Abstract
The development of adenoviral vectors based on non-human serotypes such as the chimpanzee adenovirus AdC7 may allow for their utilization in populations harboring neutralizing antibodies to common human serotypes. Because adenoviral vectors can be used to generate potent T cell responses, they may be useful as vaccines against pandemic influenza such as may be caused by the H5N1 strains that are currently endemic in avian populations. The influenza nucleoprotein (NP) is known to provide MHC Class I restricted epitopes that are effective in evoking a cytolytic response. Because there is only low sequence variation in NP sequences between different influenza strains, a T cell vaccine may provide heterosubtypic protection against a spectrum of influenza A strains. An AdC7 vector expressing the influenza A/Puerto Rico/8/34 NP was tested for its efficacy in protecting BALB/c mice against two H5N1 strains and compared to a conventional human adenovirus serotype 5 vaccine. The AdC7 NP vaccine elicited a strong anti-NP T cell response. When tested in a mouse challenge model, there was improved survival following challenge with two strains of H5N1 that have caused human outbreaks, Vietnam/1203/04 and Hong Kong/483/97, although the improved survival reached statistical significance only with the strain from Vietnam.
1. Introduction
Influenza A nucleoprotein is an attractive candidate as a component of a “universal” influenza vaccine [1], because murine [2, 3] and human [4] cytotoxic T lymphocytes (CTLs) activated to influenza A nucleoprotein (NP) epitopes are capable of cross-reacting with NP from different influenza A strains. This is due to the relatively low rate of drift in the NP sequence [5]. Active protection of mice from a lethal challenge with heterotypic influenza A following DNA vaccination with NP has been demonstrated [6]; this protection could be adoptively transferred to naïve mice by CD4+ or CD8+ T cells from immunized mice [7].
Adenoviral vaccines are especially efficient in eliciting strong T cell responses against products of the transgene. Successful cross-protection in mice against H5N1 avian influenza has been demonstrated using an adenovirus serotype 5 (Ad5) vaccine expressing NP from the H1N1 strain A/PR/8/34 [8]. This suggests that the use of genetic vaccines expressing non-variant viral genes may be used as a first line of defense during an epidemic where the correct conventional strain-matched hemagglutinin (HA) and neuraminidase (NA) based vaccine is not available. However, for the vector to have efficacy it is desirable to use an adenovirus serotype against which there is a low rate of prevalence of pre-existing neutralizing antibodies. Adenoviruses from animals are therefore being explored as possible vectors [9-12]. Adenoviruses that were originally isolated from chimpanzees have recently been shown to be useful vaccine vectors in a variety of animal studies and are now in development for HIV clinical trials [13-17]. In this report we have tested the efficacy of a single dose of a chimpanzee adenovirus vaccine expressing the influenza A NP in protecting against two different avian H5N1 strains that have caused human outbreaks (Hong Kong/97 and Vietnam/04) and compared it to a human adenovirus serotype 5 (Ad5) — based vector.
2. Materials and Methods
2.1 Adenovirus vectors
The nucleotide sequence encoding the H1N1 influenza A virus NP (A/Puerto Rico/8/34/Mount Sinai, GenBank accession number AF389119.1) was codon optimized and completely synthesized (Celtek Genes, Nashville, TN). An expression cassette (approximately 2.5 kb) composed of the human cytomegalovirus early promoter, the codon optimized influenza A NP coding sequence and the bovine growth hormone polyadenylation signal was inserted in place of an E1 deletion in Ad5 or chimpanzee adenovirus Pan 7 (simian adenovirus 24) by the construction of plasmid molecular clones as described [11]. The recombinant adenoviruses (AdH5-FluA NP and AdC7-FluA NP respectively) were rescued by transfecting the plasmids into HEK 293 cells. The adenoviruses were purified by cesium chloride density gradient centrifugation (PennVector, University of Pennsylvania, Philadelphia, PA) and the particle titer determined by measuring absorbance at 260 nm.
