Abstract
Genetically-encoded fluorescence resonance energy transfer (FRET) sensors for phosphate (Pi) (FLIPPi) were engineered by fusing a predicted Synechococcus phosphate-binding protein (PiBP) to eCFP and Venus. Purified fluorescent indicator protein for inorganic phosphate (FLIPPi), in which the fluorophores are attached to the same PiBP lobe, shows Pi-dependent increases in FRET efficiency. FLIPPi affinity mutants cover Pi changes over eight orders of magnitude. COS-7 cells co-expressing a low-affinity FLIPPi and a Na+/Pi co-transporter exhibited FRET changes when perfused with 100 µM Pi, demonstrating concentrative Pi uptake by PiT2. FLIPPi sensors are suitable for real-time monitoring of Pi metabolism in living cells, providing a new tool for fluxomics, analysis of pathophysiology or changes of Pi during cell migration.
Keywords: Fluorescence energy transfer, Phosphate starvation, Synechococcus, Biosensor
1. Introduction
Phosphate (Pi) is an essential macronutrient for all living organisms. It serves central biological functions as part of nucleic acids, phospholipids and ATP, as a metabolite involved in energy transfer, as a component of signal transduction cascades, and in the regulation of enzymes and metabolic processes. Adenosine triphosphate (ATP) is the dominant ‘energy currency’ in the cell; the hydrolysis of ATP to adenosine diphosphate (ADP) plus Pi releases energy that fuels most energy-requiring processes in the cell. Indeed, ATP is required for phosphorylation of glucose, which enables the glucose to enter the glycolytic pathway. Moreover Pi is essential for the skeleton, with 85% of the total body Pi incorporated into the bone. Therefore, Pi is a critical metabolite, and subtle concentration changes of this molecule can profoundly alter cellular growth and metabolism. Cellular uptake is mediated by concentrative Pi transporters, which in mammalian cells use the proton and sodium gradients [1]. Since Pi and phosphorylated compounds are present in all cellular compartments, and since their concentrations can differ significantly between cells in a specific tissue type, and between the compartments within a cell [2], it is important to develop tools that permit quantitative determination of changes in the Pi concentration in living cells with subcellular resolution. 31P NMR can only be used to determine subcellular distribution of Pi within tissue samples in cases, where pH differs between compartments, and Pi concentrations are in the millimolar range [2]. A high-sensitivity methodology that would allow for dynamic analysis of Pi in subcellular compartments is still lacking.
The uptake of Pi into Gram-negative bacteria, such as Escherichia coli and Salmonella, is mediated by the combined action of an outer membrane porin, a periplasmic Pi-specific binding protein (PiBP), and an inner membrane ABC transporter. PiBP binds both monobasic and dibasic phosphate [3] with an overall apparent Kd of 0.8 µM [4]; the dissociation rate constant was determined to be 21 s−1 at neutral pH and low ionic strength [5]. PiBP consists of two globular domains connected by a flexible hinge [6]. In the absence of Pi, PiBP globular domains are separated, exposing a cleft that is accessible to soluble metabolites. Binding of PiBP to Pi induces a conformational change in which the two globular domains of the protein come closer together, solvent is excluded from the binding pocket and hydrogen bonds form between the amino acids of PiBP and bound Pi [7].
PiBP from E. coli functions as an in vitro Pi sensor when coupled to the fluorophore N-[2-(1-maleimidyl)ethyl]-7-(diethylamino) coumarin-3-carboxamide (MDCC) [8,9]; E. coli PiBP was independently shown to signal Pi binding via any of six small molecule dyes [10]. However, such dyes are difficult to introduce into living cells, cannot be easily localized to subcellular compartments, and are likely toxic to cells. The availability of genetically-encoded FRET sensors for Pi would enable systematic screening of mutant collections for factors that control Pi homeostasis, or of chemical libraries to identify novel drugs.
Fluorescence resonance energy transfer (FRET) between chromatic variants of the green fluorescent protein (GFP) can be exploited to monitor conformational changes in proteins [11,12]. The allosteric coupling between ligand binding and FRET can be used to quantitatively measure analyte levels; indeed, nanosensors for sugars (e.g. maltose, ribose, and glucose [13–16]) and amino acids (e.g. glutamate [17]) have been created by fusing a bacterial periplasmic-binding protein specific for a given ligand to FRET donor and acceptor fluorophores. These ratiometric nanosensors were expressed in live cells and used for in vivo analysis of metabolite concentrations in the cytosol, in the nucleus, inside the ER lumen and at the cell surface [13–15,17–19].
The Synechococcus PiBP, by homology with the E. coli PiBP [3,7], is predicted to be a type II PBP, with both termini on the same protein lobe. It has been previously shown that such proteins, when fused to FRET donor and acceptor fluorophores, can make good sensors (as in the case of the E. coli glutamate/aspartate-binding protein [17,20]), indicating that other subtle effects (dipole orientation changes, surface interactions between PiBP and fluorophores) in addition to changes in distance between the donor and acceptor fluorophores contribute to the FRET transfer efficiency.
We tested several PiBPs linearly fused with eCFP and Venus (an eYFP variant with reduced pH and chloride sensitivity [21]), in search of a sensor for which Pi elicits a significant change in FRET. A suitable sensor was created from a Synechococcus protein predicted by sequence homology to be a Pi-binding protein; the response of the sensor was modest, saturable, and consistent with a single Pi-binding site. The sensor was optimized by site-directed mutagenesis of the binding protein–chromophore linkers and the Pi-binding pocket to improve the signal change, and to create a family of sensors with variant affinities.
When FLIPPi (fluorescent indicator protein for inorganic phosphate) with a Kd of 30 mM was expressed in Pi-starved Chinese hamster ovary (CHO) cells, it responded to extracellular Pi with a reduced Venus/eCFP fluorescence emission intensity ratio. A FLIPPi variant with a Kd of 5 µM did not respond to Pi perfusion, consistent with the hypothesis that the intracellular concentration of Pi is outside of the dynamic range of this higher-affinity sensor. Additionally, the sensor reliably detected changes in external Pi availability of unstarved COS-7 cells that were over-expressing a Na+/Pi cotransporter. This new family of FLIPPi sensors can be used to monitor Pi levels over a wide dynamic range that encompass those levels that typically occur in living cells.
