Abstract
Mutations in POLG are a major contributor to pediatric and adult mitochondrial diseases. However, the consequences of many POLG mutations are not well understood. We investigated the molecular cause of Alpers syndome in a patient harboring the POLG mutations A467T in trans with c.2157+5_+6 gc→ag in intron 12. Analysis of transcripts arising from the c.2157+5_+6 gc→ag allele revealed alternative splicing with an insertion of 30 intronic nucleotides leading to a premature termination codon. These transcripts were subsequently removed through nonsense-mediated decay, leading to haplotype insufficiency due to expression of the A467T allele and decreased expression of the c.2157+5_+6 gc→ag allele, which is likely responsible for the Alpers syndrome phenotype.
Keywords: Mitochondrial disease, mitochondrial DNA replication, Alpers syndrome, nonsense-mediated decay, de novo mutation, haplotype insufficiency
1. INTRODUCTION
Mitochondrial diseases arise because of disturbances within the mitochondria (Dimauro and Davidzon, 2005; Wallace, 1999). One subset of these diseases encompasses are due to defects in mitochondrial DNA (mtDNA) stability (Copeland, 2008). POLG encodes the mtDNA polymerase (pol γ) and is a major locus of mitochondrial disease (Chan and Copeland, 2008). Patients with POLG mitochondrial disease show mtDNA deletions or depletion in symptomatic tissues as pol γ is essential for mtDNA replication and repair. To date, approximately 150 mutations in POLG have been identified (http://tools.niehs.nih.gov/polg/), with three main disease phenotypes, that is Alpers syndrome and related mtDNA depletion disorders, ataxia-neuropathy syndromes, and progressive external ophthalmoplegia.
Alpers syndrome is a rare heritable autosomal recessive disorder affecting young children (Alpers, 1931; Harding et al., 1995; Naviaux et al., 1999). The disease is characterized by refractory seizures, psychomotor regression, hepatic dysfunction and depletion of mtDNA (Harding et al., 1995; Naviaux et al., 1999). It generally manifests during the first few weeks to years of life and symptoms develop in a stepwise manner leading eventually to early death. POLG mutations were first linked to Alpers syndrome in 2004 (Naviaux and Nguyen, 2004) when two unrelated probands were found to have compound heterozygous mutations in POLG, namely the A467T mutation on one allele and a premature termination codon (PTC) mutation, E873X, on the other allele. A467T is the most common POLG mutation, and is particularly prevalent in Alpers syndrome where ∼65% of patients carry at least one A467T allele (Nguyen et al., 2006). Previous biochemical studies in our laboratory using purified recombinant pol γ containing the A467T mutation showed that this enzyme had very low DNA polymerase activity. Furthermore, pol γ with the A467T mutation had an impaired ability to functionally associate with the pol γ accessory subunit (Chan et al., 2005a), which is needed for highly processive DNA synthesis and enhanced DNA binding (Lim et al., 1999).
We also studied the skin fibroblasts of one patient from the aforementioned Alpers POLG study (Naviaux and Nguyen, 2004) in order to understand the contribution of the E873X allele to disease (Chan et al., 2005b). The allele containing the E873X mutation in exon 17 was expected to produce a truncated protein. However, only full-length p140 protein was detected. Sequence analysis of the cDNA from the pre-spliced message showed that both alleles were represented equally. Sequence analysis of cDNA derived from mature message revealed only transcripts containing the A467T mutation indicating that full-length pol γ arose from the allele containing the A467T mutation. Further analyses revealed that transcripts arising from the E873X allele were degraded by the nonsense-mediated mRNA decay (NMD) pathway. The NMD pathway degrades transcripts containing PTCs that are at least 50–55 nucleotides upstream from at least one intron. Additionally, sequencing of a shadow band showed that exon 17 had been skipped by the nonsense-associated alternative splicing pathway (NAS), which produces a frameshift leading to another PTC (Chan et al., 2005b). NMD and NAS destroyed virtually all mRNAs produced from the allele containing the PTC. Thus, the severity of disease for this patient was most likely due to mono-allelic expression of A467T pol γ. Extrapolating to other mutations of this nature, NMD and NAS are expected to remove those POLG mRNAs containing PTCs.
There are currently 18 POLG mutations that may produce a PTC, frameshift or an alternatively spliced transcript (Table 1). The mechanisms of pathogenesis via intronic POLG mutations have not yet been described until now. We report an Alpers syndrome patient with a POLG genotype of A467T/c.2157+5_+6 gc→ag in trans. We investigated the consequence of the double intronic mutation on splicing and degradation of the POLG message. We observed alternative splicing of this allele leading to a PTC and degradation of the message by NMD. This results in a much decreased expression of the c.2157+5_+6 gc→ag allele, with normal expression of the allele containing the A467T mutation, which explains the Alpers syndrome phenotype for this patient.
Table 1.