2.2 Expression of Influenza A NP in vector — transduced A549 cells
The detection of influenza A NP expression was done following a protocol described earlier [17] with some modifications. The human lung carcinoma cell line A549 (ATCC # CCL-185) was maintained in F12K medium supplemented with antibiotics and 10% fetal bovine serum (FBS). They were transduced with recombinant adenoviral vectors at a multiplicity of 103 or 104 particles per cell. After 36 h, the cells were harvested, washed with 1% FBS in phosphate-buffered saline and counted. 106 cells were permeabilized in Cytofix/Cytoperm (BD Biosciences, San Jose, CA) for 20 min on ice. Subsequently, cells were washed and incubated with an anti-influenza A NP mouse monoclonal antibody (MAB8258, Chemicon International, Temecula, CA) for 30 min on ice. The cells were washed again and stained with a phycoerythrin — labeled goat anti — mouse antibody (Caltag Invitrogen, Carlsbad, CA) for 30 min on ice. After washing again, the cells were examined by one color flow cytometry (FC 500, Beckman Coulter, Miami, FL) and analyzed using FlowJo software (Tree Star, Ashland, OR).
2.3 Mice
BALB/c mice (6–8 weeks old) were purchased from Charles River Laboratories (Wilmington, MA) and vaccinated at the University of Pennsylvania, Philadelphia, PA. Following vaccination the mice were transported to the National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, Canada where they were challenged with H5N1 influenza strains.
2.4 Peptide mapping of CD8 T cell epitopes in BALB/c mice
BALB/c mice were immunized with 5 × 1010 particles of AdH5-NP or AdC7-NP administered by intra-muscular injection. At the peak of response (day 9, previously determined) splenocytes were isolated and stimulated with pools of overlapping peptides (15-mers with 11-amino acid overlaps) spanning the entire influenza A (PR8) NP sequence in a γ-interferon (IFN-γ) ELISpot assay using 105 splenocytes per well. Spots were counted with an ELISpot reader system (Autoimmun Diagnostika, Strassberg, Germany). The number of peptide-reactive cells were represented as spot-forming cells (SFCs) per 106 splenocytes and were calculated by subtracting spot numbers in medium-only wells from spot numbers in peptide-containing wells. A cross-referencing method was used to identify the candidate for the dominant CD8+ epitope, which was further confirmed by intra-cellular staining for IFN-γ followed by flow cytometry visualization of stained cells.
2.5 Cytokine staining of murine splenocytes
Staining was conducted as described earlier [17] with some modifications. A peptide of sequence TYQRTRALV corresponding to influenza A NP residues 147-155, which is an immunodominant MHC class I epitope (H-2Kd restricted ) [18], was synthesized by Mimotopes, Adelaide, Australia. Splenocytes from control and immunized BALB/c mice (H-2Kd haplotype), harvested 9 days after vaccination with an adenovirus vector, were stimulated with the peptide at a concentration of 2 μg/ml for 5 h at 37°C in 10% CO2 in the presence of 1 μl/ml brefeldin A (GolgiPlug; BD Biosciences). After washing, cells were stained with fluorescein-labeled anti-mouse CD8 antibody (BD Biosciences). Cells were then washed and permeabilized in Cytofix/Cytoperm for 20 min on ice. Subsequently, the cells were washed again and stained with the anti-cytokine antibodies phycoerythrin (PE)-labeled anti-mouse IFN-γ BD Biosciences), PE-Cy7-labeled anti-mouse TNF-α (BD Biosciences), and allophycocyanin-labeled mouse interleukin-2 (IL2) antibody (eBioscience, SanDiego, CA). After a final wash, the cells were examined by multi-color flow cytometry (FC 500, Beckman Coulter) and analyzed using FlowJo software (Tree Star).
2.6 Influenza viruses
A stock of influenza virus strain PR8 [A/PR/8/34(H1N1)] was a kind gift of Prof. Walter Gerhard, Wistar Institute, Philadelphia, PA. The avian viruses (influenza A/Vietnam/1203/04 H5N1 and influenza A/Hong Kong/483/97) were obtained from the Centers for Disease Control, Atlanta, GA Virus stocks were prepared by culturing in minimal essential medium supplemented with 0.3% bovine serum albumin (MEM/BSA) and antibiotics, supplemented with 1μg/ml TPCK-treated trypsin, on Madin-Darby canine kidney (MDCK) cells. Virus titer was determined by standard plaque assay on MDCK cells in the presence of 1.0 μg/ml TPCK-treated trypsin. LD50 determinations were carried out by intranasally inoculating isoflurane-anaesthetized 6-week-old female BALB/c mice with 10-fold serial dilutions (five mice/dilution) of virus in 50 μl of phosphate-buffered saline and monitoring daily for disease symptoms and survival. The LD50 was calculated by the method of Reed and Muench.
