Abstract
Although it is well established that DNA-protein crosslinks are formed as a consequence of cellular exposure to agents such as formaldehyde, transplatin, ionizing and ultraviolet radiation, the biochemical pathways that promote cellular survival via repair or tolerance of these lesions are poorly understood. To investigate the mechanisms that function to limit DNA-protein crosslink-induced cytotoxicity, the Saccharomyces cerevisiae non-essential gene deletion library was screened for increased sensitivity to formaldehyde exposure. Following low-dose, chronic exposure, strains containing deletions in genes mediating homologous recombination showed the greatest sensitivity, while under the same exposure conditions, deletions in genes associated with nucleotide excision repair conferred only low to moderate sensitivities. However, when the exposure regime was changed to a high-dose acute (short-term) formaldehyde treatment, the genes that conferred maximal survival switched to the nucleotide excision repair pathway, with little contribution of the homologous recombination genes. Data are presented which suggest that following acute formaldehyde exposure, repair and/or tolerance of DNA-protein crosslinks proceeds via formation of nucleotide excision repair-dependent single-strand break intermediates and without a detectable accumulation of double-strand breaks. These data clearly demonstrate a differential pathway response to chronic versus acute formaldehyde exposures and may have significance and implications for risk extrapolation in human exposure studies.
1. Introduction
Despite the evidence that DNA-protein crosslinks (DPCs) are a threat to genomic integrity, the biochemical pathways that promote cellular survival via repair or tolerance of these lesions are poorly understood. There have been several reports that implicate specific pathways in this process, such as nucleotide excision repair (NER) [1-3], proteasomal degradation [2, 4], Fanconi/BRCA pathway [5], and homologous recombination (HR) [1, 5]. However, the accumulated data have not given rise to a clear and defined role for each pathway, and in some instances, the findings are inconsistent concerning the relative contributions of each process. For example, transplatin-induced DPCs were found to be more persistent in NER-deficient xeroderma pigmentosum (XP) group A fibroblasts when compared with normal cells, suggesting a critical role for NER in the removal of these lesions [3]. However, an NER-centric model does not explain the data of Gantt [6], who showed that rapidly-proliferating, SV40-transformed XPA cells were proficient in the repair of transplatin-induced DPCs, whereas the slow-growing, non-transformed parental line was deficient in repair of these lesions [6]. In addition, other studies have demonstrated that sensitivity to formaldehyde-induced crosslinks was not significantly higher in XPA and XPF cells [4, 7]. Those studies suggested that a yet undefined “active repair process” might control the removal of formaldehyde-induced DPCs [4]. The complexity of DPC repair was further exemplified by studies demonstrating that inhibition of the proteasomal activity in mammalian cells resulted in decreased repair of formaldehyde-induced DPCs. This suggested that DPC removal was dependent on proteasomal degradation of the cross-linked protein [2, 4].
Biochemical studies have also implicated NER in the repair of DPCs. Reconstituted NER complexes have been shown to initiate repair of DPCs and DNA-peptide crosslinks. In the first biochemical study to investigate whether the NER pathway could remove intact covalently linked proteins, Minko et al created a site-specific ∼16 kDa protein adduct on DNA linked via a reduced abasic site [8]. The prokaryotic NER complex, UvrABC, was able to catalyze dual incisions on the DPC-containing strand, with efficiency comparable to that measured for polycyclic aromatic hydrocarbon-modified DNA substrates. Subsequent analyses of UvrABC incision on DNA-peptide crosslinks revealed increased kinetics of incision when the lesion was a small oligopeptide [9]. These data suggested that proteolytic degradation of the protein portion of the DPC was not absolutely essential for E. coli NER to initiate repair. More recently, Nakano et al examined the efficiency of the E. coli UvrABC incision as a function of oligopeptide-adduct size [1]. This investigation demonstrated that the bacterial NER system was able to repair DPCs containing cross-linked proteins less than 12-14 kDa in size. Interestingly, larger DPCs appeared to be processed by the E. coli recBCD-dependent pathway, suggesting the formation of HR intermediates in the course of repair [1]. In contrast, the reconstituted mammalian NER system was incapable of excision of an intact 16 kDa DNA crosslinked protein, while efficient removal of DNA-peptide substrates was observed [10].