2. Materials and methods
2.1. FLIPPi constructs and plasmids
A truncated PiBP (Synechococcus strain A; ORF01723), encoding the predicted mature protein without periplasmic leader sequence, was amplified by PCR from genomic DNA of the thermophilic cyanobacterium [22] using the primers 5′-ATTGGTACCGTAGGATTTCTAACAGCG-3′ and 5′-ATAGGTACCGTTAACGGTGATGGAATC-3′. The PCR fragment was cloned into the KpnI site of FLIPmal-25µ [13] in pRSET-B (Invitrogen, USA), exchanging the sequence encoding the maltose-binding protein with that of PiBP. The resulting plasmid was named pRSET-FLIPPi-840n. To improve the fluorophore maturation efficiency and the pH- and chloride-tolerance of the sensor, the sequence of eYFP in pRSET-FLIPPi-840n was replaced with that of the YFP variant Venus [21], and the composite linkers were shortened by site-directed mutagenesis to yield FLIPPi-260n. Affinity mutants of FLIPPi were created by site-directed mutagenesis [41], generating the T22A, S52A, G162A and T163A variant sensor proteins. These pRSET-FLIPPi constructs were introduced into E. coli BL21(DE3)Gold (Stratagene, USA) by electroporation, and the expressed proteins were extracted and purified as described [13]. For expression in the cytosol of CHO cells, DNA fragments containing FLIPPi-5µ and FLIPPi-30m sequences were excised from pRSET-FLIPPi-5µ and pRSET-FLIPPi-30m, respectively, with BamHI/HindIII and cloned into pCDNA3.1 (Invitrogen, USA). For the co-expression of FLIPPi-30m and the human Na+/Pi transporter PiT2, an EcoRV site was introduced at the 3′- end of the eGFP sequence in pCMS-eGFP (Clontech, USA), and the eGFP sequence was replaced by the BamHI–PmeI fragment from pcDNA3.1-FLIPPi-30m. The resulting plasmid was named pCMS-FLIPPi-30m. pcDNA-PiT2, which carries the VSV-tagged human PiT2 transporter, was kindly provided by Dr. Heard (Institut Pasteur, France). pcDNA-PiT2 was digested with HindIII, blunted, and the PiT2 sequence was excised with NotI and cloned into XhoI (blunt-ended)/NotI sites in pCMS-FLIPPi-30m.
2.2. In vitro characterization of FLIPPi
Emission spectra and ligand titration curves were obtained using a monochromator microplate reader (Safire, Tecan, Austria, excitation 433/12 nm; emission 485/12 and 528/12 nm; gain 70). Enzymatic Pi-depletion of buffers was achieved by overnight dialysis against 1 U nucleoside phosphorylase and 1 mM inosine (Sigma, USA). All assays were performed in 20 mM Tris–HCl buffer, pH 7.0. The Kd of each FLIPPi sensor was determined by fitting ligand titration curves to a single-site binding isotherm: S = (r − rapo)/(rsat − rapo) = [L]/(Kd + [L]), where S is saturation; [L], ligand concentration; r, ratio; rapo, ratio in the absence of ligand; and rsat, ratio at saturation with ligand. Measurements were performed with at least three independent protein extracts.
2.3. Cell culture and transfection
Chinese hamster CHO cells were grown in minimum essential MEM-α medium supplemented with 10% fetal bovine serum (Gibco). Cells were cultured at 37 °C and 5% CO2. For imaging, the cells were cultured in eight-well tissue culture glass slides (BD Falcon). The efficiency of transfection, performed using Lipofectamine 2000 (Invitrogen, USA) and determined by counting fluorescent cells, was at least 30%. Cell starvation was started 16 h prior to the experiment by replacement of MEM-α medium with modified Tyrode’s saline solution containing 100 mM MOPS, 2.5 mM KCl, 25 mM HEPES, 30 mM glucose, 2 mM CaCl2 and 2 mM MgCl2. COS-7 cells were grown in DMEM medium supplemented with 10% FBS (Difco, USA) on poly-d-lysine (Sigma) coated cover slips.
2.4. Imaging of cell cultures
CHO and COS-7 cells were analyzed for FRET 24–48 h after transfection, using an inverted epifluorescence microscope (DM IRE2, Leica, Germany) with a cooled CoolSnap HQ digital camera (Photometrics, USA) and a 63× water immersion lens (HCX PL APO, Leica, Germany). Dual emission intensity ratios were simultaneously recorded using a DualView with an OI-5-EM filter set (Optical Insights, USA) and Metafluor 6.3 software (Molecular Devices, USA). Depending on expression level, the exposure times varied between 0.5 and 2.5 s. Unless stated otherwise, CHO cells were perfused with Pi-free modified Tyrode’s saline solution at a flow rate of 1.8 ml/min in a chamber with a total volume of <0.6 ml. For each imaging experiment, at least three independent repetitions were performed. COS-7 cells were perfused with Pi-free Tyrode’s saline solution. Confocal analysis was performed as described [38].
3. Results
3.1. Identification of a PiPB suitable for sensor design
The PiBP protein from E. coli allosterically signals Pi binding in conjunction with a variety of small molecule dyes [10]; therefore, this protein scaffold was selected for conversion to a FRET sensor. Fusion constructs were expressed in E. coli, protein was isolated by His affinity chromatography and the FRET change was measured in a buffer solution enzymatically depleted of Pi, to which Pi was added at the desired concentration. Pi levels above 5 mM led to significant fluorescence quenching (data not shown).
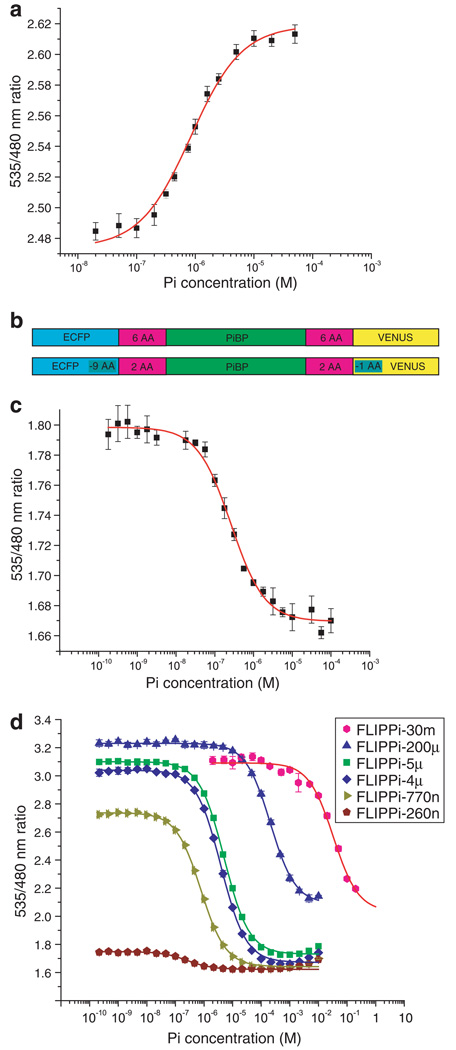
The PiBP sensor initially constructed was found to have a high FRET efficiency, but exhibited no Pi-dependent change (data not shown). We therefore sought to identify additional Pi-binding domains that could be used to construct sensor that would respond to the Pi concentration. We constructed a phylogenetic tree of putative Pi-binding proteins from several bacterial species (Supplementary Fig. 1) and used this tree to select representatives from evolutionarily divergent subclades, reasoning that this strategy might increase our sampling of potential dipole–dipole relative orientations of the donor and acceptor fluorophores. The sensors generated from PiBP genes from Pyrococcus furiosus and Sulfolobus solfataricus also failed to show a significant FRET change, but one constructed using a PiBP homologue from the thermophilic Synechococcus strain OS-A [22] did exhibit a phosphate-dependent FRET increase; this sensor was designated FLIPPi-WT. The affinity of this sensor for Pi was 840 nM (with a Hill coefficient of 1.03), consistent with the 800 nM Kd reported for the E. coli PiBP [4]. The ratio change observed for FLIPPi-WT was 0.14 (6% change; Fig. 1a). It is thus apparently possible to obtain a functional FRET sensor even for proteins in which the fluorophore attachment is adjacent on the same lobe.
