Abstract
Genome-wide association studies (GWAS) have identified seven breast cancer susceptibility loci, but these explain only a small fraction of the familial risk of the disease. Five of these loci were identified through a two-stage GWAS involving 390 familial cases and 364 controls in the first stage, and 3,990 cases and 3,916 controls in the second stage1. To identify additional loci, we tested over 800 promising associations from this GWAS in a further two stages involving 37,012 cases and 40,069 controls from 33 studies in the CGEMS collaboration and Breast Cancer Association Consortium. We found strong evidence for additional susceptibility loci on 3p (rs4973768: per-allele OR = 1.11, 95% CI = 1.08–1.13, P = 4.1 × 10−23) and 17q (rs6504950: per-allele OR = 0.95, 95% CI = 0.92–0.97, P = 1.4 × 10−8). Potential causative genes include SLC4A7 and NEK10 on 3p and COX11 on 17q.
Genome-wide association studies (GWAS) have been successful at identifying many disease susceptibility loci, including several for common cancers. We recently conducted a multistage GWAS based on 390 breast cancer cases with a strong family history of the disease and 364 controls in the first stage, and 3,990 cases and 3,916 controls in the second stage. We then genotyped the 30 most significant SNPs in a third stage involving 21,860 cases and 22,578 controls from 22 studies in the Breast Cancer Association Consortium (BCAC; see URLs section in Methods). Through this combined analysis, we identified five loci with strong statistical evidence of association1. One of these loci, FGFR2, was also identified in a second scan2, and additional susceptibility loci on 2q, 5p and 6q have been identified in subsequent scans3–5. Together, these loci explain an estimated 5.4% of the known familial aggregation of breast cancer, suggesting strongly that further loci remain to be identified.
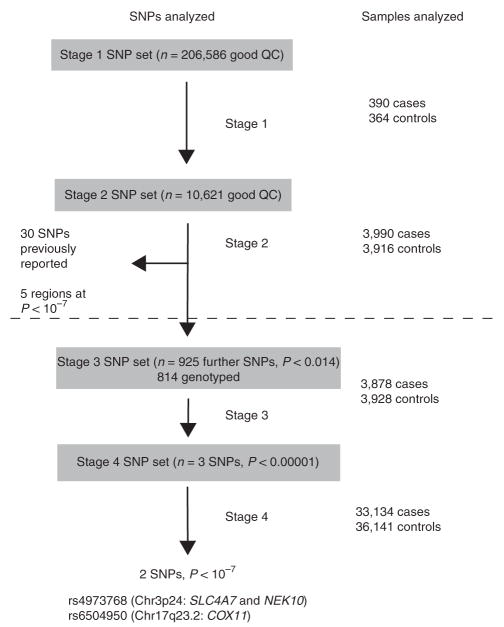
In an attempt to identify further loci at which common variants are associated with breast cancer risk, we conducted a more comprehensive evaluation of promising associations from our GWAS (Fig. 1). We identified a further 925 SNPs that showed evidence for association in the first two stages of our study (combined P trend <0.014) and attempted to genotype them in a third stage, involving a further 3,878 cases and 3,928 controls from three studies corresponding to stage 2 of the Cancer Genetic Markers of Susceptibility (CGEMS) collaboration. We successfully genotyped 814 of these SNPs as part of a 30,278 SNP custom Illumina iSelect array. After combination of these data with the original GWAS data, three SNPs had P values <10−5 (rs4973768, rs4132417, rs6504950). We then evaluated these SNPs in a fourth stage, using data from a further 27 studies in BCAC. We also incorporated data from two further studies contributing to the Cancer Genetic Markers of Susceptibility (CGEMS) collaboration2 and, for rs4973768, data from 1,143 cases and 1,141 controls obtained as part of the CGEMS GWAS2. In total, 36,141 controls and 33,134 cases of invasive breast cancer were genotyped as part of stage 4.
Figure 1.
Summary of the study design. Stages 1 and 2 are as reported in Easton et al.1. Stages 3 and 4, below the dotted line, are the stages conducted in this follow-up study.