In order to more clearly define the cellular pathways or proteins involved in repair or tolerance of DPC adducts, a genome-wide approach was utilized. Herein, results are reported on the screening of the Saccharomyces cerevisiae non-essential gene deletion library (∼5000 genes) for deletions that enhance cytotoxicity following exposure to the DPC-inducing agent, formaldehyde. As will be described, there are dramatic differences in the DNA repair and tolerance pathways that confer formaldehyde resistance following chronic versus acute exposures.
2. Materials and methods
2.1 Yeast strains and chemicals
Chemicals were purchased from Sigma unless otherwise noted. The MAT-a (BY4741) S. cerevisiae deletion strain library was obtained from the European S. cerevisiae archives for functional analysis (EUROSCARF). Additional pso2Δ S. cerevisiae strains, LBY9 (W303-1A-background) and KGY212 (A364A-background) were obtained from Peter McHugh (U. of Oxford) [11] and Robb Moses (OHSU) [12], respectively.
2.2 Parameters determining chronic and acute formaldehyde screening conditions
The formaldehyde concentration used in the chronic exposure was determined by growing wild-type, rad4Δ and rad52Δ strains in YPD (yeast extract, peptone, dextrose) media containing 0-10 mM formaldehyde for 48 hr. Additionally, exponentially growing cultures were serially diluted and aliquots spotted on agar-YPD plates containing formaldehyde concentrations ranging from 0-10 mM. Colonies grew for 48 hr and optimal concentrations for differential cytotoxicity were determined.
To determine the conditions for acute formaldehyde exposure, the same yeast strains as described above were harvested in exponential growth phase and resuspended in media containing 20-80 mM formaldehyde for 15 min. Cells were collected by centrifugation, resuspended in fresh media and aliquots of serially diluted cultures spotted on agar-YPD plates. Differential survival was assessed after 48 hr at 30°C.
2.3 Formaldehyde sensitivity screen of the non-essential gene library
The master MAT-a (BY4741) S. cerevisiae deletion strain library was stored at -80°C in YPD + 15% glycerol. Duplicates and liquid cultures were made by growing the yeast in YPD media for 48 hr at 30°C as follows: cells were transferred (∼1μl) with a 96 Floating-Pin Replicator to rich YPD agar plates containing G418 (200 μg/ml). The liquid cultures were transferred using the floating-pin replicator in triplicate onto one plate (Omnitray, NUNC plates) containing no formaldehyde and another plate containing 1.5 mM formaldehyde. Strains were grown at 30°C and imaged at 24 and 48 hr after plating using an AlphaEase FC Imaging System. It was predetermined that formaldehyde was stable under these conditions and plates could be used for at least 1 week after initial preparation as evidenced by comparable levels of formaldehyde-induced cytotoxicity. Strains were classified as sensitive when limited or no growth was observed in the two replicates of the 1.5 mM formaldehyde plating. The entire library was independently screened twice with each deletion strain assayed in triplicate per screen. Strains that exhibited sensitivity to formaldehyde relative to the wild-type strain were individually re-assayed from freezer stocks to verify formaldehyde sensitivity.
2.4 Cell survival assays
For rapid semi-quantitative survival assessments, cells were cultured in YPD overnight and diluted to 1×107 cells/ml. Aliquots (2 μl) of serial 10-fold dilutions of cells were spotted onto YPD agar plates containing the indicated concentrations of formaldehyde ranging from 0-2.0 mM. Cells were cultured at 30°C for 2 days (chronic exposure). For acute formaldehyde exposure, cells from an overnight culture were harvested by centrifugation, resuspended in 1 ml water and exposed to 60 mM formaldehyde for 15 min. After the exposure, cells were collected by centrifugation, washed twice in water, and resuspended in YPD at a concentration of 1×107 cells/ml. Each strain was serially diluted (1:10) in YPD before plating 2 μl of each suspension onto YPD agar without formaldehyde. Cells were grown for 2 days at 30°C and images captured on an AlphaEase FC imaging system.