Premature termination mutations in the POLG gene and associated disease.
| Amino Acid Substitution | DNA mutation | Disease | Genetics | Reference |
|---|---|---|---|---|
| Q68X | 202 c→t (exon 2) | Alpers | Found in trans with A467T | (Wong et al., 2008) |
| W235X | 705 g→a (exon 3) | Myocerebrohepato-pathy syndrome | Found in trans with A467T | (de Vries et al., 2006) |
| T326fsX387 | 975–976 ins c (exon 4) | Alpers | Found in trans with A467T | (Naimi et al., 2006) |
| R374X | 1120 c→t (exon 5) | Alpers | Found in trans with A467T. | (Ashley et al., 2008) |
| L424GfsX28 (CT)deletion-452X | 1270–1271 del ct (exon 7) | Alpers | Found in trans with A467T | (Wong et al., 2008) |
| PEO | Sporadic Found in trans with G431V | (Agostino et al., 2003) | ||
| T452X | 1356 t→g (exon 7) | PEO | Sporadic | (Hudson et al., 2006) |
| R709X | 2125 c→t (exon 12) | PEO | Sporadic, Found in trans with T251I–P587L | (Del Bo et al., 2003; Di Fonzo et al., 2003) |
| Q715X | 2143 c→t (exon 12) | Alpers | Found in trans with A467T | (Wong et al., 2008) |
| c. 2157+5_+6 | 2157 gc→ ag (exon 12) | Alpers | Found in trans with A467T | (Wong et al., 2008) |
| 2354Gins at G785 | 2354 ins G STOP @ codon 806 (exon 14) | arPEO | Found in trans with T251I | (Lamantea et al., 2002) |
| c. 2480+1 | 2480+1 g→a splice (exon 15) | Alpers | Found in trans with W748S–E1143G | (Wong et al., 2008) |
| IVS15-9-c.2485del 12bp | 2485 del 12bp (exon 16) | Alpers | Found in trans with A467T. Splice site mutation 3'exon15/intron junction, | (Horvath et al., 2006) |
| E873X | 2617 g→t (exon 17) | Alpers | Found in trans with A467T | (Ashley et al., 2008; Chan et al., 2005b; Naviaux and Nguyen, 2004; Naviaux and Nguyen, 2005) |
| L965X | 2894 t→g (exon 18) | arPEO | Found in cis with E1143G and in trans with R627Q–Q1236H | (Horvath et al., 2006) |
| W1020X | 3057 g→a (exon 19) | Alpers | Found in trans with A467T | (Nguyen et al., 2005) |
| 3482 +2T to C | 3482 +2t’s splice at intron 21 (exon 21) | Alpers | Exon21/Intron21 splice site mutation at a.a. 1161. Found in trans with A467T | (Ferrari et al., 2005; Horvath et al., 2006) |
| L1173fsX | 3518 ins gact, fs in exon 22 (exon 22) | Alpers | Found in trans with A467T | (Nguyen et al., 2006) |
| Y1210fs1216X | 3630 ins c (exon 22) | Alpers | Found in trans with W748S–E1143G, Mutation causes frameshift to stop at a.a. 1225 | (Ferrari et al., 2005; Spinazzola et al., 2009) |
2. MATERIALS AND METHODS
2.1. Patient history
This patient was previously described as ‘Patient 2’ or ‘Case 2’ (Lutz et al., 2009; Wong et al., 2008). The patient was an 18-month-old Caucasian boy born to healthy, unrelated parents. He was the second son of three siblings. Motor development was delayed, with crawling but not walking achieved. He was noted to have lower extremity weakness. His speech development was also delayed. MRI at 17 months showed normal brain findings, but he had a spinal cord syrinx at T11-L1. At 18 months, he experienced an acute episode of emesis while eating, followed by irritability and lethargy. The episode was not preceded by fasting, fever, or illness. During this episode, he progressed to have unresponsiveness, eye deviation, jaw clenching, and hypotonia of the trunk and extremities. During transport to the emergency department, he developed repetitive generalized tonic-clonic seizures that evolved into refractory status epilepticus. He was transferred to a tertiary care facility and placed in a pentobarbital coma for seizure control after failing standard treatment with phenobarbital, fosphenytoin, midazolam, lorazepam, and diazepam. Valproic acid was avoided. After 30 days on this treatment, his seizures resolved and pentobarbital was discontinued. He was left with brain atrophy and a severe encephalopathy characterized by choreo-athetoid movements, cortico-visual impairment, diffuse hypotonia, and severe swallowing dysfunction. Transaminases were mildly elevated during his hospitalization but normalized as his seizures resolved.
The patient became more alert and regained the ability to babble at 20 months. However, by 21 months, his seizures recurred despite treatment with topiramate, levetiracetam, and phenobarbital. At 24 months, he was noted to have non-dependent edema, ascites, and abnormal coagulation studies indicating liver biosynthetic failure. Parents elected to place the child in hospice care, and he died at 25 months. Autopsy confirmed liver fibrosis, with nests of cells with severe vacuolation, areas of bile duct proliferation, and small numbers of lymphocytes scattered throughout. The spleen showed erythrocyte sequestration and lymphoid aggregates with germinal centers. The pancreas appeared normal. Kidneys appeared normal except for hypercellularity. The heart showed diffuse hypercellularity and thin myocytes with frequent dark nuclei. There was anasarca, atelectasis of the lungs, and a large amount of cerebrospinal fluid surrounding a relatively small-sized brain.
