Abstract
Cell cycle plays a crucial role in regulating the pathway used to repair DNA double-strand breaks (DSBs). In Saccharomyces cerevisiae, homologous recombination is primarily limited to non-G1 cells as the formation of recombinogenic single-stranded DNA requires CDK1-dependent 5′ to 3′ resection of DNA ends. However, the effect of cell cycle on non-homologous end joining (NHEJ) is not yet clearly defined. Using an assay to quantitatively measure the contributions of each repair pathway to repair product formation and cellular survival after DSB induction, we found that NHEJ is most efficient at G1, and markedly repressed at G2. Repression of NHEJ at G2 is achieved by efficient end resection and by the reduced association of core NHEJ proteins with DNA breaks, both of which depend on the CDK1 activity. Importantly, repression of 5′ end resection by CDK1 inhibition at G2 alone did not fully restore either physical association of Ku/Dnl4-Lif1 with DSBs or NHEJ proficiency to the level at G1. Expression of excess Ku can partially offset the inhibition of end joining at G2. The results suggest that regulation of Ku/Dnl4-Lif1 affinity for DNA ends may contribute to the cell cycle-dependent modulation of NHEJ efficiency.
Keywords: Double strand break, End joining, Repair choice, Cell cycle
1. Introduction
Mutations, especially gross chromosome rearrangements, are frequent in cells from cancer patients, and often the consequence of failed and/or improper repair of DNA lesions (reviewed in [1]). Among DNA lesions, double-strand breaks (DSBs) pose the most serious challenge as the un-repaired DSB is highly mutagenic and frequently lethal. Two highly efficient repair pathways have evolved to deal with this type of lesion: homologous recombination and non-homologous end joining (NHEJ) [2,3]. Homologous recombination (HR) uses an undamaged template, often found in the sister chromatid or homologous chromosome to restore chromosome integrity [3]. NHEJ, in contrast, restores chromosome integrity by simple ligation of the broken chromosome ends back together following appropriate end processing [4,5]. Because end processing, catalyzed by a collection of polymerases and nucleases, inevitably alters terminal sequences, NHEJ is often considered more mutagenic than HR [4,6]. However, HR between ectopic templates can in principle lead to chromosome rearrangement type mutations [7]. The prudent choice of repair pathway will thus greatly reduce the mutation rate and improve the overall cellular fitness [3]. The fact that specific types of DSB repair such as V(D)J recombination, class switch recombination (CSR) and meiotic recombination exclusively rely on one of these two repair pathways also lends additional support to cellular regulation of repair pathway choice [8-10]. How a cell imposes the use of a specific repair pathways has begun to emerge with a finding of a role of the post-cleavage Rag1/Rag2 complex during V(D)J recombination [9] and end processing in yeast [11,12].
The cell cycle has long been recognized as a crucial factor for determining repair pathway choice [3]. Early studies in vertebrates showed that NHEJ-deficient scid cells and ku70-/- chicken DT40 cells were hypersensitive to ionizing radiation only during G1 and early S phase of the cell cycle, whereas HR-defective rad54-/- cells were radiation sensitive at late S/G2 [13,14]. In the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, recombination is also tightly linked to cell cycle such that recombination between sister chromatids predominates at S/G2 [11,12,15,16]. When sister chromatids are unavailable in G1, recombination is minimized to prevent the loss of heterozygosity or chromosome rearrangements [17]. Recombination in yeast is suppressed at G1 because end resection, the key early step that produces the 3′ single-stranded DNA (ssDNA) essential for strand invasion [18], depends on the Cdk1 kinase activity [11,12,16]. Recently, phosphorylation of Sae2 or its mammalian counterpart CtIP, at its carboxyl terminus by Cdk1 has been implicated in cell cycle dependent regulation of DSB processing and hence repair choice during the cell cycle [19,20]. The strand invasion step during gene conversion, where Rad51-coated presynaptic filaments search and anneal to the homologous template, is also controlled by the cell cycle [11]. Finally, cell cycle controls repair pathway choice by modulating expression of recombination proteins. In mammals, the expression level of Rad51 and Rad52 is the lowest at G0/G1 and gradually increases during S/G2/M [21], and in S. pombe, the resection facilitator Ctp1 protein (Sae2 in S. cerevisiae) is not expressed at G1 [22]. An equivalent expression pattern of Sae2 during the cell cycle is not detected in S. cerevisiae [23], however, suggesting that this type of regulation might be species specific.
While evidence has accumulated for cell cycle-dependent regulation of HR, it is not clear whether the cell cycle has a similar role in regulating NHEJ. In fact, considerable uncertainty exists as to the role of NHEJ in DSB repair during the S and G2 phases of the cell cycle. DNA ends with extensive 5′ degradation would not likely be favorable substrates for NHEJ [11,24,25], suggesting that Cdk1-dependent resection of DNA ends might suppress NHEJ at late S/G2. Alternatively, efficient HR at S/G2 may simply out-compete NHEJ and cells may channel DNA breaks preferentially to HR despite the lack of a clear decline in NHEJ efficiency at this cell cycle stage [26,27].