Fig. 1.
(a) In vitro ligand-dependent FRET changes of FLIPPi-WT purified nanosensors. Non-linear regression best fit and error bars are shown. (b) FLIPPi-WT construct (above); FLIPPi-260n construct (below; nine amino acids were deleted from the C-terminus of the eCFP, one from the N-terminus of Venus, and four from each binding protein-fluorophore linker). (c) and (d) In vitro ligand-dependent FRET changes of FLIPPi-260n (c) and affinity mutants of the FLIPPi-260n construct (d; Table 1).
3.2. Mutants with altered Pi affinity and improved signal change
Previous studies had shown that shortening the flexible domains linking the binding protein with the fluorophores could improve the allosteric coupling and thus the FRET signal change [20]; a total of 18 amino acids were thus shortened from the sensor sequence: 9 from the C-terminus of the eCFP, 1 from the N-terminus of Venus, and 4 from each binding protein–fluorophore linker (Fig. 1b). Titration of this modified protein (designated FLIPPi-260n) gave an increase in eCFP emission and a decrease in Venus emission, resulting in a Pi-dependent decrease in FRET. The binding constant for Pi was determined to be 260 nM, with a maximum ratio change of −0.13 (7%; Fig. 1c). Linker deletion has thus reversed the sign of the FRET ratio change without altering the magnitude; the affinity has also been slightly increased.
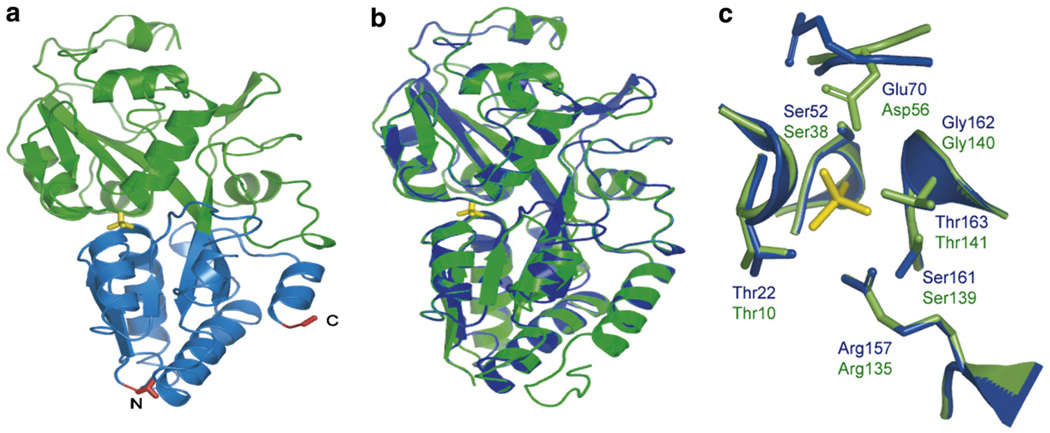
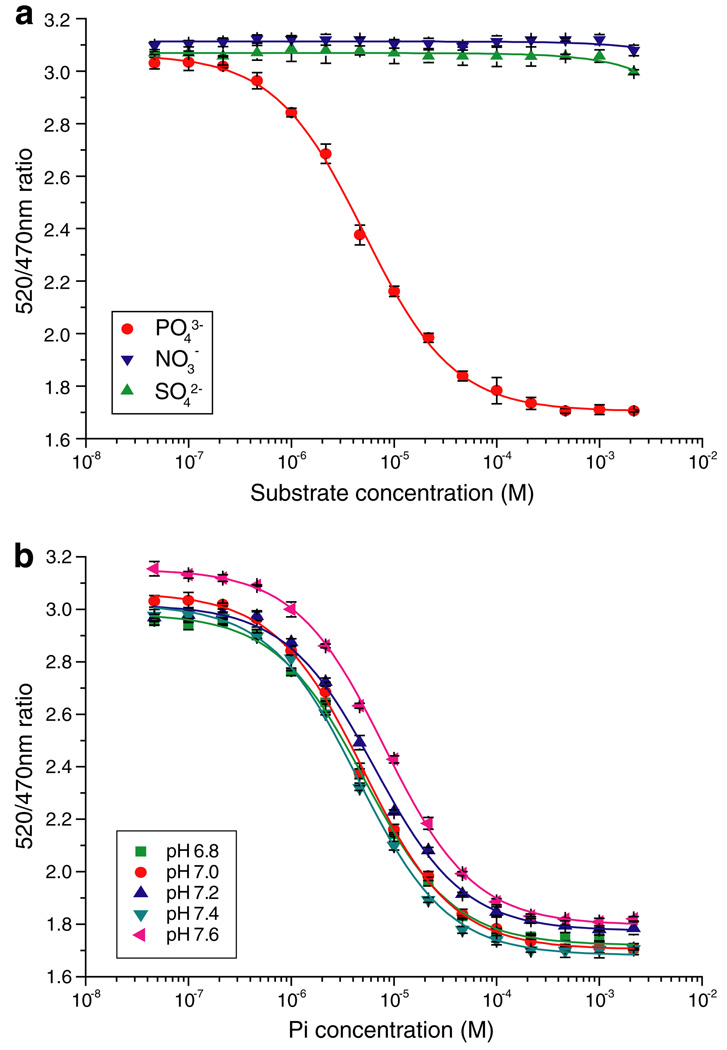
As a basis for further signal enhancement and for designing affinity mutants, the PiBP sequences of Synechococcus sp. and E. coli were aligned (identity 37%) and a homology model was generated with 3D-JIGSAW [23] (E. coli PiBP: Protein Data Bank structural identifier 1a40) (Fig. 2). To expand the dynamic range of the Pi sensor family, site-directed mutagenesis was used to decrease the Pi-binding affinity. The Pi present in the binding pocket of E. coli PiBP is held in place by strong hydrogen bonds [24]; the structural alignment of the binding pockets of the E. coli structure and the Synechococcus model shows that the interacting side-chains are completely conserved, except for the conservative substitution of Asp56 in the E. coli protein to Glu70 in the Synechococcus protein (Fig. 2c). Five residues of FLIPPi-260n were selected for site-directed mutagenesis, generating a set of sensors with a broad range of Pi-binding affinities (Fig. 1d and Table 1). All five affinity mutants showed a greatly improved FRET change upon Pi binding (8–10-fold), predominantly through an increase of the apo FRET efficiency. This is consistent with either or both of two scenarios: (i) residual Pi is bound to the high-affinity sensors and artificially depresses the signal change, or (ii) the FRET efficiency of the apo and/or ligand-bound states of the sensor is perturbed by an alteration of the precise open and closed state forms, which could impact relative dipole orientations and thus FRET efficiency [20]. The mutants were designated FLIPPi-770n, FLIPPi-4µ, FLIPPi-5µ, FLIPPi-200µ, and FLIPPi-30m, according to their Pi-binding affinity. The computed Hill coefficients of all PiBPs are very close to 1, consistent with a single Pi-binding site [4]. The improved dynamic ranges of the individual sensors (due to their greater signal-to-noise ratios) in conjunction with the distribution of their equilibrium binding constants, creates a wide composite dynamic range of the set of all six sensors, covering 30 nM–150 mM, except for the interval of 1.5–3 mM (Fig. 1d and Table 1). To determine the substrate specificity and pH sensitivity of the sensor in the physiological range, the pH dependence of phosphate binding as well as the response to sulfate and nitrate was tested for FLIPPi-5µ (Fig. 3). FLIPPi-5µ did not respond to nitrate or sulfate, moreover, the binding isotherms for phosphate were very similar between pH 6.8 and 7.6. Since the Kd did not shift significantly, despite the significant relative change in the relative amounts of and (pKa 7.2), the sensor appears independent of pH changes in this range due to similar recognition of both forms.