2.7 Vaccination
The chimpanzee adenovirus vector AdC7-FluA NP and the human Ad5 vector AdH5-FluA NP, both expressing the NP from H1N1 strain A/PR/8/34, were used to vaccinate BALB/c mice. Mice were sedated with intra-peritoneal ketamine/xylazine, and the total adenovirus dose (1011 viral particles per mouse) divided into two 25 μl injections given intra-muscularly to the hind leg tibialis anterior.
2.8 Challenge
Mice were anesthetized in an atmosphere of 5% isoflurane and challenged with influenza A/PR/8/34, influenza A/ Vietnam/1203/04 or influenza A/Hong Kong/483/1997 in 50 μl MEM/BSA, via the intranasal route. The experiments were performed under approved animal use documents and according to the guidelines of the Canadian Council on Animal Care and of the University of Pennsylvania.
2.9 Determination of anti-influenza A neutralization titer
BALB/c mice were immunized with 1011 particles of AdH5-NP or AdC7-NP administered by intra-muscular injection. Prior to administration of the influenza virus challenge, approximately 100 μl of blood was withdrawn from 10 mice from each challenge group by retro-orbital bleeding. The sera from each challenge group were pooled (4 pools) and dilutions (two fold serial dilutions, starting at 1:4) tested for the presence of neutralizing activity against influenza A PR8 using the hemagglutination inhibition assay following the WHO standardized protocol. The test was also performed with 4 times less virus to increase sensitivity and to detect low-titer activity. Positive controls were detected at a dilution 1:512.
3. Results
3.1 Expression of influenza A NP following transduction in culture
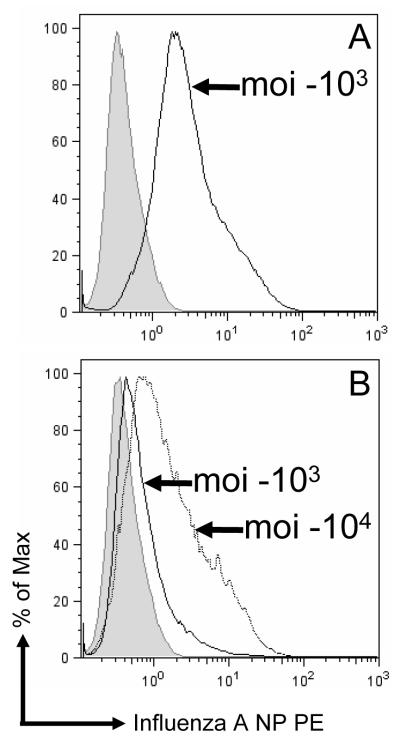
Human lung carcinoma A549 cells were infected with AdH5-FluA NP or AdC7-FluA NP as described in Materials and Methods. The cells were analyzed by flow cytometry 24 h later (Fig. 1). The level of expression was lower with the chimpanzee adenovirus vector AdC7 than with the Ad5 vector following transduction in culture, although the number of cells that could be transduced with each vector is similar. We have previously reported the much lower level of expression with the chimpanzee adenovirus vector compared to Ad5 vectors following transduction in culture [17]. However when tested in mouse muscle we have found both Ad5 and AdC7 vectors to express the marker transgenes β-galactosidase and α1-anti-trypsin at approximately comparable levels [11].
Fig. 1.