One SNP, rs4973768, showed clear evidence of association in the stage 4 replication (Table 1 and Fig. 2; per-allele OR = 1.11, 95% CI = 1.08–1.13, P = 1.4 × 10−18) and overall (P = 4.1 × 10−23). A second SNP, rs6504950, also showed evidence of replication and reached ‘genome-wide’ significance overall (Table 1 and Fig. 2; per-allele OR = 0.95, 95% CI = 0.92–0.97, P = 0.00010 in stage 4; P = 1.4 × 10−8 overall). There was no evidence of heterogeneity in the OR estimates among studies in stage 4 for either SNP. For both SNPs, the per-allele OR was very similar in populations of European and Asian descent (rs4973768: 1.11, 95% CI = 1.00–1.23 in Asians versus 1.11, 1.08–1.14 in Europeans; rs6504950: 0.96, 0.82–1.12 in Asians versus 0.95, 0.93–0.98 in Europeans; Fig. 2), and were similar between hospital-based and population-based case-control studies. rs4132417 showed no evidence of association in the replication (per-allele OR = 1.00, 95% CI = 0.97–1.03, P = 0.97 in stage 4, P = 0.016 overall) and is therefore likely to have been a false positive association in stages 1–3.
Table 1.
Estimated odds ratios and tests of association for rs4973768 and rs6504950
| Marker (chromosome, position) | Allelesa | Stage (cases/controls) | MAFb | Per-allele OR (95% CI)c | Heterozygote OR (95% CI)d | Homozygote OR (95% CI)e | P trend |
|---|---|---|---|---|---|---|---|
| rs4973768 (3p24, 27391017) | Stage 1 (388/355) | 0.46 | 1.33 (1.07–1.64) | 1.45 (1.01–2.07) | 1.76 (1.15–2.68) | 0.0087 | |
| C/T | Stage 2 (3,951/3,870) | 0.47 | 1.06 (0.99–1.03) | 0.99 (0.89–1.10) | 1.13 (0.99–1.28) | 0.081 | |
| Stage 3 (3,872/3,925) | 0.48 | 1.13 (1.06–1.20) | 1.03 (0.93–1.15) | 1.27 (1.12–1.44) | 0.00025 | ||
| Stage 4 (30,256/34,063) | 0.46 (0.21) | 1.11 (1.08–1.13) | 1.12 (1.08–1.17) | 1.23 (1.17–1.29) | 1.4 × 10−18 | ||
| Combined | 4.1 × 10−23 | ||||||
| rs6504950 (17q23, 50411470) | G/A | Stage 1 (390/357) | 0.31 | 0.76 (0.61–0.96) | 0.83 (0.61–1.13) | 0.52 (0.31–0.89) | 0.018 |
| Stage 2 (3,976/3,894) | 0.29 | 0.90 (0.84–0.96) | 0.86 (0.78–0.94) | 0.86 (0.73–1.02) | 0.0020 | ||
| Stage 3 (3,870/3,923) | 0.28 | 0.91 (0.85–0.98) | 0.89 (0.81–0.97) | 0.88 (0.73–1.04) | 0.012 | ||
| Stage 4 (30,470/33,302) | 0.27 (0.08) | 0.95 (0.92–0.97) | 0.96 (0.92–0.99) | 0.89 (0.83–0.95) | 0.00010 | ||
| Combined | 1.4 × 10−8 |
Major/minor allele (+ strand), based on the allele frequency in Europeans.
Minor allele frequency in Europeans (minor allele frequency in Southeast Asians in stage 4 shown in brackets).
Odds ratios (OR) per copy of the minor allele.
OR for heterozygotes relative to homozygotes for the common allele.
OR for homozygotes for the rare allele, relative to homozygotes for the common allele.
Figure 2.