Fig. 2.
The predicted structure of PiBP in alignment with the E. coli PiBP (PDB 1a40). (a) Synechococcus PiBP structural model, predicted by 3D-JIGSAW. Phosphate is in yellow, and the N- and C-termini in red. Terminal lobe, blue; other lobe, green. (b) Alignment of the model (blue) with the crystal structure of E. coli (green). (c) Alignment of binding sites. The only sequence difference is Glu70 in FLIPPi and Asp56 in E. coli. The Synechococcus structure has likely been poorly modeled in the region of Glu70 and Ser161 (phosphate was not present in the 3D-JIGSAW modeling step). Green: E. coli; blue: FLIPPi; yellow: phosphate.
Table 1.
FLIPPi affinity mutants
| Sensor | Sequence | Kd (M)a | ΔRmaxa,b | Hill coefficient | Dynamic rangec |
|---|---|---|---|---|---|
| FLIPPi-260n | Wild-type | 2.6 × 10−7 | −0.13 | 1.03 | 25–1200 nM |
| FLIPPi-770n | S161A | 7.7 × 10−7 | −1.07 | 1.03 | 80–5600 nM |
| FLIPPi-4µ | T163A | 3.9 × 10−6 | −1.34 | 1.02 | 0.4–25 µM |
| FLIPPi-5µ | S52A | 5.1 × 10−6 | −1.33 | 1.03 | 0.5–34 µM |
| FLIPPi-200µ | G162A | 2.1 × 10−4 | −1.13 | 1.00 | 25–1600 µM |
| FLIPPi-30m | T22A | 3.3 × 10−2 | −1.03 | 1.03 | 3–170 mM |
Binding constants determined in vitro.
R2 for the fit exceeded 0.98 in all analyses.
ΔRmax, in vitro maximum change in ratio between ligand-free and ligand-saturated sensor.
Range between 10% and 90% saturation of the sensor (the effective quantification range).
Fig. 3.
Specificity and pH sensitivity of FLIPPi-5µ. (a) Selectivity of FLIPPi-5µ for phosphate over nitrate and sulfate. Solid lines were plotted by curve fitting. (b) Phosphate binding of FLIPPi-5µ was tested in 20 mM Tris buffers adjusted to pH 6.8, 7.0, 7.2, 7.4 and 7.6, then titrated with phosphate solutions of the same respective pH. Statistical analysis (ANOVA) showed no significant difference regarding the maximal ratio change between pH 6.8 and 7.4.
Previous analyses have suggested that intracellular Pi concentrations are in the range of 1–10 mM [2,25–27], thus the single sensor FLIPPi-30m (dynamic range 3–170 mM) or the combination of FLIPPi-30m and FLIPPi-200µ offer the best starting points for monitoring in vivo Pi concentrations.
3.3. Sensor response in Pi-starved CHO cells
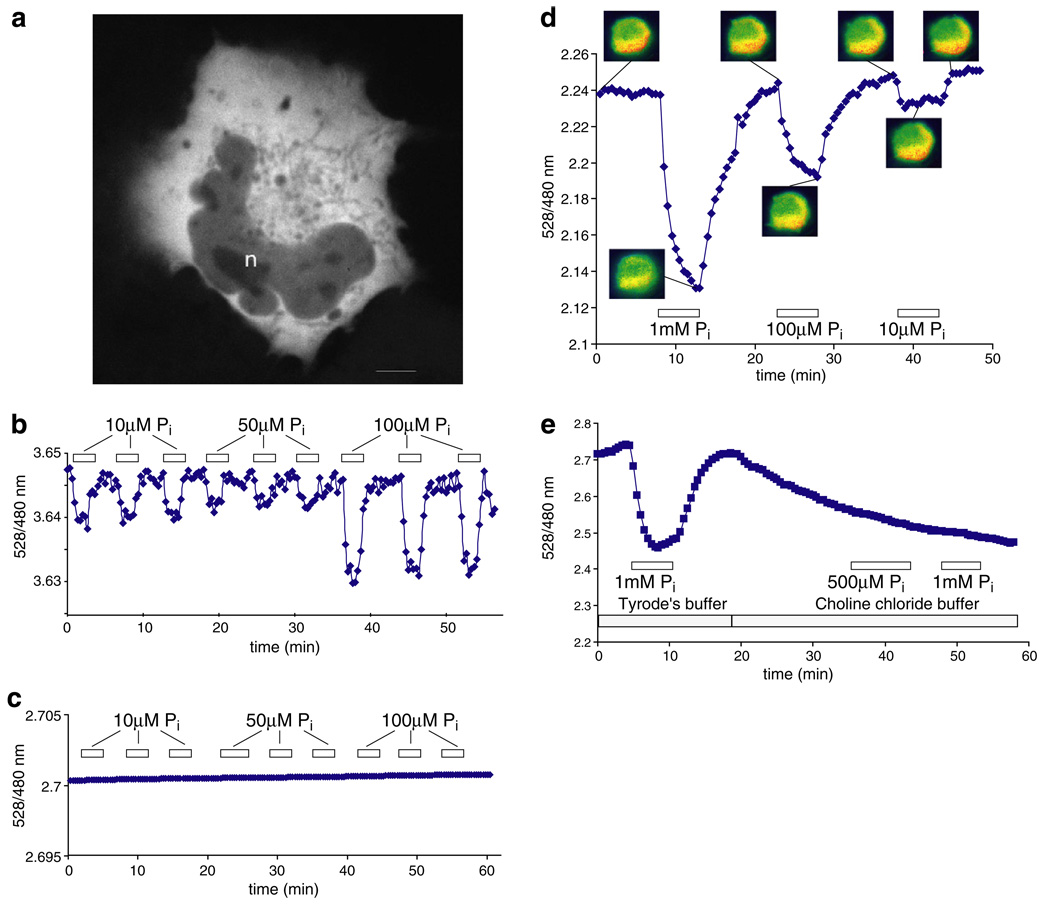
“Cytosolic” Pi levels (including the endoplasmic reticulum and all cytoplasmic organelles) have been estimated at approximately 6 mM in both plant and animal cells [25,27]. Thus, the FLIPPi-30m sensor seems well suited to monitor physiologically-relevant Pi concentrations (in contrast to the other sensors, which should be fully-saturated). CHO cells were independently transfected with either the FLIPPi-30m “working” or FLIPPi-200µ and FLIPPi-5µ “control” sensors. The chimeric protein is distributed evenly within the cytosol and is excluded from vesicular bodies and the nucleus (Fig. 4a). Initial experiments with resting CHO cells showed no FRET change for either sensor upon perfusion with up to 10 mM Pi, suggesting limiting transport rates. Pi deprivation is known to regulate Na+/Pi transporter activity [1]. Therefore, CHO cells were grown overnight after transfection and Pi-starved for 16 h by replacing Pi-replete growth medium with modified Tyrode’s saline solution lacking Pi, before making the measurements. Cells expressing FLIPPi-30m were perfused with increasing concentrations of external Pi, and changes in the fluorescence peak emission ratio were monitored (Fig. 4b). After a stable YFP/CFP emission intensity ratio was attained at each Pi concentration, Pi was removed by perfusion with Pi-free solution, returning the sensor fairly rapidly to baseline emission. FLIPPi-5µ did not show a signal change upon perfusion with even a high concentration of Pi (Fig. 4c). Similar results were obtained for FLIPPi-200µ (data not shown). Starved cells expressing FLIPPi-30m responded to solutions containing 50 µM and 100 µM Pi; the ratio change was significantly higher at 100 µM external Pi (Fig. 4b). Under the conditions used (118 mM NaCl, pH 7.3), the endogenous Na+/Pi cotransporters should theoretically be able to concentrate Pi inside the cells up to 100-fold, consistent with the increase observed using the FLIPPi-30m sensor [1]. Cells were highly sensitive after starvation, however, and either lost fluorescence or detached from the slide during the experiment. Thus, the responses were not robust enough for a more detailed analysis of flux.
Fig. 4.
FRET ratio change in response to Pi perfusion of Pi-starved CHO cells, and COS-7 cells co-expressing a Na+/Pi cotransporter. (a) Confocal image of a COS-7 cell expressing FLIPPi-30m. Fluorescence is largely excluded from the nucleus (n). The scale bar represents 5 µm. (b) The low-affinity “working” sensor FLIPPi-30m in Pi-starved CHO cells. (c) The high-affinity “control” sensor FLIPPi-5µ is saturated and does not respond to Pi perfusion. (d) The FLIPPi-30m sensor in resting COS-7 cells co-expressing the sodium-dependent phosphate cotransporter PiT2. (e) In sodium-free modified Tyrode’s choline buffer, the response of the sensor to Pi is abolished. The boxed numbers indicate the Pi concentration used for perfusion.