The initial laboratory work-up included AST 85 U/L (range 16–46 U/L), ALT 69 U/L (range 29–46), plasma lactate 3.2 mM (range 0.5–2.0), CSF lactate was 3.4 mM (range 0.6–2.2) and protein was 46 mg/dL (range 15–40). Liver transaminases remained mildly elevated throughout his hospitalization. AST peaked at 195 U/L and ALT at 200 U/L. MRI of the brain showed abnormal restricted diffusion involving the subcortical white matter in the left posterior parietal and occipital lobes consistent with multi-regional stroke-like episodes or ischemia. T2 signal was increased bilaterally in the thalami.
2.2. Molecular genetics
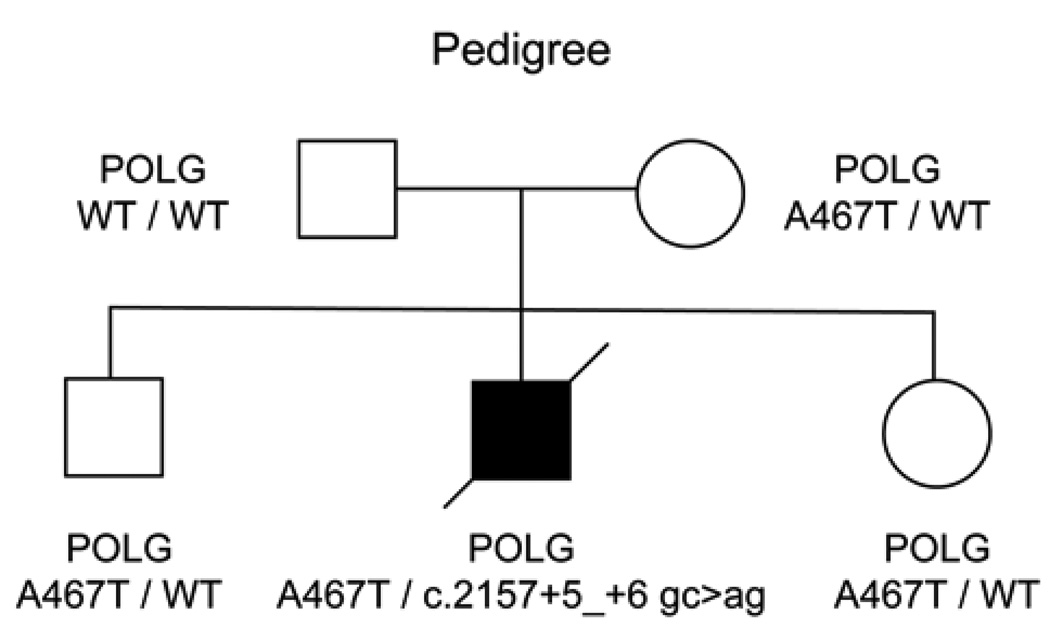
The patient had a compound heterozygous POLG genotype, with A467T on one allele, and two intronic changes in the other POLG allele (2157+5_+6 gc>ag). The proband’s mother was heterozygous for the A467T mutation (A467T/WT), as well as his siblings, however the father was wild-type for both POLG alleles (Figure 1). Neither the A467T mutation, nor 2157+5_+6 gc>ag were seen in the father’s DNA extracted from blood. Analysis of 15 unlinked microsatellite markers was consistent with stated paternity (Wong et al., 2008). This suggests a de novo mutation originating on the paternal allele.
Figure 1.
Pedigree of proband. Black symbol indicates Alpers syndrome. WT = wild-type.
2.3.Cell culture
A muscle biopsy was performed on the patient at 19 months. At this time, skin fibroblasts were obtained and cultured. Cells were grown at 37ºC in 5% CO2 in D-MEM/F-12 medium (Invitrogen) supplemented with 10% fetal calf serum, 1 mM pyruvate, non-essential amino acids, penicillin and streptomycin.
2.4. RNA extraction
Total RNA was isolated using the RNeasy kit (Qiagen). To remove any genomic DNA contamination, RNA samples were treated twice with DNaseI, first with Qiagen DNaseI for 15 min at room temperature as stated in the RNeasy protocol, and then with 6 units Ambion RNase-free DNaseI for 1 hr at 37ºC. DNaseI was then removed using the supplied DNase inactivation reagent. DNA-free status was confirmed by performing PCR on purified RNA using the same primers, without the addition of reverse transcriptase.