In this study, we examined the effect of cell cycle on the repair of DNA breaks by NHEJ and described a biochemical basis for NHEJ suppression at S/G2 that operates independently of HR events. We have uncovered a role of Cdk1 in opposing the association of Ku/Dnl4-Lif1 with DNA breaks and thus in discouraging commitment to NHEJ at S/G2. Cdk1-dependent inhibition of NHEJ factor recruitment at DNA breaks is distinct from Cdk1’s role in recombination and end processing, and represents a novel mode of pathway choice control for DSB repair.
2. Materials and Methods
2.1. Strains
Strains used in these studies are listed in Table 1. JKM161 and its mutant derivatives bearing a single HMLα donor were used to measure HR and NHEJ efficiency, while donorless JKM179 derivatives were used to detect resection and the enrichment of repair proteins at a DNA break by ChIP. Yeast cell synchrony was achieved by incubation with 10 μg/ml of α factor (G1) or 15 μg/ml of nocodazole (G2) for 4 h prior to HO induction, and confirmed by FACS analysis.
Table 1. Genotypes of strains used in the study.
| Strain | Genotype | Reference |
|---|---|---|
| JKM161 | hoΔ MATaHMLalpha hmrΔ::ADE1 ade1-100 leu2-3,112 trp1::hisG’ lys5 ura3-52 ade3::GAL::HO | [28,30] |
| JKM179 | hoΔ MATahmlΔ::ADE1 hmrΔ::ADE1 ade1-100 leu2-3,112 trp1::hisG’ lys5 ura3-52 ade3::GAL::HO | [28] |
| CY6012a | JKM179 GAL::SIC1-myc | [11] |
| tGI561 | JKM179 cdc28-as1 | [11] |
| YZY46 | JKM161 yku70Δ::URA3 | [28] |
| YZY51 | JKM161 rad52Δ::TRP1 | This work |
| YZY105 | JKM179 LIF1-MYC::KAN | [28] |
| YZY193 | JKM179 cdc28-as1 LIF1-MYC::KAN | This work |
| YZY194 | JKM179 GAL::SIC1-myc LIF1-MYC::KAN | This work |
| SLY205 | JKM179 pKu70, pKu80 | This work |
2.2 Survival rate determination and detection of repair products
HO breaks were induced by addition of galactose [final conc. 2% (w/v)] to cells grown in YEP-glycerol pre-induction media. Following 1 h incubation in the galactose-containing medium, cells were plated onto YEP-dextrose plates and the number of colonies was counted after 3 days at 30°C. The survival frequency was calculated by normalization with the number of colonies obtained without galactose addition. To detect HR or NHEJ products, genomic DNA extracted from time course samples was digested with EcoRV, separated by an agarose gel (1%), and transferred to nylon membrane (Hybond N+). The blot was probed with a 1 kb 32P-labeled MAT distal probe that recognizes the 3.4 kb (MATα) or the 5.0 kb (MATa) fragment containing the HO cleavage site on yeast chromosome III. The levels of HR and NHEJ repair products were determined after normalization to a HIS3 sequence that served as an internal loading control.
2.3. Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as described previously [28]. After immunoprecipitation and crosslink reversal, purified DNA was analyzed by real time quantitative PCR using multiple sets of primers that anneal 0.2-kb, 1-kb, and 5-kb from the DSB, as well as primers specific for the PRE1 gene situated on chromosome V as a control. The antibodies for RPA were a generous gift from Dr. S. Brill.
2.4. Ligation mediated PCR assay
Ligation mediated PCR was performed as described [25], except that real time quantitative PCR was used instead of radiolabeled PCR. Briefly, genomic DNA was extracted by a standard glass bead protocol, and subjected to ligation with a linker containing a 4 base overhang complementary to the distal side of the HO cut site. Only the unprocessed ends could be ligated with the linker. PCR was carried out using a pair of primers recognizing DNA sequence distal to the HO site and the adaptor.
2.5. Immuno blot
Whole cell extracts were prepared as described [29]. Proteins were separated by SDS-PAGE and transferred to PVDF membrane. The TAP fusion proteins were detected by Peroxidase-Anti-Peroxidase (PAP) soluble complex (Sigma). Phosphorylation of the B subunit of DNA polymerase α, a marker for Cdk1/Clb activity, was detected by monoclonal antibody 6D2 [11](a gift from Dr. Achille Pellicioli).
3. Results
3. 1. NHEJ is repressed at G2
To assess the effect of cell cycle on DSB repair, a DSB was induced at the MAT locus of strain JKM161 using a galactose-inducible HO gene integrated at the ADE3 locus. Because this strain lacks HMRa, HMLα is the sole template for the repairing the HO break (Fig. 1a)[28,30]. In JKM161, repair of HO-induced break by HR is associated with mating type switching from a to α, while NHEJ will retain the mating type as a. HO cleavage is very efficient (>99% after 1 h induction) and almost all survivors repair the DSB by one of the two repair mechanisms [28,30].
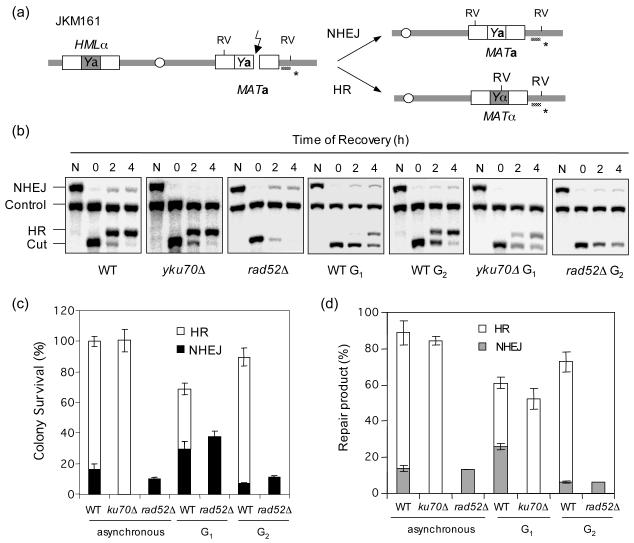
Figure 1.