Forest plots of the per-allele OR by study for rs4973768 (left) and rs6504950 (right). Squares represent the estimated OR for each study. Lines indicate the 95% confidence interval. Diamonds represent the OR estimates and confidence limits for the subgroups indicated.
rs4973768 showed clear evidence of an increasing risk with number of rarer (T) alleles, with an estimated OR = 1.12 (95% CI = 1.08–1.17) in heterozygotes and 1.23 (1.17–1.29) in homozygotes for the T allele (Table 1). There was some suggestion of a trend in OR by age, with a higher OR below age 50 y (Ptrend = 0.038; Supplementary Table 1 online). The per-allele OR was higher for ER-positive (per-allele OR = 1.12, 95% CI = 1.09–1.16) than for ER-negative breast cancer (OR = 1.06, 1.01–1.12; P = 0.022 for heterogeneity in the OR by ER status; Supplementary Table 2 online), consistent with a pattern observed for several other breast cancer susceptibility loci, notably FGFR2 and the 8q24 locus6. Contrary to the pattern seen for other susceptibility loci, there was no evidence of an association with a positive family history of breast cancer (Supplementary Table 3 online). However, the number of cases with a positive family history was limited, and the effect predicted under a multiplicative polygenic model (an approximately 50% greater effect in women with a family history, or per-allele OR = 1.16) could not be clearly excluded in this analysis. rs6504950 also showed a stronger association in ER-positive disease (OR = 0.94, 95% CI = 0.91–0.97) versus ER-negative disease (OR = 1.03, 0.98–1.09; P = 0.00078 for heterogeneity in the OR by ER status), but no association with age or family history.
In addition to the three SNPs above, we identified a further 13 SNPs that were significant at P < 10−4 (but not P < 10−5) after stages 1–3. We evaluated these associations using a further 3,777 cases and 4,171 controls from three additional studies (Supplementary Table 4 online). Only one SNP, rs1357245, showed evidence of association in this replication study, in the same direction as the original association (P = 0.0010; P = 1.9 × 10−7 overall). Notably, this SNP lies in the same 600-kb linkage disequilibrium (LD) block as rs4973768 on 3p and is correlated with it (r2 = 0.58).
To further refine the evidence for association in this 3p24 region, we identified all SNPs within the LD block that were correlated with either rs4973768 at r2 > 0.2 or rs1357245 at r2 > 0.3 according to the HapMap CEU (Caucasians of European descent from Utah) data. These SNPs could be tagged with a set of 28 SNPs (minimum r2 = 0.8; Fig. 3a). We genotyped these 28 SNPs in 2,301 cases and 2,256 controls from the UK SEARCH study (Supplementary Table 5 online). In forward stepwise logistic regression analysis, the strongest marker was rs2307032, and no SNP provided a significant improvement in fit after adjustment for rs2307032. rs2307032 is correlated with both rs4973768 and rs1357245 (r2 = 0.45 and 0.39, respectively). Haplotype analysis identified two common haplotypes (carrying the same alleles at rs2307032, rs4973768 and rs1357245) associated with disease risk (haplotypes B and J in Supplementary Table 6 online). These results suggest that the association with SNPs in this region may be driven by a single common variant correlated with rs2307032, rs4973768 and rs1357245. However, full resequencing of the region and genotyping in larger case-control studies will be required to provide clear evidence as to the likely causal variant(s).
Figure 3.
Maps of associated regions on 3p24 and 17q23.2. (a) HapMap CEU–derived ~ 600-kb LD block around the 3p loci illustrating all the common SNPs (MAF 0.05). Extent of block delimited by dotted lines. Squares on the LD block indicate the correlation (r2) between SNP genotypes on a greyscale (darker squares = higher correlations). Approximate size and transcripts of NEK10 (chromosome positions 2712398–27307988) and SLC4A7 (chromosome positions 27389218–27473249) inferred from the NCBI reference assembly (solid arrow) and Ensembl (dashed arrow) are indicated. Genotyped tag SNPs are marked by circles: black filled circles show SNPs significantly (P < 0.05) associated with breast cancer in the SEARCH study; black-outline-only circles show tag SNPs not associated with disease. The most strongly associated tag SNPs, as discussed in the text, are marked by stars (1, rs1357245; 2, rs4973768; 3, rs2307032). (b) HapMap CEU–derived ~ 300-kb LD block on 17q tagged rs6504950 (marked by star). Extent of block delimited by dotted lines. The approximate size and transcripts associated with the three known nearby genes COX11, TOM1L1 and STXBP4 as inferred by NCBI are indicated.