2.5. cDNA synthesis, PCR, sequencing and cloning
First strand cDNA synthesis was carried out on DNase1-treated RNA with the Superscript III First-Strand Synthesis system (Invitrogen), using a gene specific primer for POLG (2995R (Chan et al., 2005b)). Primer sequences for cDNA were as follows:
| Exonic primers | |
| Spanning exons 5 to 8 of POLG | |
| Forward | 5'-TTG TGA AGG GCA CCA TGA-3' |
| Reverse | 5'-CTG TGG CTG GTT CCT TCT T-3' |
| Spanning exons 11 to 14 of POLG | |
| Forward | 5'-AGT TCC TGC TCA CTG ACA ATA G-3' |
| Reverse | 5'-GCG TTC CTC CAG AAA GAA ATC-3' |
| Intronic primers | |
| Spanning exons 7 to 8 of POLG | |
| Forward | 5'-GTC TTG CCT CCT GTG GTC AT-3' |
| Reverse | 5'-CTG TGG CTG GTT CCT TCT T-3' |
| Spanning exons 11 to 14 of POLG | |
| Forward | 5'-TGT CAA TCA ATC CCT GTC TAA AAC C-3' |
| Reverse | 5'-CGG CTC CAG CAG TTA CAC CAA GAA G-3' |
PCR amplifications of cDNA were performed in a Perkin Elmer GeneAmp PCR Cycler 9700 using the following conditions: the thermal cycling program consisted of 10 min at 95ºC, followed by 20 cycles of 94ºC 30 s, 60ºC - 50ºC 30s, 72ºC 60 s (touchdown PCR, annealing temperature was decreased 0.5ºC after each cycle). This touchdown step was followed by 10 cycles of 94ºC 30 s, 50ºC 30 s, 72ºC 60 s. The samples were then cooled to 20ºC for 4 min and then kept at 4ºC. PCR products were resolved by agarose gel electrophoresis. Products were gel purified and directly sequenced using a BigDye terminator sequencing kit (Applied Biosystems) on an ABI PRISM 377 DNA sequencer. Alignments of the sequences were performed using SeqWeb Version 2 (Accelrys).
PCR products were also cloned into pCR4Blunt-TOPO plasmid using the Zero Blunt TOPO PCR Cloning Kit for Sequencing (Invitrogen). Plasmids containing inserts from individual colonies were purified using the Miniprep plasmid purification kit (Qiagen) and were sequenced using BigDye terminator sequencing and one of the primers used for the original PCR.
3. RESULTS
We investigated the molecular cause of Alpers syndrome in a patient with the A467T mutation in one POLG allele and a c.2157+5_+6gc→ag insertion mutation in intron 12 in the other POLG allele (Figure 2). While the pathogenesis of the A467T allele is well established as a recessive mutation in Alpers syndrome and other POLG mitochondrial diseases, the consequence of the c.2157+5_+6 gc>ag mutation is unclear. As this change is close to the canonical splice site, we wanted to determine whether this mutation did indeed affect splicing, which could lead to a mutant pol γ with severe defects in mtDNA replication and repair.
Figure 2.
Genomic DNA sequence of exon 12 - exon 14 from the POLG allele containing the c.2157+5_+6 gc>ag mutation. This alters a ‘gc’ to ‘ag’ at the 5th and 6th position 3' of the exon 12 - intron 12 splice junction.
3.1. Alternative splicing of POLG transcripts containing the c.2157+5_+6 gc>ag mutation
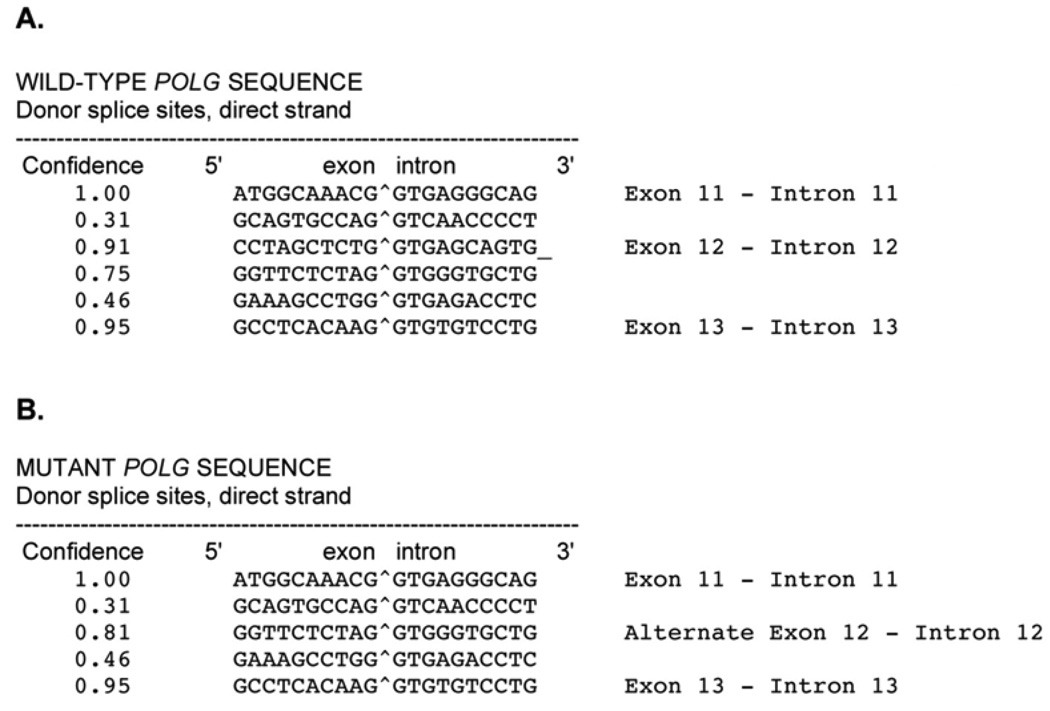
The c.2157+5_+6 gc→ag mutation alters two nucleotides at the 5th and 6th base position of POLG intron 12. To investigate whether this two-nucleotide change could alter splicing, we first used a splice site prediction program, NetGene2, which uses neural network predictions of splice sites in human DNA (http://www.cbs.dtu.dk/services/NetGene2/; (Brunak et al., 1991; Hebsgaard et al., 1996)). Analysis of the wild-type sequence in this region accurately predicts wild-type exon 11 - intron 11, exon 12 - intron 12, and exon 13 - intron 13 donor splice sites (Figure 3A). However, POLG sequence containing the c.2157+5_+6 gc→ag mutation predicts a different exon 12 - intron 12 donor splice site (Figure 3B). This predicted splice site would lead to an alternatively spliced transcript containing an extra 30 nucleotides with a UAG stop codon in the 28th to 30th position.