In order to assess the relative survival, strains representing major DNA repair pathways were chosen and colony forming assays performed. For both chronic and acute exposure, the yeast strains were grown overnight at 30°C, with vigorous shaking, resulting in a log-phase culture. Cells were diluted such that following formaldehyde exposure, the total number of surviving colonies ranged from 30-300 cells per plate under unexposed conditions. For chronic exposures, formaldehyde was added to the plates at various concentrations (0-2.0 mM), and colonies were counted after 2-3 days of growth at 30°C. For acute exposures, cells were pelleted by centrifugation, resuspended, and exposed as indicated to formaldehyde (0-80 mM) for 15 min, pelleted at 4,000 rpm for 2 min and washed twice. Cells were resuspended in media and plated onto YPD agar without formaldehyde. All experiments were repeated at least three independent times.
2.5 Pulsed field gel electrophoresis analyses (PFGE)
PFGE analyses were performed to determine if strand breaks accumulate following formaldehyde exposure. Assays were performed using the CHEF genomic DNA plug kit (Bio-Rad). Yeast cells were grown overnight at 30°C or for 48 hr in the presence of 1.5 mM formaldehyde. For acute exposure, yeast strains were incubated with 60 mM formaldehyde for 15 min. After exposure, cells were washed, pelleted and resuspended in YPD media. Additionally, PFGE was performed on cells arrested in G1 using α-mating factor. The arrest (>90%) was confirmed visually using a light microscope (Nikon, Eclipse E 200, 100× magnification). Following arrest, cells were exposed to 60 mM formaldehyde for 15 min, pelleted, washed twice, and resuspended in YPD containing α-mating factor (20 μg/ml) in order to maintain G1 arrest. From the inoculated media, 2×108 cells were used for each plug. Plugs were made of unexposed cells and from aliquots harvested at 0, 4, 6, and 8 hr after exposure. The cells were processed for PFGE analyses according to the manufacturer's protocol. The yeast chromosomes were separated on a 2% pulsed field certified agarose gel in 0.5× TBE (90 mM Tris base, 89 mM boric acid, 10 mM EDTA pH 8.0), recirculating at 14°C, for 20 hr at 6.5 V/cm with a 60 to 120 sec switch time ramp at an included angle of 120°. Images were captured on an AlphaEase FC imaging system.
2.6 SDS/KCl precipitation to detect DPCs
After chronic or acute treatment, 1×107 cells were pelleted by centrifugation, resuspended and washed with 1 ml of TE (10 mM Tris-HCl, 1 mM EDTA, pH 7.8). Cells were resuspended in 0.25 ml of lyticase buffer (BioRad), 8 μl of lyticase (BioRad), and 1 μl of yeast protease inhibitor cocktail for 30 min at 30°C. After confirming spheroplast formation by microscopy, 0.25 ml of 4% SDS was added and cells were frozen and thawed at 55°C for 5 min. To the suspension, 0.5 ml of 200 mM KCl, 20 mM Tris-HCl (pH 7.5) was added and the DNA sheared by repeated pipetting. The solution was cooled on ice for 5 min. The precipitate was pelleted at 3500 × g for 5 min and the supernatant saved to measure unbound DNA. The pellet was washed in 20 mM Tris-HCl (pH 7.5), 200 mM KCl, incubated at 55°C for 5 min, cooled on ice for 5 min, followed by centrifugation at 3600 × g for 4 min. This wash procedure was repeated 3 times before a final resuspension in 0.5 ml of the same buffer. Proteins were digested by adding 0.2 mg/ml of proteinase K and incubating at 55°C for 45 min. The solution was cooled on ice and 50 μl of 10 mg/ml bovine serum albumin (BSA) (New England BioLabs) was added and placed on ice for 5 min. The precipitate was pelleted at 3500 × g for 5 min. Individually, the final supernatant (10 μl) and the supernatant from the first wash (5 μl) were separated on a 0.7% agarose gel stained with either ethidium bromide (0.001%) or a 1× solution of Sybr Gold™ (Invitrogen). Images were captured on an AlphaEase FC imaging system and the amount of DNA in each lane was quantified with ImageQuant software (GE Healthcare Lifesciences). To control for potential differential efficiencies in the extraction of DNA from formaldehyde-treated cells, the amount of cross-linked DNA was calculated as a ratio of SDS/KCl precipitated DNA to unbound DNA. Due to the limited quantities of cross-linked DNA, the amounts of recoverable unbound DNAs were always at least in 100-fold excess over cross-linked DNAs.