NHEJ is repressed at G2. (a) Schematic diagram showing relevant genomic structure of JKM161. The location of a MAT specific probe (*) and the restriction endonuclease cleavage sites (EcoRV: RV) used for Southern blot analysis to detect repair product formation are indicated. (b) Southern blot analysis of the repair product formation. N represents no galactose. (c) Plot demonstrating percent colony survival by HR and NHEJ. (d) Plot demonstrating percentage of repair products by HR and NHEJ. Percent repair product was calculated by dividing the repair product signal (HR or NHEJ in (b)) with the signal of the HIS3 control (control) after 4 h of recovery in the glucose containing medium. Data represents the mean ± s.d. of three or more independent experiments.
When HO was induced for 1 h in logarithmically growing wild type cells, almost all cells survived the break. Among survivors, 85% repaired the break by HR (α mating type), whereas 15% repaired the break by NHEJ (a mating type) (Fig. 1c). Deletion of RAD52 completely eliminated HR leaving only ∼10% survivors, all of which were repaired by NHEJ (a mating type). In contrast, deletion of YKU70 had no influence on the survival frequency apparently because more survivors form by HR (switched to α type), consistent with the previous reports that HR is enhanced in the absence of Yku70 [28,31-33](Fig. 1c). We also used Southern blot hybridization to measure the amount of repair product generated as a function of time after HO expression (Figs. 1b, 1d). The amount of HR and NHEJ products in the Southern blot analysis was in an excellent agreement with the survival data in wild type, yku70Δ and rad52Δ strains.
The effect of cell cycle on the DSB repair pathway choice was determined by arresting cells at G1 with α factor or at G2 with nocodazole treatment, and then inducing HO for 1 h before turning it off by addition of glucose. The HO enzyme is rapidly (a half life of ∼10 min) degraded by the ubiquitin-26S proteasome system after glucose addition [34]. Following 3 hours of incubation in glucose containing media at the indicated cell cycle stage, cells were then plated onto glucose containing plates to score the number of survivors (Fig. 1c). At G1 far less of the survivors (41% vs. 85% in asynchronous cells) repaired the DSB by HR, while more repaired the break by NHEJ (35% vs. 15% in asynchronous cells; Fig. 1c). In contrast, over 85% of G2 cells repaired the break by HR but a mere 7% did so by NHEJ (Fig. 1c). Detection of repair products by Southern blot hybridization confirmed this result; over 25% of the G1 vs. only 5% of the G2 arrested cells repair the DSB by NHEJ, whereas 10% of the G1 vs. 95% of the G2 cells repaired the break by HR (Figs. 1b, 1d). The results confirm the previous reports that HR is less efficient at G1 than G2 [11,12,16], and further demonstrate that NHEJ is less efficient at G2 than G1. The results are highly congruent with the reduced NHEJ at S/G2 previously observed using a plasmid based NHEJ assay in cells synchronously progressing through the cell cycle [35].
3.2. NHEJ repression at G2 does not depend on homologous recombination
NHEJ and HR likely compete for the same DSB substrate [3] and therefore the reduced NHEJ at G2 may simply reflect the increased effectiveness of HR at this stage of the cell cycle. To address this issue, we determined the NHEJ efficiency in the G2 arrested rad52Δ cells by scoring the number of survivors with the a mating type after HO expression. We also quantitated the NHEJ or HR product using Southern blot hybridization. We also examined, the effect of NHEJ on HR at G1 by measuring the HR efficiency in the G1 arrested yku70Δ cells. We found that deletion of YKU70 dramatically improved HR at G1, suggesting that the reduced HR at G1 is partly due to the elevated NHEJ at this cell cycle stage (Fig. 1b, 1d). In contrast, deletion of RAD52 did not increase the amount of NHEJ product nor the number of MATa survivors, indicating that HR does not compete with NHEJ at G2 (Figs. 1b, 1c, and 1d). Deletion of HMLα in JKM161 to eliminate the only remaining HR template also did not influence the NHEJ frequency at G2 (data not shown). Together, we conclude that NHEJ repression at G2 is distinct from efficient HR at this stage of the cell cycle.