Figure 3.
Splice sites as predicted by NetGene2 server, which produces neural network prediction of splice sites in human DNA (http://www.cbs.dtu.dk/services/NetGene2/) (Brunak et al., 1991; Hebsgaard et al., 1996). (A) Wild-type POLG sequence. (B)POLG sequence containing the c2157+5_+6 gc>ag mutation.
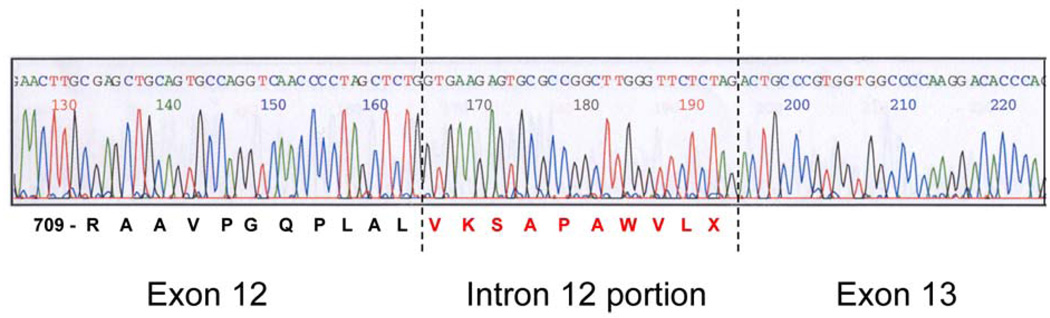
To assess whether the predicted scenario occurs in vivo, we developed a fibroblast cell line from this patient using cells from a muscle biopsy taken at 19 months. Whole cell RNA was isolated from cultured fibroblasts. First-strand POLG cDNA was synthesized and used for all experiments described. PCR was performed in order to amplify mature message using exonic primers spanning exons 11 to 14. PCR products were ligated into pCR4Blunt-TOPO plasmids and individual clones isolated. Sequence analysis of 21 individual clones revealed that 81% (95% CI = 58–95%) of the transcripts were correctly spliced, while 19% (95% CI = 5–42%) were alternatively spliced. No exon skipping was observed. The sequence of the alternatively spliced cDNAs indicated bypass of the normal exon 12 - intron 12 splice site, with mature message containing an extra 30 nucleotides of intron 12 sequence (Figure 4). The new splice site corresponds with the predicted splice site by NetGene2 analysis (Figure 3B). All alternatively spliced transcripts, representing 19% of the total transcripts, contained the two intronic c.2157+5_+6 gc→ag mutations.
Figure 4.
Sequence analysis of individual clones from cDNA spanning exons 11 – 14 reveals an inclusion of part of intron 12 in the final fully-spliced transcript. The extra 30 nucleotide insert contains an in-frame PTC at nucleotides 28–30 just before the alternative splice site.
3.2. Both alleles are represented equally in pre-spliced transcripts
We determined the proportion of both POLG alleles prior to splicing. Amplification of prespliced message was performed using intronic primers spanning codon 467 (exon 7). Clones containing these PCR products were constructed as described above and individual clones were sequenced. Sequence analysis of these pre-spliced clones revealed an equal distribution (1:1) of each allele (Ala467:Thr467).