2.7 Velocity sedimentation ultracentrifugation
Overnight cultures of yeast cells were diluted 1:2 in fresh media and grown for 4 hr. Cells were arrested with α-mating factor (20 μg/ml) 2 hr prior to formaldehyde treatment. Cells were treated with 60 mM formaldehyde for 15 min, washed twice and resuspended in YPD and grown for an additional 4 hr in YPD containing α-mating factor. β-mercaptoethanol (0.2%) and 45 mg of lyticase were added and the cells incubated at 30°C for 45 min. Cells were centrifuged at 1500 × g for 10 minutes and resuspended in 2 ml TE. Sucrose gradients, 15 to 30%, were made by mixing the appropriate amount of sucrose with a buffer consisting of 15 mM EDTA, 10 mM Tris-HCl pH 7.4, 0.9 M NaCl, and 0.1 N NaOH. Linear gradients were poured with a gradient mixer into Sorvall PA ultracentrifuge tubes.
Lyticase digested cells were poured on top of the gradient and 50 μl of 10% SDS was added. The gradients were placed at 4°C for 10 min to lyse cells, and centrifuged at 4°C, 11,500 rpm for 20 hr in a Sorvall AH627 rotor. After centrifugation, fractions (800 μl) were removed from the bottom of the tubes with an automated fractionator and 24 μl of 2N HCl was added to neutralize the solution. A total of 50 μl of each fraction was added to 100 μl of TE containing 1.5× Sybr Gold™ in a 96-well plate. The plate was scanned with a Spectramax Gemini XS plate reader using an excitation wavelength of 490 nm and an emission wavelength of 540 nm.
3. Results
3.1 Formaldehyde sensitivity screen
In order to identify cellular pathways involved in enhancing cellular resistance to DPC-inducing agents, the S. cerevisiae MAT-a non-essential gene library consisting of ∼5000 individual strains [13] was screened for formaldehyde sensitivity under conditions of continuous (chronic), low-dose exposure. Prior to screening the entire library, it was necessary to establish appropriate assay conditions in which a differential in cytotoxicity was observed between the wild-type and representative strains from the major DNA repair pathways, such as NER deficient rad4Δ and HR deficient rad52Δ. Aliquots of exponentially growing cultures were spotted on agar plates that contained concentrations of formaldehyde ranging from 0-10 mM. Wild-type cells showed a marked decrease in survival at the 2.0 mM concentration, with virtually no survival observed at concentrations > 3 mM (data not shown). In contrast, repair-deficient strains showed greater cytotoxicity as compared to the wild-type strain at 1.0-1.5 mM formaldehyde (data not shown).
In order to address whether the formaldehyde was stable under the conditions used, plates containing 1.5 mM formaldehyde were prepared and stored for various times and subsequently used in screening assays. These data revealed that highly reproducible results could be obtained using plates that had been stored up to 7 days, suggesting that the effective concentration of formaldehyde does not significantly change due to interactions with the YPD media. To ensure that these doses of formaldehyde were cytotoxic, agar plugs containing the yeast spots lacking colonies were removed from the formaldehyde-containing plates and placed in liquid YPD and allowed to grow for 48 hr. No growth was observed (data not shown).