3.4. Co-expression of a Na+/Pi transporter and the FRET sensor in COS-7 cells
Similarly, COS-7 cells expressing the FLIPPi-30m sensor also did not reproducibly respond to perfused Pi (data not shown). It is known that phosphate transport activities are down-regulated by external Pi [28,29]. As an alternative means of increasing Pi transport rates of the COS-7 cells, the human Na+/Pi cotransporter PiT2 was co-expressed along with the FLIPPi-30m sensor. When cells co-expressing FLIPPi-30m and PiT2 were perfused with Pi, changes in the fluorescence ratio consistent with increased steady-state levels of Pi in the cytosol were observed (Fig. 4d). On rare occasions, a similar response was obtained in cells expressing only FLIPPi-30m (data not shown). The response to the external Pi supply was greater in COS-7 cells than in starved CHO cells, possibly as a consequence of the robust Pi transport activity of PiT2, as well as increased cell viability. To confirm that the changes in emission intensity ratio were a consequence of changes in the cytosolic Pi level, and not of secondary effects induced by external Pi, the response of COS-7 cells was measured in a buffer in which the sodium in Tyrode’s saline solution was replaced with choline (since Pi uptake is mediated by Na+/Pi co-transporters, the absence of a sodium gradient should prevent uptake of Pi). Equilibration of the cells into the Tyrode’s choline buffer yielded a lower FRET ratio (perhaps through solution effects on the fluorophore dipoles), but also abolished Pi-dependent changes (Fig. 4e), suggesting that the response of the sensor is indeed specifically due to Pi import by the Na+/Pi cotransporter. Taken together, these results suggest that in the presence of sufficient transport activity, Pi metabolism may be monitored in a variety of mammalian cell types.
4. Discussion
Pi plays a number of important roles in all cellular compartments. 31P-NMR allows the determination of cellular Pi levels, and can distinguish between different Pi pools in cases where pH differs between compartments [2]. The methodology allows dynamic measurements, but is of insufficient sensitivity for single cell studies. Pi concentrations may be tracked in large compartments such as the plant vacuole, which is more acidic than the cytoplasm [27], but organelles such as the ER, peroxisomes, plastids, and mitochondria either have too small Pi pools and/or a pH near the cytosolic pH value, which makes the signal indistinguishable from the background noise or superimposed on the cytosolic Pi signal, respectively. In addition, a Pi signal from a specific compartment may be NMR invisible because of fast spin–spin relaxation caused by compounds present in the compartment. Variant Pi transporters are present in the inner mitochondrial membrane [30] and in plastids [31], suggesting that both steady-state and dynamic levels of Pi may be quite different for the various sub-cellular compartments.
Therefore, novel methods and reagents are required for the detection of Pi concentrations with sufficient spatial and temporal resolution to address the complex issue of Pi homeostasis. Recently, genetically-encoded FRET sensors have been developed for monitoring calcium, various sugars and amino acids [13,18,32,33]. These sensors make use of a designed allosteric linkage between an analyte-binding recognition element (ideally with a significant binding-induced conformational change; calmodulin was employed for the calcium sensor, bacterial periplasmic-binding proteins for the others) and a macroscopic signal-producing reporter element (in this case, FRET-capable cyan and yellow variants of GFP).
Bacterial periplasmic-binding proteins (PBPs) exhibit a conserved domain structure of two (in rare examples, three) α/β lobes connected by β-strand “hinge” elements [34]. A ligand-binding cleft is formed at the interface between the two lobes; upon binding, a hinge-bending motion sequesters the ligand, excluding solvent and presenting functionally-compatible binding site groups from both protein lobes [34,35]. It is this hinge-bending motion that allosterically reorients the reporter fluorophores (in this report, linearly fused to the N- and C-termini of the PBP sequence), giving rise to a change in FRET. Relative fluorophore separation is generally considered to be the dominant feature in resonance energy transfer, reflected in its inverse-sixth power dependence in the Förster equation [36]. Relative fluorophore angular orientation has also been shown to be of great importance [37], and in complex systems such as a chimeric multi-domain protein, these terms are likely to be highly inter-dependent.