3.3. The bulk of mature POLG transcripts contain the A467T mutation
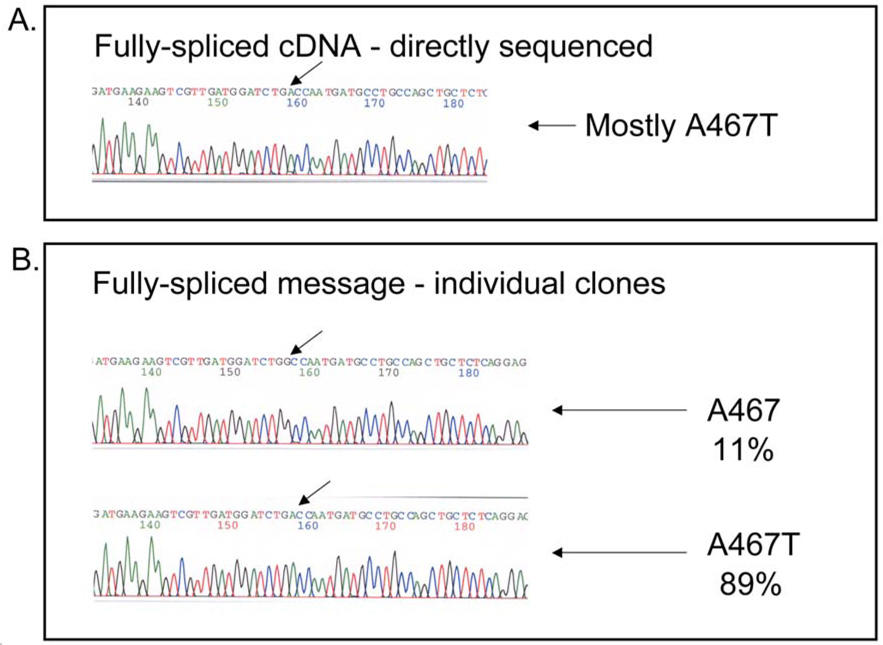
As we saw an equal representation of transcripts arising from each POLG allele prior to splicing, we wanted to determine the proportion of mature transcripts containing the A467T mutation. Direct sequencing of this mature, fully-spliced message showed that the bulk of this message contained Thr467, indicative of the A467T mutation (Figure 5A). To gauge a quantitative level of steady-state A467T message we isolated individual cDNA clones and subjected them to sequencing. Using the exonic primers to exons 5 and 8, PCR products were ligated into the pCR4Blunt-TOPO plasmids. Forty-six individual clones were picked for sequencing analysis (Figure 5B). Sequence analysis of individual cDNA clones confirm that 89% (95% CI = 76–96%) of the mature message contained the A467T mutation, while only 11% (95% CI = 4–24%) of this population arises from the allele containing the c.2157+5_+6 gc>ag mutation. Thus, the bulk of the transcripts arise from the allele containing the A467T mutation. By comparing the results from 3.1. and by determining the binomial 95% confidence intervals, we conclude that the two populations (that is, those that contain the A467T mutation and those that are spliced correctly) are one and the same (p = 0.44). This suggests that the mature POLG message containing the c.2157+5_+6 gc>ag are spliced differently and are degraded.
Figure 5.
Sequence analysis of genomic DNA and fully-spliced mRNA fragments. (A) Fully-spliced message from fibroblasts (direct sequencing after amplification from cDNA) showed that most transcripts contain the A467T mutation. (B) Sequencing of individual clones revealed both populations, with the A467T allele predominating.
4. DISCUSSION
Here we show that the c.2157+5_+6 gc>ag POLG allele is indeed pathogenic by affecting splicing of the POLG mRNA. This mutation causes the exon 12 - intron 12 splice junction to be bypassed, allowing the cell to utilize the next likely splice junction at c.2157+30. Within this 30 nucleotide insertion is an in-frame UAG PTC at c.2157+28–30. Transcripts with PTCs that are at least 50–55 nucleotides upstream of an intron are subjected to NMD. The message arising from the c.2157+5_+6 gc>ag POLG allele fits this criterion as POLG contains 23 exons, and the double mutation occurs in intron 12.
The degradation of the c.2157+5_+6 gc>ag POLG message was confirmed by sequencing individual mature cDNA clones. Analysis revealed that 19% of the fully spliced transcripts were alternatively spliced. These alternatively spliced transcripts all contained the two intronic mutations. Sequence analysis of prespliced POLG transcripts in the exon 7 region showed an equal proportion of wild-type message and message containing A467T. The discrepancy between the prespliced transcripts and the relatively low amount of c.2157+5_+6 gc>ag POLG message indicates that the c.2157+5_+6 gc>ag message was degraded, due to the PTC which triggered the NMD pathway. This caused an effective haplotype insufficiency, where the bulk of the surviving transcripts contain the A467T mutation. Our previous biochemical analysis showed that the A467T mutation results in a pol γ with low intrinsic polymerase activity and a defect in functional association with the pol γ accessory subunit leading to poor mtDNA replication (Chan et al., 2005a). However, this does not address why patients who are homozygous for the A467T mutation can develop one of the three major POLG disease phenotypes, and have a longer survival time as compared with compound A467T/W748S heterozygotes (Tzoulis et al., 2006). Our previous study of the first genotyped Alpers patient (A467T/E873X) (Chan et al., 2005b), and this current study take us closer to answering this question. In the previous study, we showed that for a patient with A467T/E873X POLG, most transcripts arise from the allele containing the A467T mutation, leading to mono-allelic expression of A467T pol γ, and is the most likely cause of early onset Alpers syndome in this patient (Chan et al., 2005b). Table 1 reveals a striking correlation of PTC mutations in trans with either A467T or W748S and early onset Alpers or myocerebrohepatopathy syndrome, while other mutations in trans with PTCs are associated with later onset PEO disease. This suggests that functional gene dose of the A467T allele could dictate age of onset.
We present the first analysis of the consequence of an intronic mutation that leads to alternative splicing in a POLG disease patient. Our analysis represents the second POLG case that causes one of the POLG alleles to be removed by NMD. Transcripts derived from POLG alleles containing any of the 18 known PTCs or splice-site mutations are also expected to undergo NMD. Thus, transcripts arising from alleles containing POLG PTC, frame-shift, or intronic mutations that are predicted to affect splicing should be regarded as possible targets for NMD. Consequently for patients and physicians, the mutation found in the other POLG allele may be a better predictor of disease state.