Having established the assay conditions that gave a robust signal-to-noise ratio, the complete library was screened two independent times with each strain assayed in triplicate per screen. A representative set of plates from the screen is shown in Supplementary Fig. 1. Increased cytotoxicity under the chronic exposure conditions was observed in 44 deletion strains, with the majority being genes involved in cell cycle and DNA repair. Genes involved in metabolism, transcription, protein fate and cellular transport were also identified (Table 1). All strains listed as sensitive in Table 1 were independently confirmed by re-assaying individual strains using serial dilution spot assays under chronic formaldehyde exposure. Only those deletion strains showing significant sensitivity are listed in Table 1. Interestingly, the genome-wide screen suggested that for strains involved in DNA repair and tolerance mechanisms, the strains deleted in HR genes, not NER, appeared to be the most sensitive to formaldehyde.
TABLE 1. Categorization of Yeast Strains that Display Increased Sensitivity Following Chronic Formaldehyde Exposure.
Formaldehyde sensitive strains were categorized in functional groups according to the MIPS functional database (http://mips.gsf.de/proj/funcatDB/). These strains were identified in the two independent screens and sensitivity was confirmed by spot assay. Strains that failed these criteria were not included.
Genes Required for Formaldehyde Resistance
| Functional Category | Genes/ORFs |
|---|---|
| Metabolism…………………………….. | SFA1 ERG3 ERG6 ERG5 PSD1 ADH1 |
| Cell Cycle and DNA Processing……………………………. | SPT7 CDC50 RAD55 XRS2 RAD51 RAD4 CDC26 RAD54 MSH1 MGM101 RAD5 TOP3 MMS22 RAD52 SGS1 RAD14 MRE11 RAD50 RAD1 CTF4 |
| Transcription………………………….. | RPN4 SNF6 DAL81 LSM1 SWI3 SNF2 MED1 |
| Miscellaneous………………………… | NBP2 VID22 ARP5 NUP84 VPS9 ECM30 OPI11 NRP1 YLR235C TMA23 BEM4 |
3.2 Homologous recombination protects against chronic formaldehyde exposure
Following the initial genome-wide chronic screen, the relative contributions of each DNA repair and tolerance pathway in protecting the cell against formaldehyde-induced toxicity were assessed in more detail. A subset of strains that had been previously characterized to be involved in DNA damage repair and tolerance mechanisms was chosen for more detailed cytotoxicity analyses based on their sensitivity to formaldehyde. In addition to those strains identified in the screen, additional strains from the NER, base excision repair (BER), HR and non-homologous end-joining (NHEJ) pathways that were not identified as sensitive in the genome-wide screen were selected as controls (Fig. 1 and Supplementary Fig. 2).
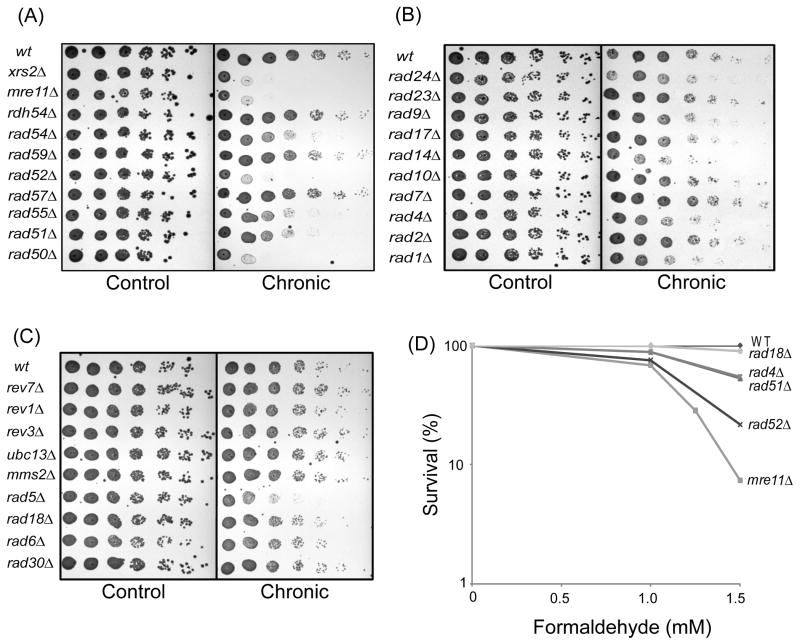
Fig. 1. Survival Analyses of Yeast Deletion Strains Exposed to Chronic Low Dose Formaldehyde.