3.3. Recruitment of NHEJ proteins to DSBs is suppressed at G2
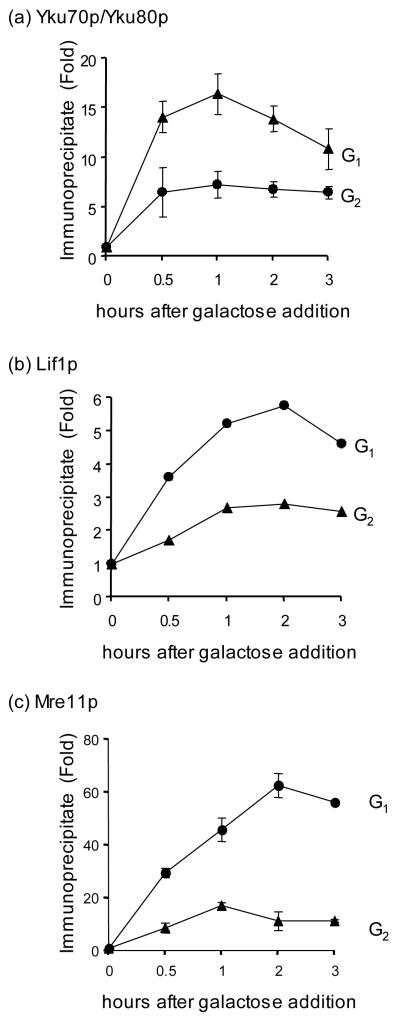
The recruitment of enzymes to the proper DNA substrate is an active process in many DNA transactions including transcription and DNA replication and is frequently used to control the corresponding enzymatic reactions. To elucidate the molecular basis of the cell cycle dependent regulation of NHEJ, we used ChIP to examine if core NHEJ proteins (Yku, Lif1, and Mre11) are recruited to DSBs differently at G1 and G2 in a donorless yeast strain. We found that all three NHEJ proteins were enriched at the HO break more markedly at G1 than G2 (Fig. 2). Reduction in recruitment of NHEJ proteins at the DSB at G2 is due neither to protein level decline at this cell cycle stage as the amount of Yku, Mre11 and Lif1 proteins remained constant at both G1 and G2 with or without DNA damage (Supplemental Fig. 2 and data not shown) nor to the presence of active HR at G2 (Supplemental Fig. 3; the slight reduction in the recruitment of YKu at a DSB in rad52Δ may be attributed to reduced HO induction efficiency in this mutant). Previously, both Rad51 and Rad52 were shown to be recruited to a DSB more efficiently at G2, consistent with the suppression of HR at G1 [11,12,16,36]. Together, these results suggest that the effect of cell cycle on DSB repair pathway choice may at least partly depend on the efficient recruitment of the corresponding repair proteins to the site of DNA breaks.
Figure 2.
The recruitment of NHEJ proteins at DSBs is reduced at G2. Kinetics of Yku (a), Lif1 (b), and Mre11 (c) recruitment to the HO induced DSB in G1 and G2 were determined by ChIP assays. Fold immunoprecipitate represents the ratio of the Yku, Lif1, or Mre11 IP PCR signal before and after HO induction, normalized by the PCR signal of the PRE1 control. Data represent the mean ± s.d. of three or more independent experiments.
3.4. Recruitment of Ku at DSBs is not controlled solely by 5′ end resection
A key question is how the cell cycle modulates the recruitment of recombination and NHEJ factors to DSB sites. Cdk1-dependent resection of DNA ends produces single stranded DNA near a DNA break at the S/G2 phase of the cell cycle [11,12], and HR proteins such as RPA and Rad51 bind preferentially to ssDNA at S/G2. We reasoned that core NHEJ proteins, Yku70/80, Mre11/Rad50/Xrs2, and Dnl4/Lif1, might bind DNA ends more efficiently at G1 because of their stronger affinity for unprocessed DNA ends, whereas HR proteins such as RPA and Rad51 bind preferentially to single strand DNA at S/G2. The model is particularly appealing in explaining the cell cycle dependent enrichment of Yku proteins, as they are already well known for their high affinity to double stranded DNA ends [37]. Likewise, RPA and Rad51 are known single strand DNA binding proteins [38].
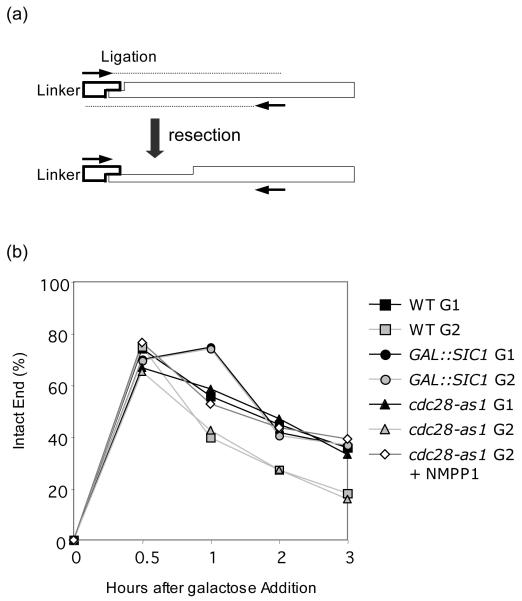
To test this hypothesis, we inhibited the Cdk1 activity crucial for end resection either by overexpressing the Sic1 protein, a Cdk1 inhibitor or by expressing Cdc28-as1, a hypomorphic Cdk1 sensitive to the ATP analogue inhibitor 1-NMPP1 [11]. We then monitored the extent of end processing by measuring the level of DSBs carrying an unresected 4 base pair overhang by ligation-mediated PCR (LM-PCR) (Fig. 3a)[25]. As expected, overproduction of Sic1 or inhibition of cdc28-as1 by 1-NMPP1 treatment at G2 almost completely blocked processing of DNA ends, so that the level of intact DSB ends was comparable to that in G1, reaffirming that resection was dependent on Cdk1 activity (Fig. 3b).
Figure 3.