The associated region on 3p24 contains two known genes, NEK10 and SLC4A7. NEK10 (Never-in mitosis related kinase 10) is one of a family of 11 NIMA (never in mitosis a) related kinases that are involved in cell cycle control7. No function has been ascribed to NEK10, but NEK2, NEK6, NEK7 and NEK9 seem to be involved in regulation of mitosis, whereas NEK1 and NEK8 have been associated with polycystic kidney disease8. SLC4A7 (solute carrier family 4, sodium bicarbonate cotransporter, member 7) is a potential tyrosine kinase substrate that has been shown to have reduced expression in breast tumor sections and cell lines9. The protein is located in the cell membrane and has been predicted to affect the pH of the micro-environment around breast tumor cells9.
rs6504950 lies in a 300-kb LD block on 17q23.2 (Fig. 3b). The SNP itself lies in intron 1 of STXBP4 (syntaxin binding protein 4), an insulin-regulated STX4-binding protein involved in the control of glucose transport and GLUT4 vesicle translocation10. Other genes in the block include COX11 (cytochrome C assembly protein 11, approximately 10 kb upstream of rs6504950) and TOM1L1 (target of myb1-like1). Of interest, the risk allele of rs6504950 is associated with higher levels of COX11 expression in lymphocytes in the HapMap samples (P = 0.000014)11, but not with expression levels of either STXBP4 or TOM1L1.
Given the OR and allele frequency estimates for European populations, rs4973768 would explain approximately 0.4% of the familial risk of breast cancer, and rs6504950 would explain approximately 0.07% (although the true strength of the associations at these loci might be stronger if the causal variant(s) are not in strong LD with the marker SNP). Taking these together with previously identified loci, we estimate the fraction of the familial risk explained by all known common susceptibility alleles to be 5.9%.
This analysis emphasizes that follow-up of tentative associations in GWAS through large replication studies (such as the ~ 40,000 cases and ~ 40,000 controls in the current study) can reliably identify additional susceptibility loci. However, the power to have detected these associations with this strategy was still limited (37% for rs4973768, and less than 1% for rs6504960, assuming a perfect tag in the initial scan), suggesting that other breast cancer loci should be detectable by further large GWAS, together with combined analyses of GWAS and large-scale replication.
METHODS
Study design
Subjects, genotyping methods and analysis of the stages 1 and 2 of the GWAS have been described previously1. The studies that participated in stages 3 and 4 are summarized in Supplementary Table 7 online. Stage 3 comprised three studies participating in phase 2 of the CGEMS collaboration. Stage 4 comprised 27 studies from BCAC, two further studies from CGEMS phase 3 and data from the NHS obtained from the CGEMS GWAS. BCAC studies provided individual-level data on disease status (invasive breast cancer case, carcinoma-in-situ case or control), age at diagnosis or interview, ancestry group, first-degree family history of breast cancer and bilaterality of breast cancer. Twenty-one studies provided data on estrogen receptor (ER) status of the primary tumor. CGEMS studies provided summary-level data on disease status and (for five studies) ER status of the tumor. All but two studies (TWBCS and SEBCS) were conducted in Europe, North America or Australia and were comprised primarily of subjects of European ancestry. In this analysis, subjects identified as belonging to minority ancestry groups (non-Thai for TWBCS, non-Korean for SEBCS, non-European for other studies) by questionnaire or genotyping were excluded.