(A) HR deletion strains, (B) NER and cell cycle-associated deletion strains, (C) Translesion synthesis (post replication repair) strains show differential growth following formaldehyde exposure. The strains shown were grown exponentially in YPD media and 2 μl aliquots of serially diluted cells (10-fold, left to right) were spotted onto an agar plate and grown for 2 days at 30°C. In each case, the left panel is unexposed (no formaldehyde) and the right panel shows the same strains grown in the presence of (A) 1.25 mM, (B, C) 1.5 mM formaldehyde. (D) The colony forming ability of mre11Δ, rad4Δ, rad52Δ, rad51Δ, rad18Δ and wild-type strains after chronic exposure for 48 hr.
Cells were cultured in YPD and serial dilutions of these exponentially growing cells were spotted onto YPD agar plates containing 1.25 mM or 1.5 mM formaldehyde and grown at 30°C for 48 hr. Consistent with the library screen, the highest sensitivities were observed in many of the strains with gene deletions in the HR pathway, including rad50Δ, rad52Δ, mre11Δ and xrs2Δ. These strains showed high sensitivities when exposed to only 1.25 mM formaldehyde (Fig. 1A). In contrast, strains deleted for rad51, rad54 and rad55 displayed moderate sensitivity, while rdh54Δ, rad59Δ and rad57Δ were not sensitive under these conditions. Comparatively, strains deleted for the NER mutants, rad1, rad4, and rad14 demonstrated only low to moderate sensitivities to low dose formaldehyde, while strains deleted in rad2, rad7, rad1, rad17, rad23, and rad24 were closely comparable with the wild-type strain (Fig. 1B). Strains with deletions in genes traditionally defined as the RAD6 epistasis group (post-replication repair) were not sensitive to formaldehyde under these conditions, with the exception being the moderate to high sensitivity of the rad5 strain (Fig. 1C). To confirm these data, colony-forming assays were carried out on a subset of these strains (Fig. 1D). As anticipated based on the data in Fig. 1A-C, rad18Δ was comparable to the wild-type strain, while rad4Δ and rad51Δ were moderately sensitive and mre11Δ and rad52Δ showed marked sensitivity.
3.3 Nucleotide excision repair protects cells from acute formaldehyde exposure
To determine if the length of time and concentration of formaldehyde exposure affected the pathway used to repair the DPCs, strains representing the major DNA repair or damage tolerance pathways were assayed for sensitivity to formaldehyde at an acute exposure (60 mM,15 min) and compared with a chronic exposure (1.5 mM, 48 hr) (Fig. 2A). It had been noted in the original screen that the Δsgs1 and Δtop3 strains were highly sensitive to chronic formaldehyde exposure. The Sgs1/Top3 complex is involved in maintenance of replisome stability and the Sgs1 RecQ-family helicase has been implicated in double-strand break processing [14, 15]. Thus, it was also of interest to examine the relative sensitivity of these two deletion strains following chronic versus acute exposure.
Fig. 2. Survival Analyses of Yeast Deletion Strains Following Exposure to Acute High Dose Formaldehyde.
(A) Selected yeast deletion strains representing DNA damage response pathways were cultured for exponential growth. For the control (left) and chronic 1.5 mM formaldehyde exposure (middle), each culture was serially diluted and aliquots spotted. For the acute exposure (right), following exponential growth in liquid media, cells were exposed to 60 mM formaldehyde for 15 min and washed twice prior to diluting and spotting on the plates. (B) The colony forming abilities of WT, mre11Δ, rad4Δ, top3Δ, rev3Δ and rad52Δ after acute exposure for 15 min are plotted. The cells were grown for 2 days at 30°C.