Cdk1 activity suppresses end resection. (a) Scheme of the ligation-mediated PCR (LM-PCR). The adaptor, fully complementary to the 4 nucleotide 3′ overhang generated by HO induced cleavage, is ligated to genomic DNA. The two primers (arrows) anneal to the adaptor and 0.4 kb distal to HO cut site and will only amplify DNA ends with intact, unresected 5′ chromosomal ends. Even a single base pair of resection will prevent the ligation and leave no signal in the PCR reaction. (b) Percentage of intact DNA ends remaining at HO cleavage site at the indicated time after HO induction. Genomic DNA isolated at each time interval after HO expression was subjected to LM-PCR as described in the Materials and Methods. Percent intact ends is shown as the ratio of the PCR signal from each ligation reaction sample before and after HO induction, normalized by the PCR signal of the PRE1 control. To calculate percent intact ends, the ratio of the LM-PCR signal from Lev473 strain carrying HO recognition sequences modified to a BstXI site, whose digestion generates the same 4 nucleotide 3′ overhang as HO cleavage, normalized by the PCR signal of the PRE1 control after BstXI cleavage, was set to 100% [25].
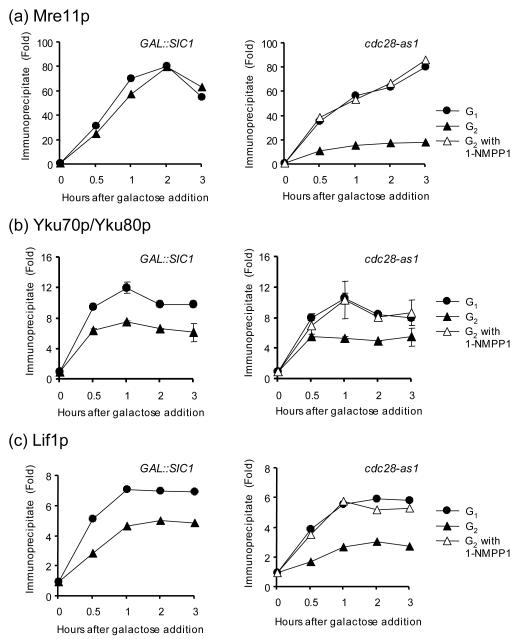
We then examined the enrichment of NHEJ proteins at the HO break in G2 when Cdk1was inhibited. We found that Mre11 was enriched to similar levels in G1 and G2 when Cdk1 activity was inhibited (Fig. 4a). Similarly, enrichment of Yku70/80 and Lif1 at a DSB was markedly improved when cdc28-as1 activity was inhibited by 1-NMPP1 treatment at G2 (Fig. 4b and 4c). However, Cdk1 inhibition by overproduction of Sic1 failed to fully de-repress recruitment of Yku and Lif1 to the HO-induced break at G2, even though resection of DNA ends and formation of ssDNA were reduced to a level indistinguishable from that at G1 (Fig. 3b, 4b, and 4c). These results suggest that resection is not the sole factor regulating the binding of NHEJ factors to DSBs.
Figure 4.
The recruitment of NHEJ proteins at DSBs is suppressed by the Cdk1 activity but not by end resection. Kinetics of Mre11 (a), Yku (b), and Lif1 (c) recruitment to the HO induced DSB in G1 and G2 from strains expressing galactose inducible Sic1 (left panel) or harboring cdc28-as1 allele supplemented with 1-NMPP1 inhibitor (right panel) were determined by ChIP assays. Fold immunoprecipitate was calculated as described in the legend for Fig. 2. Data represent the mean ± s.d. of two or more independent experiments.
3.5. Repression of NHEJ at G2 requires Cdk1 activity that modulates end resection and the recruitment of NHEJ factors to DNA break
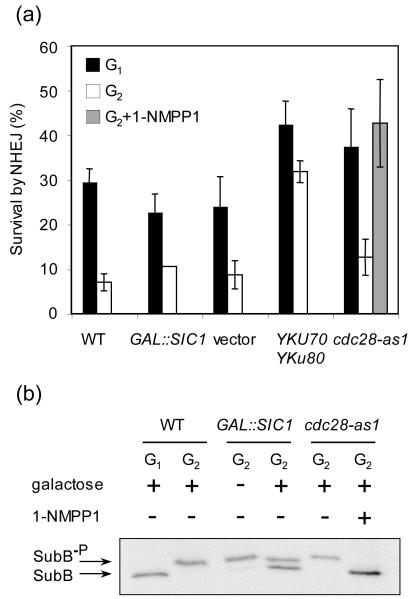
We next examined whether the elevated recruitment of Yku and Lif1 in the 1-NMPP1 treated cdc28-as1 strain was accompanied by increased NHEJ of a DSB at G2 using both the survival assay and Southern blot analysis of repair product formation. In parallel, the level of NHEJ in cells expressing excess Sic1 was determined to define the effect of Cdk1-dependent end processing on NHEJ repression at G2. We found that only 1-NMPP1-mediated cdc28-as1 inhibition increased NHEJ. While Sic1 overexpression reduced end resection and HR, it failed to enhance NHEJ mediated repair of an HO-induced DSB (Fig. 5a and [11]). These results suggest that Cdk1-dependent suppression of NHEJ protein recruitment to a DNA break contributes to the inefficient NHEJ at G2.