Expression of influenza A NP in A549 cells. A549 cells were infected with AdH5-FluA NP (A) or AdC7-FluA NP (B) at a multiplicity (moi) of 103 or104 viral particles per cell as indicated. The cells were stained for NP expression using a monoclonal anti-NP antibody as described in Materials and Methods and analyzed by flow cytometry. The graph shows histograms where the cell numbers (ordinate) are plotted against the fluorescence intensity (abscissa). The shaded curve corresponds to control uninfected cells. (For AdH5-FluA NP, only the data from the 103 multiplicity infection is shown)
3.2 Analysis of CD8+ T lymphocytes from immunized BALB/c mice
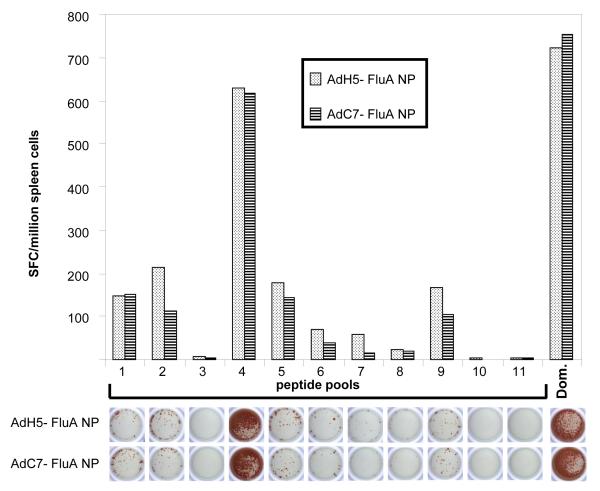
Mice vaccinated with either AdH5-FluA NP or AdC7-FluA NP were sacrificed nine days following immunization and their CD8+ T cells were analyzed. (We have previously determined the peak T cell response following immunization with Ad5 or AdC7 vectors is observed 8 and 10 days following immunization respectively, data not shown). In order to determine the breadth of the CD8+ T cell response, splenocytes were isolated and stimulated with pools of overlapping peptides (15-mers with 11-amino acid overlaps) spanning the entire influenza A (PR8) NP sequence in an IFN-γ ELISpot assay (Fig. 2). The recorded CD8+ T cell response against eleven pools (each pool containing eleven individual overlapping peptides each) encompassing the complete NP sequence is shown in Fig. 2. The response against pool 4 was clearly dominant; low level reactivity was also observed against the other pools, chiefly pools 1, 2, 5 and 9. Both the magnitude and the quality of the response was the same for the two vectors AdH5-FluA NP and AdC7-FluA NP. A cross-referencing method [19] was used to identify the candidate for the dominant CD8 epitope present in pool 4. The cross-referencing data (not shown) indicated that two peptides contained in pool 4 (SNLNDATYQRTRALV and DATYQRTRALVRTGM) were responsible for CD8+ T cell stimulation in the ELIspot assay, which was further confirmed by intra-cellular staining for IFNγ of stimulated CD8+ T cells and flow cytometry analysis. The two candidate peptides present both contain the sequence TYQRTRALV, the known dominant CD8+ T cell epitope in BALB/c mice for influenza A NP.
Fig. 2.
Breadth of CD8+ T cell response against influenza strain A/PR/8/34 NP generated in BALB/c mice immunized with 5 × 1010 particles of AdH5-NP or AdC7-NP. Splenocytes were isolated and stimulated with pools of overlapping peptides spanning the entire influenza A (PR8) NP sequence in a IFN-γ ELISpot assay. A picture of a representative ELIspot well from each assay is shown below the histograms from each peptide pool.
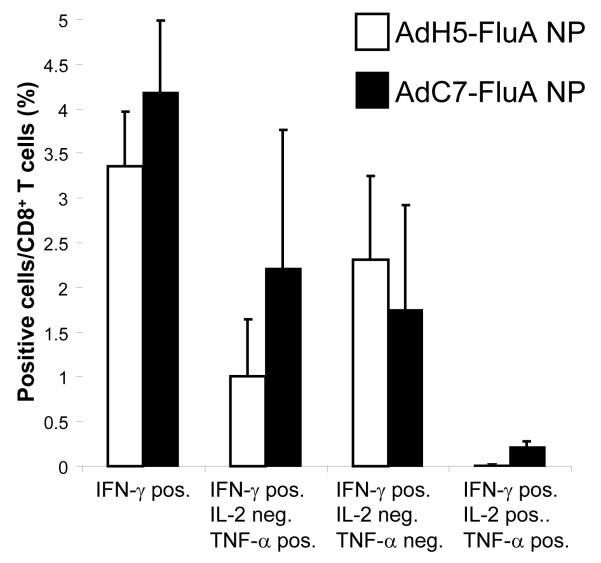