ACKNOWLEDGMENTS
We would like to thank Drs. Janssen Daly and Raghuvar Dronamraju for critically reviewing this manuscript. Funding was provided from a National Institutes of Health Career Development Award to SSLC (1K99-ES015555-01), and Intramural Research funds (Z01-ES065078) from the National Institute of Environmental Health Sciences, NIH to WCC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agostino A, Valletta L, Chinnery PF, Ferrari G, Carrara F, Taylor RW, Schaefer AM, Turnbull DM, Tiranti V, Zeviani M. Mutations of ANT1, Twinkle, and POLG1 in sporadic progressive external ophthalmoplegia (PEO) Neurology. 2003;60:1354–1356. doi: 10.1212/01.wnl.0000056088.09408.3c. [DOI] [PubMed] [Google Scholar]
- Alpers BJ. Diffuse progressive degeneration of the gray matter of the cerebrum. Archives of Neurology and Psychiatry. 1931;25:469–505. [Google Scholar]
- Ashley N, O’Rourke A, Smith C, Adams S, Gowda V, Zeviani M, Brown GK, Fratter C, Poulton J. Depletion of mitochondrial DNA in fibroblast cultures from patients with POLG1 mutations is a consequence of catalytic mutations. Hum Mol Genet. 2008;17:2496–2506. doi: 10.1093/hmg/ddn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunak S, Engelbrecht J, Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol. 1991;220:49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- Chan SS, Copeland WC. DNA polymerase gamma and mitochondrial disease: Understanding the consequence of POLG mutations. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbabio.2008.10.007. [Epub 2008 Oct 28] doi:10.1016/j.bbabio.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SSL, Longley MJ, Copeland WC. The common A467T mutation in the human mitochondrial DNA polymerase (POLG) compromises catalytic efficiency and interaction with the accessory subunit. J Biol Chem. 2005a;280:31341–31346. doi: 10.1074/jbc.M506762200. [DOI] [PubMed] [Google Scholar]
- Chan SSL, Longley MJ, Naviaux RK, Copeland WC. Mono-allelic POLG expression resulting from nonsense-mediated decay and alternative splicing in a patient with Alpers syndrome. DNA Repair. 2005b;4:1381–1389. doi: 10.1016/j.dnarep.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Copeland WC. Inherited mitochondrial diseases of DNA replication. Annu Rev Med. 2008;59:131–146. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries MC, Rodenburg RJ, Morava E, van Kaauwen EP, Ter Laak H, Mullaart RA, Snoeck IN, van Hasselt PM, Harding P, van den Heuvel LP, Smeitink JA. Multiple oxidative phosphorylation deficiencies in severe childhood multi-system disorders due to polymerase gamma (POLG1) mutations. Eur J Pediatr. 2006;166:229–234. doi: 10.1007/s00431-006-0234-9. [DOI] [PubMed] [Google Scholar]
- Del Bo R, Bordoni A, Sciacco M, Di Fonzo A, Galbiati S, Crimi M, Bresolin N, Comi GP. Remarkable infidelity of polymerase gammaA associated with mutations in POLG1 exonuclease domain. Neurology. 2003;61:903–908. doi: 10.1212/01.wnl.0000092303.13864.be. [DOI] [PubMed] [Google Scholar]
- Di Fonzo A, Bordoni A, Crimi M, Sara G, Bo RD, Bresolin N, Comi GP. POLG mutations in sporadic mitochondrial disorders with multiple mtDNA deletions. Hum Mutat. 2003;22:498–499. doi: 10.1002/humu.9203. [DOI] [PubMed] [Google Scholar]
- Dimauro S, Davidzon G. Mitochondrial DNA and disease. Ann Med. 2005;37:222–232. doi: 10.1080/07853890510007368. [DOI] [PubMed] [Google Scholar]
- Ferrari G, Lamantea E, Donati A, Filosto M, Briem E, Carrara F, Parini R, Simonati A, Santer R, Zeviani M. Infantile hepatocerebral syndromes associated with mutations in the mitochondrial DNA polymerase-{gamma}A. Brain. 2005;128:723–731. doi: 10.1093/brain/awh410. [DOI] [PubMed] [Google Scholar]
- Harding BN, Alsanjari N, Smith SJ, Wiles CM, Thrush D, Miller DH, Scaravilli F, Harding AE. Progressive neuronal degeneration of childhood with liver disease (Alpers’ disease) presenting in young adults. J Neurol Neurosurg Psychiatry. 1995;58:320–325. doi: 10.1136/jnnp.58.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouze P, Brunak S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996;24:3439–3452. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath R, Hudson G, Ferrari G, Futterer N, Ahola S, Lamantea E, Prokisch H, Lochmuller H, McFarland R, Ramesh V, Klopstock T, Freisinger P, Salvi F, Mayr JA, Santer R, Tesarova M, Zeman J, Udd B, Taylor RW, Turnbull D, Suomalainen A, Zeviani M, Chinnery PF. Phenotypic spectrum associated with mutations of the mitochondrial polymerase {gamma} gene. Brain. 2006;129:1674–1684. doi: 10.1093/brain/awl088. [DOI] [PubMed] [Google Scholar]