Analyses of these data revealed dramatic differences in the cellular pathway responses under the two conditions. As shown in Fig. 2A, left panel (control) all strains grew similar to the wild-type strain under non-exposed conditions, with only a modest slow growth phenotype shown in the mre11Δ strain. When exposed under chronic conditions (1.5 mM; middle panel), increased formaldehyde sensitivity was observed in the strains carrying deletions of genes in the HR pathway, including rad52Δ, rad50Δ, mre11Δ, and xrs2Δ. In addition, the sgs1Δ and top3Δ showed marked sensitivities, thus implicating these proteins in the processing of DPCs. Consistent with the data above, only modest sensitivity was seen for the NER mutants, rad1Δ and rad4Δ, or no sensitivity for the post-replication repair mutants, rev3Δ, rad6Δ, or rad18Δ.
In contrast, following acute exposures, the NER strains (rad1Δ and rad4Δ) demonstrated the highest sensitivity, whereas the HR deletion strains (mre11Δ, xrs2Δ, rad50Δ, rad52Δ) and the sgs1Δ and top3Δ strains exhibited moderate sensitivity (Fig. 2A; right panel). These data suggest that the relative contribution of DNA repair pathways to protection against formaldehyde-induced DPCs is dependent on the exposure conditions. In order to confirm these findings, colony forming assays were carried out on a subset of these strains under acute conditions. In agreement with the data shown in Fig. 2A, Δrad4 showed a marked sensitivity, whereas the colony forming ability of rad52Δ, mre11Δ and top3Δ were comparable to or slightly better than that of the wild-type strain.
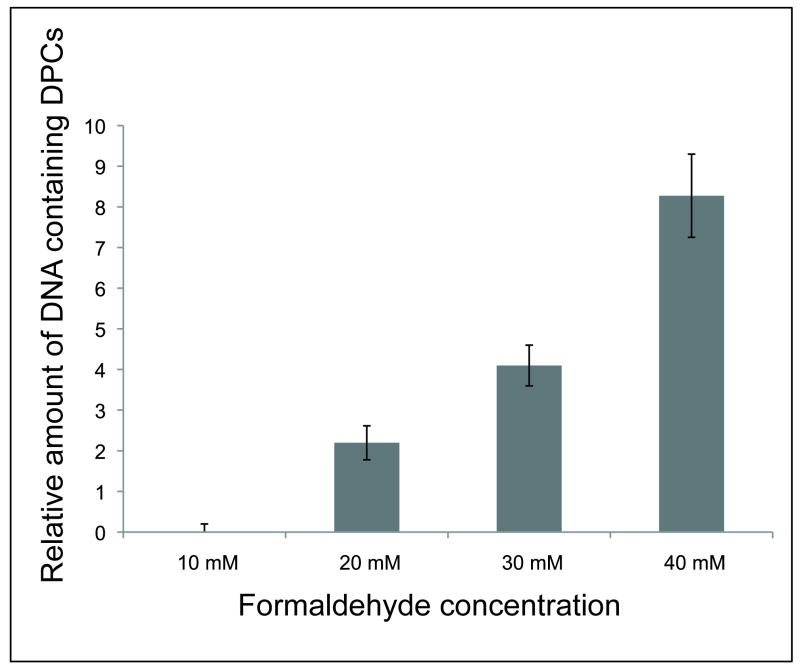
Due to the differences seen in the response pathways following formaldehyde exposure, it was of interest to directly detect DPCs in yeast DNA following chronic and acute formaldehyde exposures. To assess the dose-dependent formation of DPCs, SDS/KCl precipitation of proteins from whole cell lysates was adapted for the measurement of DPC formation as previously described [16]. This assay relies on the ability of cross-linked DNA to co-precipitate with proteins. Following a proteinase digestion, the extent of co-precipitation of DNA was measured by agarose gel electrophoresis of the DNA. As shown in Fig. 3, wild-type cells exposed to 20-40 mM formaldehyde for 15 min showed a dose-dependent accumulation of DPCs. No DPCs were detected in cells exposed to chronic doses (1.5-2 mM) of formaldehyde (data not shown). Based on the acute dose experiments, it is apparent that the lower limit of detection for this assay is between 10-20 mM exposures. Thus, DPCs may be forming at chronic doses, but are not detectable by this method.
Fig. 3. Accumulation of DPCs in Wild-Type Yeast Following Acute Formaldehyde Exposure.