Genotyping
Genotyping for stage 3 was conducted using a custom-designed Illumina iSelect array, as part of the replication phase 2 of CGEMS. Twenty-eight studies in stage 4 performed genotyping as part of BCAC (genotyping round VII). Twenty-seven studies genotyped the SNPs using a 5′ endonuclease assay (Taqman), using reagents supplied by Applied Biosystems and tested centrally. Five studies genotyped SNPs using MALDI-TOF mass spectrometry using Sequenom’s MassARRAY system and iPLEX technology. Each study also provided genotypes for at least 2% of samples in duplicate, genotypes for a standard test plate (94 samples) and sample cluster plots. We excluded individuals that failed on two or more SNPs, or 20% of the total if more than ten SNPs were typed by that study. We excluded the data on a SNP for a given study that failed to achieve prespecified quality control criteria: these included an overall call rate of > 95%, duplicate concordance and concordance of test plate genotypes of > 98%, and no evidence of deviation from Hardy-Weinberg equilibrium at P < 0.005. Two further studies (NOR and RADT) were genotyped as part of CGEMS replication phase 3, using Taq-man. Data on the NHS were taken from their GWAS, conducted using an Illumina Infinium 550k array (these data were included in stage 4 since they were not used in the analysis of stage 3 and the selection of the three SNPs for stage 4)2.
Analysis
Analyses were based on the risk of invasive breast cancer (cases of carcinoma-in-situ were also genotyped in stage 4 but are not reported here). Odds ratios (ORs) were estimated using unconditional logistic regression, adjusted for study. The ORs quoted in the text are based on the final replication phase (stage 4), as these will be least affected by ‘winner’s curse’. Significance levels were based on the Mantel extension test, stratified by study. Significance levels for stage 4 only and for all stages combined are emphasized in the text. In the latter, scores from stage 1 were given a weight of 2 to allow for the selection of cases for a strong family history, consistent with previous analyses1. Differences in the SNP associations by ER status were assessed using multivariate logistic regression, allowing a three-level outcome (ER-positive, ER-negative and control), and testing for the difference in the risk estimates for ER-positive versus ER-negative disease using a likelihood ratio test. The effect of family history was assessed using an equivalent test. Modification by age at diagnosis was tested by fitting a SNP by age-group interaction term in a logistic regression model. To estimate the power to detect each of the associations found, we computed the noncentrality parameter for the test statistic at each stage using the per-allele relative risk and allele frequency. This was used to estimate power on the basis of a simulated tetravariate normal distribution for the score statistics after each stage to allow for the correlations in the test statistics. We assumed significance thresholds of P < 0.05, P < 0.014, P < 10−5 and P < 10−7 after stages 1–4.
URLs
Breast Cancer Association Consortium, http://www.srl.cam.ac.uk/consortia/bcac/.
Supplementary Material
Acknowledgments
The initial GWAS, SEARCH, the replication genotyping through BCAC and the main analysis of this study were supported by Cancer Research UK grants C1287/A7497, C490/A11021, C1287/A10118 and C1287/A5260. D.F.E. and P.D.P.P. are supported by Cancer Research UK. Meetings of BCAC have been supported in part by ESF COST action BM0606.
The Nurses’ Health Studies are supported by US National Institutes of Health grants CA65725, CA87969, CA49449, CA67262, CA50385 and 5UO1CA098233. The WHI program is supported by contracts from the National Heart, Lung, and Blood Institute, NIH. We thank the WHI investigators and staff for their dedication and the study participants for making the program possible. A full listing of WHI investigators can be found at http://www.whiscience.org/publications/WHI_investigators_shortlist.pdf. The ACS study is supported by UO1 CA098710. We thank C. Lichtman for data management and the participants on the CPS-II. The PLCO study is supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics and contracts from the Division of Cancer Prevention, National Cancer Institute, NIH and DHHS. We thank P. Prorok, Division of Cancer Prevention, National Cancer Institute, the Screening Center investigators and staff of the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO) and