Figure 5.
NHEJ is repressed by Cdk1 activity. (a) Effect of Cdk1 inactivation on colony survival by NHEJ of an HO-induced DNA break. Cultures were induced to express HO for 1 h and plated onto YEPD medium, which shuts off HO expression. Percent survival by NHEJ is shown as the number of a type colonies growing onto the YEPD plates after 1 h of HO induction, normalized by the number of colonies growing before HO induction. Data represent the mean ± s.d. of three or more independent experiments. (b) Cdk1 activity was measured by the phosphorylation of the B subunit of DNA polymerase-alpha, a marker for Cdc28/Clb activity [11].
Recruitment of Yku and Dnl4/Lif1 to DNA breaks is mutually dependent [28,39,40]. We expressed excess Yku70 and Yku80 from plasmids harboring the YKU70 and YKU80 genes under constitutively active promoters, and asked if expression of excess Ku proteins can override NHEJ repression at G2 (Fig. 5a). We found that expression of excess Yku proteins partially rescued NHEJ at G2, increasing NHEJ to a level even higher than that at G1 when Yku was endogenously expressed. Nevertheless, neither Yku overexpression nor Sic1-mediated repression of end resection fully de-repressed NHEJ at G2; the NHEJ level at G2 failed to match that at G1 under the same conditions (Fig. 5a). The results support a model in which at least two distinct factors contribute to the NHEJ repression at G2: elevated end resection and reduced association of Yku and/or Dnl4/Lif1 to DNA breaks.
Why do two Cdk1 inhibition methods elicit very different responses to NHEJ factor recruitment and NHEJ repair, even though they are equally efficient in suppressing end resection? To gain insights into this question, we monitored CDK1 activity in cells subjected to each of these two Cdk1 inhibition methods by examining the phosphorylation status of the B subunit of DNA polymerase α, a well-known substrate for the Cdk1 kinase [41]. The data revealed that the 1-NMPP1-treated cdc28-as1 cells exhibited a far greater inhibition of Cdk1 activity than cells with Sic1 overexpression (Fig. 5b). Therefore, the residual Cdk1 activity in Sic1 overexpressing cells is insufficient to activate resection of DNA ends, but still enough to repress the recruitment of NHEJ proteins at DSBs and NHEJ activity at G2.
4. Discussion
Both NHEJ and HR are efficient in repairing DSBs, but they produce products with distinctly different mutational potentials. Proper choice of repair mechanism for a given DNA lesion will minimize cellular mutational load and is pivotal to maintain genome integrity. Previously, evidence indicated that HR efficiency fluctuates throughout cell cycle, with the key recombination step(s) being under control of the Cdk1 kinase [11,12,16]. Here, we confirmed the effect of cell cycle on HR and demonstrated an effect on NHEJ as well; NHEJ is most active at G1 but substantially attenuated at G2. Repression of NHEJ at G2 depends on Cdk1-dependent modulation of end processing and on Yku and Lif1 recruitment to DNA breaks. Importantly, elimination of HR was not sufficient to increase NHEJ at G2. We propose that regulating the affinity of Yku and Dnl4/Lif1 proteins to DNA breaks defines a novel way to enforce cellular repair pathway choice during the cell cycle in budding yeast.
Being the main engine for cell cycling, Cdk1 is an ideal molecule to coordinate the choice of DSB repair pathway. This insures the availability of the most desirable recombination template: an identical sister-chromatid. HR and NHEJ are oppositely affected by Cdk1 activity: Cdk1 activates HR, but represses NHEJ. Cdk1 controls cell cycle dependent repair pathway choice at multiple steps. First, Cdk1 activates nucleolytic processing of DNA ends to produce recombinogenic ssDNA, an essential intermediate for HR that is also inhibitory to NHEJ [11,12]. Secondly, Cdk1 is important for a later step in homologous recombination, after strand invasion and before the initiation of new DNA synthesis [11]. Thirdly, expression/post-translational modifications of Sae2/CtIP also fluctuate throughout cell cycle [22], with the protein being shown recently to be a target of Cdk1 in yeast and mammals. In this study we demonstrated that Cdk1 additionally represses NHEJ at G2 by inhibiting the recruitment of at least two NHEJ factors, Yku and Lif1, to DNA breaks.
Since the recruitment of core NHEJ proteins is inhibited by Cdk1 activity, an open question is whether Cdk1 directly phosphorylates one or more core NHEJ proteins to modulate binding affinity to DNA breaks. Direct phosphorylation of Ku by Cdk1 is highly attractive as Ku70 was previously identified as a target of cyclin dependent kinase in mouse [42]. However, removal of all the putative Cdk1 phosphorylation sites (S/TP) on Yku70 and 3 out of 4 sites on Yku80 failed to elicit any DSB repair phenotype, suggesting that the regulatory role Cdk1 on Yku is likely indirect (Y.Z. and S.L., unpublished observations). Since Dnl4/Lif1 can positively influence the binding of Ku at DSBs, it will be interesting to test whether Dnl4/Lif1 are the relevant substrate(s) of Cdk1 [28]. Alternatively, other post-translational modification of Ku (e.g. sumoylation)[43] might be required for DNA binding and regulated by Cdk1.