- Hudson G, Deschauer M, Taylor RW, Hanna MG, Fialho D, Schaefer AM, He L-P, Blakely E, Turnbull DM, Chinnery PF. POLG1, C10ORF2 & ANT1 mutations are uncommon in sporadic PEO with multiple mtDNA deletions. Neurology. 2006;66:1439–1441. doi: 10.1212/01.wnl.0000210486.32196.24. [DOI] [PubMed] [Google Scholar]
- Lamantea E, Tiranti V, Bordoni A, Toscano A, Bono F, Servidei S, Papadimitriou A, Spelbrink H, Silvestri L, Casari G, Comi G, Zeviani M. Mutations of mitochondrial DNA polymerase gamma are a frequent cause of autosomal dominant or recessive Progressive External Ophthalmoplegia. Ann Neurol. 2002;52:211–219. doi: 10.1002/ana.10278. [DOI] [PubMed] [Google Scholar]
- Lim SE, Longley MJ, Copeland WC. The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J Biol Chem. 1999;274:38197–38203. doi: 10.1074/jbc.274.53.38197. [DOI] [PubMed] [Google Scholar]
- Lutz RE, Dimmock D, Schmitt ES, Zhang Q, Tang LY, Reyes C, Truemper E, McComb RD, Hernandez A, Basinger A, Wong LJ. De Novo Mutations in POLG Presenting with Acute Liver Failure or Encephalopathy. J Pediatr Gastroenterol Nutr. 2009 Feb 25; doi: 10.1097/MPG.0b013e31817d9cad. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Naimi M, Bannwarth S, Procaccio V, Pouget J, Desnuelle C, Pellissier JF, Rotig A, Munnich A, Calvas P, Richelme C, Jonveaux P, Castelnovo G, Simon M, Clanet M, Wallace D, Paquis-Flucklinger V. Molecular analysis of ANT1, TWINKLE and POLG in patients with multiple deletions or depletion of mitochondrial DNA by a dHPLC-based assay. Eur J Hum Genet. 2006;14:917–922. doi: 10.1038/sj.ejhg.5201627. [DOI] [PubMed] [Google Scholar]
- Naviaux RK, Nguyen KV. POLG Mutations associated with Alpers’ Syndrome and Mitochondrial DNA Depletion. Ann Neurol. 2004;55:706–712. doi: 10.1002/ana.20079. [DOI] [PubMed] [Google Scholar]
- Naviaux RK, Nguyen KV. POLG Mutations associated with Alpers’ Syndrome and Mitochondrial DNA Depletion. Ann Neurol. 2005;58:491. doi: 10.1002/ana.20544. [DOI] [PubMed] [Google Scholar]
- Naviaux RK, Nyhan WL, Barshop BA, Poulton J, Markusic D, Karpinski NC, Haas RH. Mitochondrial DNA polymerase gamma deficiency and mtDNA depletion in a child with Alpers’ syndrome. Ann Neurol. 1999;45:54–58. doi: 10.1002/1531-8249(199901)45:1<54::aid-art10>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Nguyen KV, Ostergaard E, Ravn SH, Balslev T, Danielsen ER, Vardag A, McKiernan PJ, Gray G, Naviaux RK. POLG Mutations in Alpers Syndrome. Neurology. 2005;65:1493–1495. doi: 10.1212/01.wnl.0000182814.55361.70. [DOI] [PubMed] [Google Scholar]
- Nguyen KV, Sharief F, Chan SSL, Copeland WC, Naviaux RK. Molecular Diagnosis of Alpers Syndrome. J Hepatology. 2006;45:108–116. doi: 10.1016/j.jhep.2005.12.026. [DOI] [PubMed] [Google Scholar]
- Spinazzola A, Invernizzi F, Carrara F, Lamantea E, Donati A, Dirocco M, Giordano I, Meznaric-Petrusa M, Baruffini E, Ferrero I, Zeviani M. Clinical and molecular features of mitochondrial DNA depletion syndromes. J Inherit Metab Dis. 2009;265:174–192. doi: 10.1007/s10545-008-1038-z. [DOI] [PubMed] [Google Scholar]
- Tzoulis C, Engelsen BA, Telstad W, Aasly J, Zeviani M, Winterthun S, Ferrari G, Aarseth JH, Bindoff LA. The spectrum of clinical disease caused by the A467T and W748S POLG mutations: a study of 26 cases. Brain. 2006;129:1685–1692. doi: 10.1093/brain/awl097. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Wong LJ, Naviaux RK, Brunetti-Pierri N, Zhang Q, Schmitt ES, Truong C, Milone M, Cohen BH, Wical B, Ganesh J, Basinger AA, Burton BK, Swoboda K, Gilbert DL, Vanderver A, Saneto RP, Maranda B, Arnold G, Abdenur JE, Waters PJ, Copeland WC. Molecular and clinical genetics of mitochondrial diseases due to POLG mutations. Hum Mutat. 2008;29:E150–E172. doi: 10.1002/humu.20824. [DOI] [PMC free article] [PubMed] [Google Scholar]