In order to further analyze the cytokine secretion profile of stimulated CD8+ T cells, splenocytes were isolated thirty days following vaccination with either AdH5-FluA NP or AdC7-FluA NP (i.e., on the same day that the identical cohorts were challenged with the avian influenza isolates) and their CD8+ T cells were analyzed by stimulating with the immunodominant peptide TYQRTRALV and staining for the cytokines IFN—γ interleukin-2 (IL-2) and tumor necrosis factor-α (TNF-α). The data are shown in Fig. 3. Splenocytes from each of three mice that had received either of the two vectors showed a robust antigen-specific T cell response. It was of interest to determine the subset of the CD8+ T cells that were polyfunctional, i.e. capable of secreting cytokines such as IL-2 and TNF-α, which may be an indication of the quality of the T cell response [20] especially with respect to the formation of a memory population [21]. There was no significant difference in the proportion of cytokine secreting CD8+ T cells between mice administered either the AdH5-FluA NP and AdC7-FluA NP vectors except for the number of CD8+ T cells that were capable of secreting all three cytokines; there were significantly more (P < 0.01) triple positive cells from the mice that had been administered the AdC7-FluA NP compared to those vaccinated with AdH5-FluA NP. It was also of interest to determine the status of CD8+ T cells in the lung because these cells would presumably constitute the first line of defense against an airway infection. The cells from the lungs of each of the three mice vaccinated with either vector were pooled and analyzed. The fraction of cells from the lungs that could be specifically stimulated was also found to be high, 14% and 25% of all CD8+ respectively for the AdH5-FluA NP and AdC7-FluA NP vectors.
Fig. 3.
Analysis of CD8+ T cells from BALB/c mice vaccinated with AdH5-FluA NP or AdC7-FluA NP. Splenocytes were harvested from 3 mice for each vector 30 days following vaccination and the percentage of CD8+ T cells that could be stimulated by the peptide TYQRTRALV (corresponding to influenza A NP residues 147-155) to secrete IFN-γ IL-2, and TNF-α was determined. The subset of IFN-γ positive cells which were additionally positive for TNF-α and/or IL-2 in each of the samples is also shown.
In order to determine whether the adenovirus vaccines had generated any humoral response against influenza virus, sera from immunized mice obtained at the time of challenge were tested for the presence of neutralizing activity against influenza A strain PR/8/34 as described in Materials and Methods. No neutralizing activity could be detected in any of the NP expressing adenovirus vector vaccinated mice.
3.3 Challenge of the influenza A NP vaccinated mice with influenza virus H1N1 (strain A/PR/8/34)
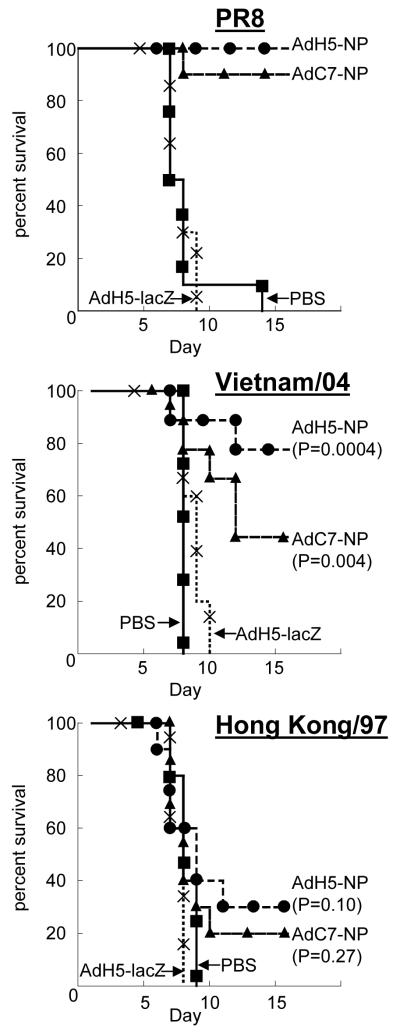
Forty BALB/c mice (6-8 weeks old) were divided into two groups of ten mice each that were dosed intra-muscularly with 1011 viral particles of either AdH5-FluA NP or AdC7-FluA NP. Twenty mice served as controls, ten dosed with saline and ten with an irrelevant E1 and E3 deleted adenovirus vector, AdH5-lacZ (Ad5 expressing E. coli β-galactosidase). Thirty-four days later all the mice were challenged intra-nasally with 10 LD50 of with the H1N1 (strain A/PR8/34) strain of influenza that carries the NP with sequence identical to the NP present in the adenovirus transgene. The survival data are shown in Fig. 4 (upper graph). All the control mice succumbed to the challenge but all but one of the mice that had been vaccinated with either the AdH5-FluA NP or AdC7-FluA NP survived the challenge.