In this study, we provide compelling evidence that NHEJ is actively suppressed at G2 by direct inhibition of NHEJ factor binding to DSBs. However, it still remains possible that commitment to NHEJ is compromised at this cell cycle stage due to elevated end resection rather than to direct modulation of NHEJ factors. For instance, Sic1 might cause a gross resection rate decline by selectively repressing certain steps of end resection, but fail to inhibit step(s) germane to NHEJ suppression. As evidence emerges that resection is a multi-step process governed by functionally overlapping nucleases and the associated protein complexes, it remains formally possible that Sic1 may inhibit distinct resection step(s) but leave others intact. In this regard, we would like to emphasize that the earliest step of resection is effectively inhibited by Sic1 overexpression in the PCR-based resection assay.
Interestingly, Clerici et al. previously reported no apparent difference in the amount of MYC epitope tagged Yku proteins at an HO-induced DSB at G1 versus G2 by ChIP assays [44]. Although the reason for this disparity is unknown, some of the difference can be explained by the non-identical behavior between MYC-tagged Yku70 and an untagged Yku protein complex in ChIP assays. In fact, very different outcomes were reported between ChIP assays using an epitope tagged version of Yku and those using a polyclonal antibody raised against Yku to examine association of Yku with telomeres and the HM loci [45].
The cell cycle dependent fluctuation of repair pathway efficiency likely reflects evolutionary pressure to preferentially use recombination for DSB repair when sister-chromatids are available, as recombination between sister-chromatids will most faithfully restore the integrity of broken chromosomes. Indeed, DSB repair mutants in other organisms exhibit radiation sensitivity profiles highly suggestive of cell cycle influence [13-15]. We speculate that a similar strategy is applied to DSB repair in mammalian cells where NHEJ plays a more prominent role in repairing DNA breaks.
In summary, the results described here have uncovered a novel role of Saccharomyces cerevisiae Cdk1 in the suppression of NHEJ at G2. Importantly, this regulation minimizes genetic mutation during repair. Our results shed light on how the two major pathways of DSB repair are coordinated during the cell cycle and reveal additional details of the molecular basis of this control.
Supplementary Material
Supplementary Figure 1. FACS profiles of JKM161 (a, b) and the RAD52 (c, d) deleted mutant arrested at G1 or G2. The time point when the glucose was added to the medium to turn off HO was marked as 0. The galactose was added 1 h prior to the glucose addition and marked as U. Flow cytometry analyses were performed on a Becton Dickinson FACSCalibur™ system.
Supplementary Figure 2. Yku70 and Yku80 (a), Mre11, Xrs2, and Lif1 (b) protein levels were measured in G1 and G2 by immunoblot analysis using PAP (Sigma). Immunoblotting with an anti-Rpt5 (a) or anti-Saw1 antibody (b) is shown as an input control.
Supplementary Figure 3. The recruitment of Yku proteins to the HO induced DSB in rad52Δ were determined by ChIP assays. Fold immunoprecipitate represents the ratio of the Yku IP PCR signal before and after HO induction, normalized by the PCR signal of the PRE1 control. Data for Yku in rad52Δ represent the mean of two independent experiments.
Acknowledgment
We thank J. Haber, G. Ira, A. Pellicioli, P. Sung, and A. Tomkinson for gifts of strains, plasmids, and antibodies. We also thank J. Haber, P. Sung, S. Jinks-Robertson, A. Tomkinson, and members of the S.E.L. laboratory for helpful discussion. This work is funded by grants from NIH (GM083010) to S.E.L. S.E.L. is a scholar of the Leukemia and Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
5. References
- [1].Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- [2].Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- [3].Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- [4].Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- [5].Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- [6].Gu J, Lieber MR. Mechanistic flexibility as a conserved theme across 3 billion years of nonhomologous DNA end-joining. Genes Dev. 2008;22:411–415. doi: 10.1101/gad.1646608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Haber JE. Transpositions and translocations induced by site-specific double-strand breaks in budding yeast. DNA Repair (Amst) 2006;5:998–1009. doi: 10.1016/j.dnarep.2006.05.025. [DOI] [PubMed] [Google Scholar]
- [8].Franco S, Alt FW, Manis JP. Pathways that suppress programmed DNA breaks from progressing to chromosomal breaks and translocations. DNA Repair (Amst) 2006;5:1030–1041. doi: 10.1016/j.dnarep.2006.05.024. [DOI] [PubMed] [Google Scholar]
- [9].Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- [10].Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- [11].Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lee SE, Mitchell RA, Cheng A, Hendrickson EA. Evidence for DNA-PK-dependent and -independent DNA double-strand break repair pathways in mammalian cells as a function of the cell cycle. Mol Cell Biol. 1997;17:1425–1433. doi: 10.1128/mcb.17.3.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bressan DA, Baxter BK, Petrini JH. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7681–7687. doi: 10.1128/mcb.19.11.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ferreira MG, Cooper JP. Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev. 2004;18:2249–2254. doi: 10.1101/gad.315804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- [18].Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chen F, Nastasi A, Shen Z, Brenneman M, Crissman H, Chen DJ. Cell cycle-dependent protein expression of mammalian homologs of yeast DNA double-strand break repair genes Rad51 and Rad52. Mutat Res. 1997;384:205–211. doi: 10.1016/s0921-8777(97)00020-7. [DOI] [PubMed] [Google Scholar]