T. Sheehy and staff at SAIC-Frederick. We acknowledge the study participants for their contributions to making this study possible. The ABCFS was supported by the National Health and Medical Research Council (NHMRC) of Australia (#145604), the US NIH (RO1 CA102740-01A2) and by the National Cancer Institute, NIH under RFA #CA-95-011 through cooperative agreements with members of the Breast Cancer Family Registry (Breast CFR) and principal investigators from Cancer Care Ontario (UO1 CA69467), Columbia University (U01 CA69398), Fox Chase Cancer Center (U01 CA69631), Huntsman Cancer Institute (U01 CA69446), Northern California Cancer Center (U01 CA69417) and University of Melbourne (U01 CA69638). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of collaborating centers in the Breast CFR, nor does mention of trade names, commercial products or organizations imply endorsement by the US government or the Breast CFR. The ABCFS was initially supported by the NHMRC, the New South Wales Cancer Council and the Victorian Health Promotion Foundation. J.L.H. and M.C.S. are supported by NHMRC. We thank M. Angelakos, J. Maskiell and G. Dite. We thank X. Chen for genotyping the ABCFS, kConFab and AOCS samples, and H. Thorne, E. Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics and the Clinical Follow Up Study (funded by NHMRC grants 145684, 288704 and 454508) for their contributions to this resource, and the many families who contribute to kConFab. kConFab is supported by grants from the National Breast Cancer Foundation, the National Health and Medical Research Council (NHMRC) and by the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. The AOCS Management Group (D. Bowtell, G.C.-T., A. deFazio, D. Gertig, A. Green and P. Webb) gratefully acknowledges the contribution of all the clinical and scientific collaborators (see http://www.aocstudy.org/). The ACS Management Group (A. Green, P. Parsons, N. Hayward, P. Webb, D. Whiteman) thank all of the project staff, collaborating institutions and study participants. Financial support was provided by US Army Medical Research and Materiel Command under DAMD17-01-1-0729, the Cancer Council Tasmania and Cancer Foundation of Western Australia (AOCS study) and The National Health and Medical Research Council of Australia (199600) (ACS study). G.C.-T. is supported by NHMRC. Funding of the ABCS study was provided by the Dutch Cancer Society (grants NKI 2001-2423; 2007-3839) and the Dutch National Genomics Initiative. ABCS acknowledges L. Braaf, L. Van’t Veer, F. Van Leeuwen, R. Tollenaar and other contributors to the ‘BOSOM’ study and the support of H.B. Bueno-de-Mesquita for organizing the release of control DNA. The British Breast Cancer Study is funded by Cancer Research UK and Breakthrough Breast Cancer. We acknowledge NHS funding to the NIHR Biomedical Research Centre, and the National Cancer Research Network (NCRN). The CGPS was supported by the Danish Medical Research Council and Copenhagen County. The CNIO-BCS was supported by the Genome Spain Foundation and grants from the Asociación Española Contra Cáncer and the Fondo de Investigación Sanitario (PI081120 and PI081583). We thank J.I. Arias from the Hospital Monte Naranco, P. Zamora from the Hospital La Paz and C. Alonso and T. Moreno from the CNIO. The GENICA study was supported by the German Human Genome Project and funded by the Federal Ministry of Education and Research (BMBF) Germany grants 01KW9975/5, 01KW9976/8, 01KW9977/0 and 01KW0114. Genotyping analysis was supported by the Robert Bosch Foundation of Medical Research. B. Pesch, V. Harth and T. Brüning were involved in the recruitment of study subjects and responsible for the collection of epidemiological data. The Genetic Epidemiology Study of Breast Cancer by Age 50 (GESBC) was supported by the Deutsche Krebshilfe e.V. (project number 70492) and the genotyping in part by the state of Baden-Württemberg through Medical Faculty of the University of Ulm (P.685). We thank U. Eilber and T. Koehler for their technical support. We cordially thank M. Bremer, A. Scharf and C. Sohn for their support of the breast cancer studies in Hannover. HABCS and HMBCS received funding through a Hannelore-Munke stipend to N.V.B. HUBCS was supported by a grant from the German Federal Ministry of Education and Research (RUS 08/017). HEBCS wishes to thank H. Jäntti and K. Aaltonen for help with the subject data. The Finnish Cancer registry is gratefully acknowledged for