded in biodegradable polymers for use in mammalian cell-based screens. PNAS. 2004;101(46):16144–16149. doi: 10.1073/pnas.0404425101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat. Methods. 2006;3(9):715–9. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- 45.Bailey SN, Ali SM, Carpenter AE, Higgins CO, Sabatini DM. Microarrays of lentiviruses for gene function screens in immortalized and primary cells. Nat. Methods. 2006;3(2):117–122. doi: 10.1038/nmeth848. [DOI] [PubMed] [Google Scholar]
- 46.Chen M, Du Q, Zhang HY, Wang X, Liang Z. High-throughput screening using siRNA (RNAi) libraries. Exp. Rev. Mol. Diagn. 2007;7(3):281–91. doi: 10.1586/14737159.7.3.281. [DOI] [PubMed] [Google Scholar]
- 47.Storrie H, Mooney DJ. Sustained delivery of plasmid DNA from polymeric scaffolds for tissue engineering. Adv. Drug. Deliv. Rev. 2006;58(4):500–14. doi: 10.1016/j.addr.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 48.Partridge KA, Oreffo RO. Gene delivery in bone tissue engineering: progress and prospects using viral and nonviral strategies. Tissue Eng. 2004;10(12):295–307. doi: 10.1089/107632704322791934. [DOI] [PubMed] [Google Scholar]
- 49.Merdan T, Kopecek J, Kissel T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Adv. Drug. Deliv. Rev. 2002;54(5):715–58. doi: 10.1016/s0169-409x(02)00046-7. [DOI] [PubMed] [Google Scholar]
- 50.Wagner E, Kircheis R, Walker GF. Targeted nucleic acid delivery into tumors: new avenues for cancer therapy. Biomed. Pharmacother. 2004;58(3):152–61. doi: 10.1016/j.biopha.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 51.Anderson DG, Lynn DM, Langer R. Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angew. Chem. Int. Ed. Engl. 2003;42(27):3153–8. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- 52.Anderson DG, Akinc A, Hossain N, Langer R. Structure/property studies of polymeric gene delivery using a library of poly(beta-amino esters) Mol. Ther. 2005;11(3):426–34. doi: 10.1016/j.ymthe.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 53.Anderson DG, Peng W, Akinc A, Hossain N, Kohn A, Padera R, Langer R, Sawicki JA. A polymer library approach to suicide gene therapy for cancer. Proc. Natl. Acad. Sci. USA. 2004;101(45):16028–33. doi: 10.1073/pnas.0407218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng W, Anderson DG, Bao Y, Padera RF, Jr., Langer R, Sawicki JA. Nanoparticulate delivery of suicide DNA to murine prostate and prostate tumors. Prostate. 2007;67(8):855–62. doi: 10.1002/pros.20576. [DOI] [PubMed] [Google Scholar]
- 55.Green JJ, Shi J, Chiu E, Leshchiner ES, Langer R, Anderson DG. Biodegradable polymeric vectors for gene delivery to human endothelial cells. Bioconjug. Chem. 2006;17(5):1162–9. doi: 10.1021/bc0600968. [DOI] [PubMed] [Google Scholar]
- 56.Zugates GT, Peng W, Zumbuehl A, Jhunjhunwala S, Huang YH, Langer R, Sawicki JA, Anderson DG. Rapid Optimization of Gene Delivery by Parallel End-modification of Poly(beta-amino ester)s. Mol. Ther. 2007;15(7):1306–12. doi: 10.1038/mt.sj.6300132. [DOI] [PubMed] [Google Scholar]