The E. coli PiBP protein is a type II periplasmic-binding protein [6], with both N- and C-termini on the same protein lobe. A naive first-order approximation of fluorophore position and conformation suggests that the absence of a clear binding-driven lever-arm displacement of the fluorophores relative to one another would produce similar fluorophore relative positional and orientational ensembles in the ligand-free open and ligand-bound closed states, yielding a sensor with a negligible FRET signal change. When the E. coli PiBP was converted to a FRET sensor, it did indeed fail to exhibit a significant ligand-dependent FRET change. Hypothesizing that the positioning of protein termini is a weakly selected trait, we pursued a number of other putative Pi-binding proteins as recognition elements in FRET constructs, as a way to maximize the chance of finding a combination of terminal positions that would enable the construction of a viable sensor. Six additional putative Pi-binding proteins were tested (all predicted to be type II PBPs), and of these, five failed to show a significant Pi-dependent response, suggesting that the first-order model is a powerful discriminator of functional and non-functional sensors. However, a single predicted PiBP of the seven tested (from the thermophilic cyanobacterium Synechococcus strain OS-A; three other highly homologous Synechococcus proteins failed to signal) produced a sensor with an acceptable signal change. This result, combined with the prior observations that both the type II E. coli glutamate/aspartate-binding protein [17] and sensors in which one of the fluorophores has been inserted into the binding protein sequence on the same lobe as the other terminally-fused fluorophore [20] may also produce viable FRET sensors, implies that allosteric propagation more subtle than the lever-arm motion may also give rise to fluorophore reorientation sufficient to produce a FRET change (Fig. 5). To this end, we have performed simple modeling of fluorophore position which suggests that the ligand-driven movement of the second protein lobe of a type II-binding protein may sterically re-position one or both of the fluorophores attached to the terminal lobe (data not shown). Furthermore, it is possible that the binding protein and fluorophores form nascent protein–protein interactions at the site of each chimeric attachment point, through which conformational changes may be allosterically propagated even in the absence of a dramatic re-positioning by the other protein lobe.
Fig. 5.
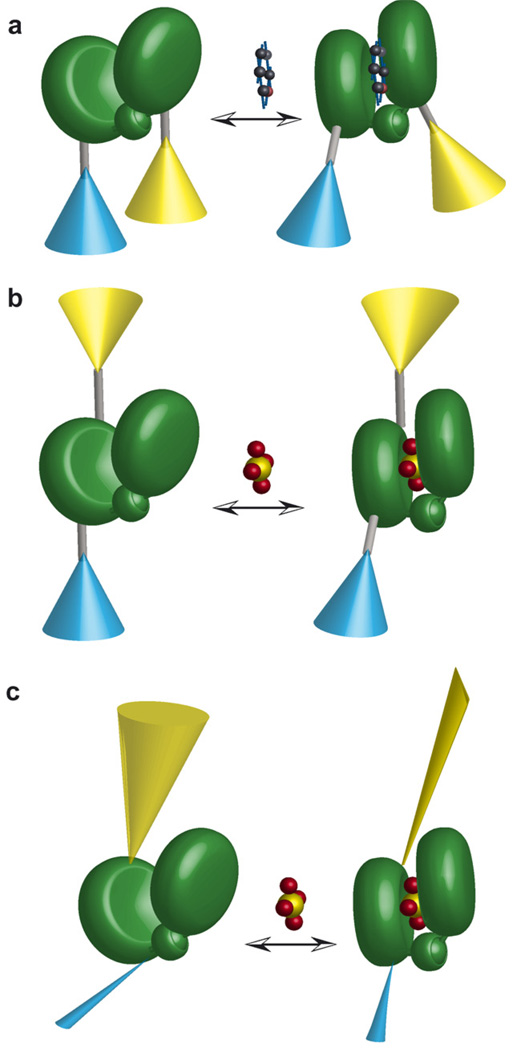
Cartoon models of FLIPPi sensor function [20]. (a) Model of FLIP sensor developed from type I periplasmic binding proteins (e.g. FLIPglu glucose binding protein [14,15]). Fluorophores (eCFP, cyan; Venus, yellow) attached on two different lobes of a binding protein (consisting of two lobes connected by a hinge, green) via linkers (grey rods). Ligand binding transduces a hinge-binding motion propagated through a lever arm to macroscopically reorient the relative excitation–emission dipole. (b) Model of a FLIP sensor developed from type II PiBPs, in which both fluorophores are attached to the same lobe. Allosteric effects of domain closure propagate into relative dipole reorientation. (c) Shortening or rigidification of a binding protein–fluorophore linker increases the degree of allosteric coupling between ligand binding and fluorophore reorientation. One possible scenario is shown here, in which the closure of the PiBP leads to a restriction in the rotational freedom of eYFP, illustrated as a change in the cone diameter.
The affinity of the Synechococcus PiBP proved to be close to that of the E. coli PiBP, and the signal change of the initial FRET construct was of similar magnitude to the other PBP-based FRET sensors constructed thus far. Site-directed mutagenesis produced a family of Pi sensors with improved signal change and Pi affinities varying over almost eight orders of magnitude.
This set of Pi sensors should be suitable for monitoring Pi transport and metabolism, such as that previously reported for glucose detection using an analogous FLIPglu sensor in COS-7 and HepG2 cells [14], and for ribose detection using a FLIPrib sensor [15]. FLIPPi sensors (both high-affinity “control” and low-affinity “working” sensors) expressed in the cytosol of non-starved CHO cells failed to show a signal change upon perfusion with Pi up to 10 mM (data not shown). This failure of the sensors to signal may have a number of explanations: the Pi concentration is much lower than previously thought, and the sensors are not seeing their ligand (unlikely, as a sensor with 5 µM affinity shows no response); the Pi concentration is much higher than previously thought, and the sensors are fully-saturated (unlikely, as a sensor with 30 mM affinity shows no response); the Pi concentration is maintained within an interval above the dynamic range of the 5 µM sensor and below that of the 30 mM sensor, i.e. the low millimolar range; or that net Pi influx into the cytosol is small compared to the rates of metabolic conversion and compartmentalization. In an attempt to distinguish between these potential explanations, CHO cells were Pi-starved for 14 h, and the measurement was repeated with the low-affinity sensor. The Pi-starved cells now showed a small but significant signal change upon Pi perfusion; this argues against the maintenance of Pi levels in the low millimolar range, as the Pi levels would have been decreased further below the dynamic range of the sensor. Rather, it is most likely that Pi transport rates increased following starvation, and that the level of uptake exceeded the net effect of metabolism and compartmentalization. Hence, Pi perfusion of the Pi-starved cells resulted in a change in the cytosolic Pi concentration.