Fig 4.
Survival plots of BALB/c mice vaccinated with the chimpanzee adenovirus vector AdC7-FluA NP and AdH5-FluA NP and challenged with the H1N1 influenza virus strain A/PR/8/34 (upper graph), and the H5N1 strains A/Vietnam/1203/04 (middle graph) and A/Hong Kong/483/97. Vaccination and challenge were carried out as described in Materials and Methods. The data were analyzed by the logrank test using MedCalc software. The statistical significance of each of the NP vaccinated groups challenged with the heterosubtypic H5N1 strains is indicated on the graph.
3.4 Heterosubtype challenge of the influenza A NP (H1N1 strain A/PR/8/34) vaccinated mice with avian H5N1 strains Hong Kong/97 and Vietnam/04
Two challenge experiments using either Hong Kong/97 or Vietnam/04 avian influenza H5N1 strains were carried out. Both strains of virus have been associated with human outbreaks. For each experiment, thirty BALB/c mice (6-8 weeks old) were divided into two groups of ten mice each that were dosed intra-muscularly with 1011 viral particles of either AdH5-FluA NP or AdC7-FluA NP. Ten mice served as controls, five dosed with PBS and five with an irrelevant E1 and E3 deleted adenovirus vector, AdH5-lacZ. Thirty days later all the mice were challenged intra-nasally with 100 LD50 of the appropriate H5N1 influenza virus. The NP present in the challenge viruses, H5N1 strains Hong Kong/97 and Vietnam/04 have sequence identities of 94% and 93% respectively with the H1N1 PR/8/34 NP and all include the immunodominant class I epitope spanning amino acids 147 -155 (TYQRTRALV). The survival data are shown in Fig. 4 (middle and lower graphs). Both AdH5 and AdC7 NP vectors resulted in improved survival versus the controls following challenge with either H5N1 strain. However. this apparent improved survival reached statistical significance only for the Vietnam/04 strain..
4. Discussion
The antigenic components of the standard trivalent influenza vaccine are HA and NA from the circulating influenza strains. The efficacy of this vaccine is contingent upon the degree of match that exists between the vaccine and challenge strains with respect to these viral surface glycoproteins that are the most prone to antigenic drift. This presents a challenge in the event of influenza pandemics where high morbidity and mortality is feared, as is possible with re-assortment strains derived from the avian H5N1 viruses or with strains similar to those that have caused sporadic outbreaks in human subjects; the rapidity of viral dissemination may not allow time for the production of large amounts of the appropriate HA and NA based vaccine.
In contemplating strategies to combat this eventuality, vaccines that provide protection by eliciting a strong cytotoxic T cell response may be useful when the T cell epitopes are derived from mostly invariant proteins such as NP or M2. However NP-based vaccines delivered using recombinant vaccinia [22], fowlpox [23] and DNA [24] have not been found to afford adequate protection. In contrast, an Ad5 recombinant NP vaccine reduced the duration of illness in pigs and in combination with an HA vaccine, was found to improve protection over that afforded by HA alone [25]. Similarly, DNA vaccination followed by a recombinant Ad5 boost using the A/PR/8/34 NP was able to protect mice against a heterosubtypic challenge with two H5N1 strains [8] showing the promise of adenoviruses as useful vectors for generating T cell immunity. However, because the activity of human based adenovirus vectors is considerably attenuated by pre-existing immunity to the vector, the availability of adenovirus serotypes against which the prevalence of pre-existing antibodies is low, is desirable. The chimpanzee adenovirus vector AdC7 is useful in this respect [14]. Although the level of transgene expression following transduction with the AdC7 vector is low following transduction in culture, we have found that expression levels with the two vectors are comparable in mouse muscle tissue in case of transgenes such as β-galactosidase or α1-anti-trypsin [11]. This is supported by the similar magnitude of the measured T cell response as well as the degree of protection from challenge afforded by the two vectors. Moreover, this protection was evident in response to the priming dose, which may be important in the event where some level of immediate protection is desirable in the face of a quickly spreading epidemic and where there may not be time for a boost, although that would clearly be desirable.