the cancer data. The HEBCS study has been financially supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (110663), Finnish Cancer Society and the Sigrid Juselius Foundation. KARBAC was supported by The Swedish Cancer Foundation, The Gustav V Jubilee Foundation and the Bert von Kantzow Foundation. KBCP is grateful to E. Myöhänen for her assistance. KBCP was supported by special Government Funding (EVO) of Kuopio University Hospital, The Finnish Cancer Society, University of Kuopio and by the Northern Savo Cancer Society. MCBCS was supported by grants from the NIH (P50 CA116201 and R01 CA122340). We thank all the participants in the MCCS and the team of investigators, project and data managers and project assistants. The MCCS is supported by the Australian National Health and Medical Research Council (grants 209057, 251533, 396414 and 504711). Cohort recruitment and follow up is funded by The Cancer Council Victoria. The OFBCR was supported by the National Cancer Institute, NIH under RFA #CA-06-503 and through cooperative agreements with members of the Breast Cancer Family Registry and Cancer Care Ontario (U01 CA69467). We thank N. Weerasooriya, M. Gill, L. Collins and N. Gokgoz for their assistance. The ORIGO study was supported by the Dutch Cancer Society; we thank P.E.A. Huijts, E. Krol-Warmerdam and J. Blom for recruiting subjects, administering questionnaires and managing clinical information. RBCS was supported by the Dutch Cancer Society. SASBAC study was supported by funding from the Agency for Science, Technology and Research of Singapore (A*STAR), the US NIH and the Susan G. Komen Breast Cancer Foundation. The SBCS was supported by Yorkshire Cancer Research and the Breast Cancer Campaign. We thank S. Higham, H. Cramp, D. Connley and S. Balasubramanian for their contribution to the SBCS. The PBCS was funded by Intramural Research Funds of the US National Cancer Institute, DHHS. The PBCS thanks N. Szeszenia-Dabrowska, B. Peplonska, W. Zatonski, M. Sherman and P. Chao for their valuable contributions to the study. SEBCS was supported by a grant of the Korea Health 21 R&D Project, Ministry of Health & Welfare (R.O.K.) (AO30001) and by a grant from the National R&D program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (0620410-1). The UCIBCS component of this research was supported by the US NIH, National Cancer Institute grants CA-58860 and CA-92044 and the Lon V. Smith Foundation grant LVS-39420.
Footnotes
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
D.F.E., A.M.D., P.D.P.P. and B.A.J.P. designed the study and obtained financial support. D.F.E. and P.D.P.P. conducted the statistical analysis. G.T., R.N.H., D.J.H. and S.J.C. directed the CGEMS study and designed and conducted the stage 3 experiment with D.F.E., S.A., M.G., C.S.H. and M.M. conducting the fine-scale mapping. M.K.H., J.M. and R.L. provided bioinformatics support. D.E., D.G.E., O.F., N.J., I.d.S.S., J.P., M.R.S. and N.R. co-ordinated the studies used in stage 1. The remaining authors coordinated the studies in stage 4 and/or undertook genotyping in those studies. D.F.E. drafted the manuscript, with substantial input from other authors. All authors contributed to the final paper.
References
- 1.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stacey SN, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 4.Stacey SN, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40:703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 5.Zheng W, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Closas M, et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin MJ, Shao L, Voehringer D, Smeal T, Jallal B. The serine/threonine kinase Nek6 is required for cell cycle progression through mitosis. J Biol Chem. 2003;278:52454–52460. doi: 10.1074/jbc.M308080200. [DOI] [PubMed] [Google Scholar]
- 8.Quarmby LM, Mahjoub MR. Caught Nek-ing: cilia and centrioles. J Cell Sci. 2005;118:5161–5169. doi: 10.1242/jcs.02681. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, et al. Differential expression of novel tyrosine kinase substrates during breast cancer development. Mol Cell Proteomics. 2007;6:2072–2087. doi: 10.1074/mcp.M700395-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Min J, et al. Synip: a novel insulin-regulated syntaxin 4-binding protein mediating GLUT4 translocation in adipocytes. Mol Cell. 1999;3:751–760. doi: 10.1016/s1097-2765(01)80007-1. [DOI] [PubMed] [Google Scholar]
- 11.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.