- [22].Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Baroni E, Viscardi V, Cartagena-Lirola H, Lucchini G, Longhese MP. The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Mol Cell Biol. 2004;24:4151–4165. doi: 10.1128/MCB.24.10.4151-4165.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Daley JM, Wilson TE. Rejoining of DNA double-strand breaks as a function of overhang length. Mol Cell Biol. 2005;25:896–906. doi: 10.1128/MCB.25.3.896-906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Frank-Vaillant M, Marcand S. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol Cell. 2002;10:1189–1199. doi: 10.1016/s1097-2765(02)00705-0. [DOI] [PubMed] [Google Scholar]
- [26].Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hinz JM, Yamada NA, Salazar EP, Tebbs RS, Thompson LH. Influence of double-strand-break repair pathways on radiosensitivity throughout the cell cycle in CHO cells. DNA Repair (Amst) 2005;4:782–792. doi: 10.1016/j.dnarep.2005.03.005. [DOI] [PubMed] [Google Scholar]
- [28].Zhang Y, Hefferin ML, Chen L, Shim EY, Tseng HM, Kwon Y, Sung P, Lee SE, Tomkinson AE. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat Struct Mol Biol. 2007;14:639–646. doi: 10.1038/nsmb1261. [DOI] [PubMed] [Google Scholar]
- [29].Shim EY, Ma JL, Oum JH, Yanez Y, Lee SE. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol Cell Biol. 2005;25:3934–3944. doi: 10.1128/MCB.25.10.3934-3944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Valencia M, Bentele M, Vaze MB, Herrmann G, Kraus E, Lee SE, Schar P, Haber JE. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature. 2001;414:666–669. doi: 10.1038/414666a. [DOI] [PubMed] [Google Scholar]
- [31].Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Allen C, Kurimasa A, Brenneman MA, Chen DJ, Nickoloff JA. DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc Natl Acad Sci U S A. 2002;99:3758–3763. doi: 10.1073/pnas.052545899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Clikeman JA, Khalsa GJ, Barton SL, Nickoloff JA. Homologous recombinational repair of double-strand breaks in yeast is enhanced by MAT heterozygosity through yKU-dependent and -independent mechanisms. Genetics. 2001;157:579–589. doi: 10.1093/genetics/157.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kaplun L, Ivantsiv Y, Kornitzer D, Raveh D. Functions of the DNA damage response pathway target Ho endonuclease of yeast for degradation via the ubiquitin-26S proteasome system. Proc Natl Acad Sci U S A. 2000;97:10077–10082. doi: 10.1073/pnas.97.18.10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Karathanasis E, Wilson TE. Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics. 2002;161:1015–1027. doi: 10.1093/genetics/161.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- [37].Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- [38].Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- [39].Palmbos PL, Wu D, Daley JM, Wilson TE. Recruitment of Saccharomyces cerevisiae Dnl4-Lif1 complex to a double-strand break requires interactions with Yku80 and the Xrs2 FHA domain. Genetics. 2008;180:1809–1819. doi: 10.1534/genetics.108.095539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Teo SH, Jackson SP. Lif1p targets the DNA ligase Lig4p to sites of DNA double-strand breaks. Curr Biol. 2000;10:165–168. doi: 10.1016/s0960-9822(00)00317-1. [DOI] [PubMed] [Google Scholar]
- [41].Foiani M, Liberi G, Lucchini G, Plevani P. Cell cycle-dependent phosphorylation and dephosphorylation of the yeast DNA polymerase alpha-primase B subunit. Mol Cell Biol. 1995;15:883–891. doi: 10.1128/mcb.15.2.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Muller-Tidow C, Ji P, Diederichs S, Potratz J, Baumer N, Kohler G, Cauvet T, Choudary C, van der Meer T, Chan WY, Nieduszynski C, Colledge WH, Carrington M, Koeffler HP, Restle A, Wiesmuller L, Sobczak-Thepot J, Berdel WE, Serve H. The cyclin A1-CDK2 complex regulates DNA double-strand break repair. Mol Cell Biol. 2004;24:8917–8928. doi: 10.1128/MCB.24.20.8917-8928.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci U S A. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Clerici M, Mantiero D, Guerini I, Lucchini G, Longhese MP. The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep. 2008;9:810–818. doi: 10.1038/embor.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Patterson EE, Fox CA. The Ku complex in silencing the cryptic mating-type loci of Saccharomyces cerevisiae. Genetics. 2008;180:771–783. doi: 10.1534/genetics.108.091710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. FACS profiles of JKM161 (a, b) and the RAD52 (c, d) deleted mutant arrested at G1 or G2. The time point when the glucose was added to the medium to turn off HO was marked as 0. The galactose was added 1 h prior to the glucose addition and marked as U. Flow cytometry analyses were performed on a Becton Dickinson FACSCalibur™ system.
Supplementary Figure 2. Yku70 and Yku80 (a), Mre11, Xrs2, and Lif1 (b) protein levels were measured in G1 and G2 by immunoblot analysis using PAP (Sigma). Immunoblotting with an anti-Rpt5 (a) or anti-Saw1 antibody (b) is shown as an input control.
Supplementary Figure 3. The recruitment of Yku proteins to the HO induced DSB in rad52Δ were determined by ChIP assays. Fold immunoprecipitate represents the ratio of the Yku IP PCR signal before and after HO induction, normalized by the PCR signal of the PRE1 control. Data for Yku in rad52Δ represent the mean of two independent experiments.