The results presented herein show that it is possible to obtain a measure of cross-protection using adenovirus vaccine expressing NP. It would also be interesting to determine whether this protection may be improved by adding other protein components of influenza virus such as the matrix protein M2 that are also known to be less susceptible to sequence drift.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Kaiser J. A one-size-fits-all flu vaccine? Science. 2006;312(5772):380–2. doi: 10.1126/science.312.5772.380. [DOI] [PubMed] [Google Scholar]
- [2].Townsend AR, McMichael AJ, Carter NP, Huddleston JA, Brownlee GG. Cytotoxic T cell recognition of the influenza nucleoprotein and hemagglutinin expressed in transfected mouse L cells. Cell. 1984;39(1):13–25. doi: 10.1016/0092-8674(84)90187-9. [DOI] [PubMed] [Google Scholar]
- [3].Yewdell JW, Bennink JR, Smith GL, Moss B. Influenza A Virus Nucleoprotein is a Major Target Antigen for Cross-Reactive Anti-Influenza A Virus Cytotoxic T Lymphocytes. PNAS. 1985;82(6):1785–89. doi: 10.1073/pnas.82.6.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Boon ACM, de Mutsert G, van Baarle D, Smith DJ, Lapedes AS, Fouchier RAM, et al. Recognition of Homo- and Heterosubtypic Variants of Influenza A Viruses by Human CD8+ T Lymphocytes. J Immunol. 2004;172(4):2453–60. doi: 10.4049/jimmunol.172.4.2453. [DOI] [PubMed] [Google Scholar]
- [5].Gorman OT, Bean WJ, Kawaoka Y, Donatelli I, Guo YJ, Webster RG. Evolution of influenza A virus nucleoprotein genes: implications for the origins of H1N1 human and classical swine viruses. J Virol. 1991;65(7):3704–14. doi: 10.1128/jvi.65.7.3704-3714.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ulmer JB, Donnelly JJ, Parker SE, Rhodes GH, Felgner PL, Dwarki VJ, et al. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science. 1993;259(5102):1745–9. doi: 10.1126/science.8456302. [see comment] [DOI] [PubMed] [Google Scholar]
- [7].Ulmer JB, Fu TM, Deck RR, Friedman A, Guan L, DeWitt C, et al. Protective CD4+ and CD8+ T cells against influenza virus induced by vaccination with nucleoprotein DNA. Journal of Virology. 1998;72(7):5648–53. doi: 10.1128/jvi.72.7.5648-5653.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Epstein SL, Kong WP, Misplon JA, Lo CY, Tumpey TM, Xu L, et al. Protection against multiple influenza A subtypes by vaccination with highly conserved nucleoprotein. Vaccine. 2005;23(4647):5404–10. doi: 10.1016/j.vaccine.2005.04.047. [DOI] [PubMed] [Google Scholar]
- [9].Kremer EJ, Boutin S, Chillon M, Danos O. Canine adenovirus vectors: an alternative for adenovirus-mediated gene transfer. Journal of Virology. 2000;74(1):505–12. doi: 10.1128/jvi.74.1.505-512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mittal SK, Middleton DM, Tikoo SK, Babiuk LA. Pathogenesis and immunogenicity of bovine adenovirus type 3 in cotton rats (Sigmodon hispidus) Virology. 1995;213(1):131–9. doi: 10.1006/viro.1995.1553. [DOI] [PubMed] [Google Scholar]
- [11].Roy S, Gao G, Lu Y, Zhou X, Lock M, Calcedo R, et al. Characterization of a family of chimpanzee adenoviruses and development of molecular clones for gene transfer vectors. Human Gene Therapy. 2004;15(5):519–30. doi: 10.1089/10430340460745838. [DOI] [PubMed] [Google Scholar]
- [12].Xu ZZ, Hyatt A, Boyle DB, Both GW. Construction of ovine adenovirus recombinants by gene insertion or deletion of related terminal region sequences. Virology. 1997;230(1):62–71. doi: 10.1006/viro.1997.8452. [DOI] [PubMed] [Google Scholar]