The Pi-starved CHO cells proved to be rather fragile, making long-term monitoring impossible. As a second demonstration that the response of the sensors is Pi transport-limited, we co-expressed the low-affinity FLIPPi sensor along with the Na+/Pi transporter PiT2 in COS-7 cells. Much larger FRET changes were then observed, indicating that the introduced Pi transporter contributed more to net Pi influx than the presumed up-regulation of endogenous Pi transporters following Pi starvation. Interestingly, Pi levels in the cytosol of COS-7 cells co-expressing PiT2 promptly decreased when the external Pi supply was removed; it is not known whether this is due to metabolism or to a net efflux of Pi into the bulk medium in response to the strong Pi gradient (or to both).
The currently available FLIPPi sensors should be sufficient to follow Pi metabolism in non-transport-limited cellular environments. Further rounds of protein engineering should produce a sensor with an affinity in the low millimolar range (thus closing the gap between the FLIPPi-200µ and FLIPPi-30m sensors), as well as a sensor with improved signal-to-noise ratio (in vivo FRET changes are invariably lower than those seen in vitro [14,15,17,38], perhaps due to ionic strength, pH, and quenching effects in the cellular milieu). We can at present not exclude that the FLIPPi-30m recognizes other phosphate-containing analytes. Thus, it will be important to compare the results obtained with the sensors in parallel with 31P NMR and other methods.
Production of a sensor for glucose-6-phosphate, combined with the Pi sensor reported here and the previously reported glucose sensor [38], should facilitate monitoring of the principal players in glucose metabolism. A battery of these sensors targeted to the cytosol and the ER should shed light on the complex metabolism and transport events involved in gluconeogenesis and glycolysis [39].
5. Conclusions
A putative PiBP from a thermophilic Synechococcus strain was successfully converted into a genetically-encoded Pi sensor. Site-directed mutagenesis facilitated the generation of affinity mutants covering a wide range of physiologically relevant Pi concentrations. So far an extensive mutagenesis has not yielded a suitable mutant (Hong Gu, unpublished). The sensors were used to measure Pi homeostasis in cultured cells in real time. A major advantage of genetically-encoded nanosensors is the ability to target the proteins to subcellular compartments, as demonstrated for the FLIPglu glucose sensor in nuclei [19] and the FLIPE glutamate sensor on the surface of PC12 neuronal cells [17]. The Pi sensors will be useful for a wide range of applications, e.g. to study Pi uptake and translocation, and regulatory mechanisms controlling compartmental Pi homeostasis. The sensors may also be useful tools to study relevant physiological or pathophysiological cellular conditions, e.g., cell motility, by analysis of potential differences between motile and sessile cells or compartmentalized responses in lamellipodia or ruffles. Furthermore, transgenic organisms expressing FLIPPi should allow the visualization of Pi activity directly in organ slices or whole organisms, such as that demonstrated for the calcium FRET indicator cameleon in Caenorhabditis elegans neurons [40].
Supplementary Material
Acknowledgements
We are grateful to Dr. Heard (Institute Pasteur, France) for providing the PiT2 clone. This work was made possible by a grant from HFSP (RGP0041/2004C) to Kristoffer Almdal (Risø, Denmark), I.J., J.K. and W.B.F. H.G. was supported in part by the Danish Research School for Biotechnology FOBI. This work was also partially funded by the Frontiers in Integrative Biology program at NSF (GC036/04/Z3423).
Abbreviations
- FLIPPi
fluorescent indicator protein for inorganic phosphate
- FRET
fluorescence resonance energy transfer
- FP
fluorescent protein
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.febslet.2006.09.048.
References
- 1.Collins JF, Bai L, Ghishan FK. The SLC20 family of proteins: dual functions as sodium-phosphate cotransporters and viral receptors. Pflügers Arch. 2004;447:647–652. doi: 10.1007/s00424-003-1088-x. [DOI] [PubMed] [Google Scholar]
- 2.Vidal G, Gallis JL, Dufour S, Canioni P. NMR studies of inorganic phosphate compartmentation in the isolated rat liver during acidic perfusion. Arch. Biochem. Biophys. 1997;337:317–325. doi: 10.1006/abbi.1996.9775. [DOI] [PubMed] [Google Scholar]
- 3.Wang ZM, Choudhary A, Ledvina PS, Quiocho FA. Fine-tuning the specificity of the periplasmic phosphate transport receptor – site-directed mutagenesis, ligand-binding, and crystallographic studies. J. Biol. Chem. 1994;269:25091–25094. doi: 10.2210/pdb1pbp/pdb. [DOI] [PubMed] [Google Scholar]
- 4.Medveczky N, Rosenberg H. Phosphate-binding protein of Escherichia coli. Biochim. Biophys. Acta. 1970;211:158. [Google Scholar]
- 5.Brune M, Hunter JL, Corrie JET, Webb MR. Direct, real-time measurement of rapid inorganic-phosphate release using a novel fluorescent probe and its application to actomyosin subfragment-1 ATPase. Biochemistry. 1994;33:8262–8271. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- 6.Ledvina PS, Yao NH, Choudhary A, Quiocho FA. Negative electrostatic surface potential of protein sites specific for anionic ligands. Proc. Natl. Acad. Sci. USA. 1996;93:6786–6791. doi: 10.1073/pnas.93.13.6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ledvina PS, Tsai AL, Wang ZM, Koehl E, Quiocho FA. Dominant role of local dipolar interactions in phosphate binding to a receptor cleft with an electronegative charge surface: equilibrium, kinetic, and crystallographic studies. Protein Sci. 1998;7:2550–2559. doi: 10.1002/pro.5560071208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salins LLE, Deo SK, Daunert S. Phosphate binding protein as the biorecognition element in a biosensor for phosphate. Sensor. Actuat. B – Chem. 2004;97:81–89. doi: 10.1016/j.snb.2003.07.019. [DOI] [PubMed] [Google Scholar]
- 9.Hirshberg M, Henrick K, Haire LL, Vasisht N, Brune M, Corrie JET, Webb MR. Crystal structure of phosphate binding protein labeled with a coumarin fluorophore, a probe for inorganic phosphate. Biochemistry. 1998;37:10381–10385. doi: 10.1021/bi980428z. [DOI] [PubMed] [Google Scholar]
- 10.De Lorimier RM, et al. Construction of a fluorescent biosensor family. Protein Sci. 2002;11:2655–2675. doi: 10.1110/ps.021860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaccolo M. Use of chimeric fluorescent proteins and fluorescence resonance energy transfer to monitor cellular responses. Circ. Res. 2004;94:866–873. doi: 10.1161/01.RES.0000123825.83803.CD. [DOI] [PubMed] [Google Scholar]