- [13].Kobinger GP, Feldmann H, Zhi Y, Schumer G, Gao G, Feldmann F, et al. Chimpanzee adenovirus vaccine protects against Zaire Ebola virus. Virology. 2006;346(2):394–401. doi: 10.1016/j.virol.2005.10.042. [DOI] [PubMed] [Google Scholar]
- [14].Zhi Y, Figueredo J, Kobinger GP, Hagan H, Calcedo R, Miller JR, et al. Efficacy of severe acute respiratory syndrome vaccine based on a nonhuman primate adenovirus in the presence of immunity against human adenovirus. Human Gene Therapy. 2006;17(5):500–6. doi: 10.1089/hum.2006.17.500. [DOI] [PubMed] [Google Scholar]
- [15].Reyes-Sandoval A, Fitzgerald JC, Grant R, Roy S, Xiang ZQ, Li Y, et al. Human immunodeficiency virus type 1-specific immune responses in primates upon sequential immunization with adenoviral vaccine carriers of human and simian serotypes. Journal of Virology. 2004;78(14):7392–9. doi: 10.1128/JVI.78.14.7392-7399.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tatsis N, Tesema L, Robinson ER, Giles-Davis W, McCoy K, Gao GP, et al. Chimpanzee-origin adenovirus vectors as vaccine carriers. Gene Therapy. 2006;13(5):421–9. doi: 10.1038/sj.gt.3302675. [DOI] [PubMed] [Google Scholar]
- [17].Roy S, Zhi Y, Kobinger GP, Figueredo J, Calcedo R, Miller JR, et al. Generation of an adenoviral vaccine vector based on simian adenovirus 21. Journal of General Virology. 2006;87(Pt 9):2477–85. doi: 10.1099/vir.0.81989-0. [DOI] [PubMed] [Google Scholar]
- [18].Rotzschke O, Falk K, Deres K, Schild H, Norda M, Metzger J, et al. Isolation and analysis of naturally processed viral peptides as recognized by cytotoxic T cells. Nature. 1990;348(6298):252–4. doi: 10.1038/348252a0. [DOI] [PubMed] [Google Scholar]
- [19].van der Most RG, Harrington LE, Giuggio V, Mahar PL, Ahmed R. Yellow fever virus 17D envelope and NS3 proteins are major targets of the antiviral T cell response in mice. Virology. 2002;296(1):117–24. doi: 10.1006/viro.2002.1432. [DOI] [PubMed] [Google Scholar]
- [20].Makedonas G, Betts MR. Polyfunctional analysis of human t cell responses: importance in vaccine immunogenicity and natural infection. Springer Semin Immunopathol. 2006;28(3):209–19. doi: 10.1007/s00281-006-0025-4. [DOI] [PubMed] [Google Scholar]
- [21].Harari A, Dutoit V, Cellerai C, Bart PA, Du Pasquier RA, Pantaleo G. Functional signatures of protective antiviral T-cell immunity in human virus infections. Immunological Reviews. 2006;211:236–54. doi: 10.1111/j.0105-2896.2006.00395.x. [DOI] [PubMed] [Google Scholar]
- [22].Andrew ME, Coupar BE, Boyle DB, Ada GL. The roles of influenza virus haemagglutinin and nucleoprotein in protection: analysis using vaccinia virus recombinants. Scandinavian Journal of Immunology. 1987;25(1):21–8. doi: 10.1111/j.1365-3083.1987.tb01042.x. [DOI] [PubMed] [Google Scholar]
- [23].Webster RG, Kawaoka Y, Taylor J, Weinberg R, Paoletti E. Efficacy of nucleoprotein and haemagglutinin antigens expressed in fowlpox virus as vaccine for influenza in chickens. Vaccine. 1991;9(5):303–8. doi: 10.1016/0264-410x(91)90055-b. [DOI] [PubMed] [Google Scholar]
- [24].Macklin MD, McCabe D, McGregor MW, Neumann V, Meyer T, Callan R, et al. Immunization of pigs with a particle-mediated DNA vaccine to influenza A virus protects against challenge with homologous virus. Journal of Virology. 1998;72(2):1491–6. doi: 10.1128/jvi.72.2.1491-1496.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wesley RD, Tang M, Lager KM. Protection of weaned pigs by vaccination with human adenovirus 5 recombinant viruses expressing the hemagglutinin and the nucleoprotein of H3N2 swine influenza virus. Vaccine. 2004;22(2526):3427–34. doi: 10.1016/j.vaccine.2004.02.040. [DOI] [PubMed] [Google Scholar]