- 12.Lalonde S, Ehrhardt DW, Frommer WB. Shining light on signaling and metabolic networks by genetically encoded biosensors. Curr. Opin. Plant Biol. 2005;8:574–581. doi: 10.1016/j.pbi.2005.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fehr M, Frommer WB, Lalonde S. Visualization of maltose uptake in living yeast cells by fluorescent nanosensors. Proc. Natl. Acad. Sci. USA. 2002;99:9846–9851. doi: 10.1073/pnas.142089199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fehr M, Lalonde S, Lager I, Wolff MW, Frommer WB. In vivo imaging of the dynamics of glucose uptake in the cytosol of COS-7 cells by fluorescent nanosensors. J. Biol. Chem. 2003;278:19127–19133. doi: 10.1074/jbc.M301333200. [DOI] [PubMed] [Google Scholar]
- 15.Lager I, Fehr M, Frommer WB, Lalonde S. Development of a fluorescent nanosensor for ribose. FEBS Lett. 2003;553:85–89. doi: 10.1016/s0014-5793(03)00976-1. [DOI] [PubMed] [Google Scholar]
- 16.Ye K, Schultz JS. Genetic engineering of an allosterically based glucose indicator protein for continuous glucose monitoring by fluorescence resonance energy transfer. Anal. Chem. 2003;75:3451–3459. doi: 10.1021/ac034022q. [DOI] [PubMed] [Google Scholar]
- 17.Okumoto S, Looger LL, Micheva KD, Reimer RJ, Smith SJ, Frommer WB. Detection of glutamate release from neurons by genetically encoded surface-displayed FRET nanosensors. Proc. Natl. Acad. Sci. USA. 2005;102:8740–8745. doi: 10.1073/pnas.0503274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fehr M, et al. Development and use of fluorescent nanosensors for metabolite imaging in living cells. Biochem. Soc. Trans. 2005;33:287–290. doi: 10.1042/BST0330287. [DOI] [PubMed] [Google Scholar]
- 19.Fehr M, Lalonde S, Ehrhardt DW, Frommer WB. Live imaging of glucose homeostasis in nuclei of COS-7 cells. J. Fluoresc. 2004;14:603–609. doi: 10.1023/b:jofl.0000039347.94943.99. [DOI] [PubMed] [Google Scholar]
- 20.Deuschle K, Okumoto S, Fehr M, Looger LL, Kozhukh L, Frommer WB. Construction and optimization of a family of genetically encoded metabolite sensors by semirational protein engineering. Protein Sci. 2005;14:2304–2314. doi: 10.1110/ps.051508105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nature Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 22.Steunou AS, et al. In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proc. Natl. Acad. Sci. USA. 2006;103:2398–2403. doi: 10.1073/pnas.0507513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Contreras-Moreira B, Bates PA. Domain fishing: a first step in protein comparative modelling. Bioinformatics. 2002;18:1141–1142. doi: 10.1093/bioinformatics/18.8.1141. [DOI] [PubMed] [Google Scholar]
- 24.Luecke H, Quiocho FA. High specificity of a phosphate transport protein determined by hydrogen-bonds. Nature. 1990;347:402–406. doi: 10.1038/347402a0. [DOI] [PubMed] [Google Scholar]
- 25.Desmoulin F, Cozzone PJ, Canioni P. Phosphorus-31 nuclear-magnetic-resonance study of phosphorylated metabolites compartmentation, intracellular pH and phosphorylation state during normoxia, hypoxia and ethanol perfusion, in the perfused rat liver. Eur. J. Biochem. 1987;162:151–159. doi: 10.1111/j.1432-1033.1987.tb10555.x. [DOI] [PubMed] [Google Scholar]
- 26.Chen KY. Study of polyphosphate metabolism in intact cells by 31-P nuclear magnetic resonance spectroscopy. Prog. Mol. Subcell. Biol. 1999;23:1. doi: 10.1007/978-3-642-58444-2_13. [DOI] [PubMed] [Google Scholar]
- 27.Rebeille F, Bligny R, Martin JB, Douce R. Relationship between the cytoplasm and the vacuole phosphate pool in Acer pseudoplatanus cells. Arch. Biochem. Biophys. 1983;225:143–148. doi: 10.1016/0003-9861(83)90017-6. [DOI] [PubMed] [Google Scholar]
- 28.Pfister MF, Hilfiker H, Forgo J, Lederer E, Biber J, Murer H. Cellular mechanisms involved in the acute adaptation of OK cell Na+/Pi-cotransport to high- or low-Pi medium. Pflügers Arch. 1998;435:713–719. doi: 10.1007/s004240050573. [DOI] [PubMed] [Google Scholar]
- 29.Rodrigues P, Heard JM. Modulation of phosphate uptake and amphotropic murine leukemia virus entry by posttranslational modifications of PIT-2. J. Virol. 1999;73:3789–3799. doi: 10.1128/jvi.73.5.3789-3799.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krämer R. Structural and functional aspects of the phosphate carrier from mitochondria. Kidney Int. 1996;49:947–952. doi: 10.1038/ki.1996.133. [DOI] [PubMed] [Google Scholar]
- 31.Knappe S, Flügge UI, Fischer K. Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol. 2003;131:1178–1190. doi: 10.1104/pp.016519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romoser VA, Hinkle PM, Persechini A. Detection in living cells of Ca2+-dependent changes in the fluorescence emission of an indicator composed of two green fluorescent protein variants linked by a calmodulin-binding sequence – a new class of fluorescent indicators. J. Biol. Chem. 1997;272:13270–13274. doi: 10.1074/jbc.272.20.13270. [DOI] [PubMed] [Google Scholar]
- 33.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 34.Fukami-Kobayashi K, Tateno Y, Nishikawa K. Domain dislocation: a change of core structure in periplasmic binding proteins in their evolutionary history. J. Mol. Biol. 1999;286:279–290. doi: 10.1006/jmbi.1998.2454. [DOI] [PubMed] [Google Scholar]
- 35.Shilton BH, Flocco MM, Nilsson M, Mowbray SL. Conformational changes of three periplasmic receptors for bacterial chemotaxis and transport: the maltose-, glucose/galactose- and ribose-binding proteins. J. Mol. Biol. 1996;264:350–363. doi: 10.1006/jmbi.1996.0645. [DOI] [PubMed] [Google Scholar]
- 36.Förster T. Intermolecular energy migration and fluorescence. Ann. Phys. 1948;2:55–75. [Google Scholar]
- 37.Van Der Meer BW, Cooker GI, Chen SYS. Resonance Energy Transfer: Theory and Data. New York: VCH Publishers; 1994. [Google Scholar]
- 38.Fehr M, Takanaga H, Ehrhardt DW, Frommer WB. Evidence for high-capacity bidirectional glucose transport across the endoplasmic reticulum membrane by genetically encoded fluorescence resonance energy transfer nanosensors. Mol. Cell. Biol. 2005;25:11102–11112. doi: 10.1128/MCB.25.24.11102-11112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arion WJ, Canfield WK. Glucose-6-phosphatase and type 1 glycogen storage disease: some critical considerations. Eur. J. Pediatr. 1993;152 Suppl. 1:S7–S13. doi: 10.1007/BF02072080. [DOI] [PubMed] [Google Scholar]
- 40.Kerr R, Lev-Ram V, Baird G, Vincent P, Tsien RY, Schafer WR. Optical imaging of calcium transients in neurons and pharyngeal muscle of C. elegans. Neuron. 2000;26:583–594. doi: 10.1016/s0896-6273(00)81196-4. [DOI] [PubMed] [Google Scholar]
- 41.Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Meth. Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.