Wild-type yeast cells were exposed to the indicated concentrations of formaldehyde for 15 min. DNA containing DPCs was isolated and analyzed as described in the methods. The data show the ratio of DNA containing DPCs compared to non-crosslinked DNA as determined by the SDS/KCl precipitation method. Each concentration was repeated in triplicate and non-treated control values were subtracted as background.
3.4 The role of nucleotide excision repair in the removal of formaldehyde-induced DPCs
After establishing that DPCs accumulate in a dose-dependant manner in cells treated with formaldehyde, the removal of these DPCs was observed at 0, 4 and 6 hr post-exposure in wild-type and NER-deficient cells. Using the SDS/KCl assay described above, it was observed that DPCs decrease at a linear rate in cells following acute exposure to formaldehyde, reaching the level of background approximately 6 hr post-exposure. Similar to previous results in mammalian cells [4], NER-deficient yeast cells show a similar rate of removal of DPCs as wild-type cells despite the large difference in sensitivity (Fig. 4).
Fig. 4. DPC Removal Following Acute Formaldehyde Exposure.
Wild-type (circles), rad1Δ (squares), and rad4Δ (triangles) yeast were exposed to 60 mM formaldehyde for 15 min. DNA containing DPCs was isolated and analyzed as described in the methods. Each concentration was independently repeated four times and non-treated control values were subtracted as background. Values were normalized so that the zero time point was 100%.
3.5 Formation of single-strand breaks (SSBs) following acute formaldehyde exposure is NER-dependent
Since the NER-deficient cells showed no differences in the rate of DPC removal by the SDS/KCl assay described above, the accumulation of SSBs after acute formaldehyde exposure was measured by alkaline sucrose gradient sedimentation analyses. DNAs isolated from wild-type yeast 4 hr after a 15 min exposure to 60 mM formaldehyde show an accumulation of SSBs as evidenced by the loss of high molecular weight DNA in the lower fractions compared to the untreated controls (Fig. 5A). The NER deficient strains, rad1Δ and rad4Δ, do not show an accumulation of SSBs under the same conditions (Fig. 5B and 5C, respectively).
Fig. 5. NER-Dependent Accumulation of Single-Strand Breaks Following Formaldehyde Exposure Assayed.
Yeast cells were exposed to 60 mM formaldehyde (squares) and allowed 4 hr recovery. Unexposed controls are shown (circles). The samples, wild-type yeast (A), rad1Δ (B) or rad4Δ (C), were analyzed by alkaline sucrose gradient sedimentation. Fractions were taken from the bottom of a 15-30% sucrose gradient that had been centrifuged for 20 hr at 11,500 rpm.
3.6 DPCs are repaired by a different pathway than DNA-DNA interstrand crosslinks
Our findings that HR and NER pathways, or components of these pathways, are involved in the repair and/or tolerance of DPCs are reminiscent of the potential interplay of these two pathways in repair of DNA-DNA interstrand crosslinks (ICL). In fact, there was significant overlap between the strains implicated in the repair of ICLs and those identified in the formaldehyde screen. Several strains sensitive to mitomycin C-induced ICLs were also sensitive to formaldehyde (rad1Δ, rad4Δ, rad51Δ, and rad52Δ) [11, 12]; however, the mitomycin C-sensitive deletion strains rev3Δ and exo1Δ were not sensitive to formaldehyde (Fig. 1, Fig. 2 and Supplementary Fig. 2, respectively). Processing of ICLs in eukaryotic systems is associated with the formation of double-strand breaks (DSBs) that are hypothesized to be formed at sites of stalled replication forks [17]. PSO2, a 5′ to 3′ exonuclease, significantly contributes to cellular resistance to ICL damage. Interestingly, pso2Δ mutants appear to be proficient for the initial incision events for ICL repair, but fail to repair ICL-induced DSBs [11]. Additionally, these mutants show decreased rates of homologous recombination, suggesting that the PSO2 nuclease acts downstream of NER in the processing of ICLs to provide a suitable substrate for HR [11, 12]. Since current models for DPC repair also predict that DSB intermediates would be formed due to replication fork collapse and since strains deleted in genes for NER and HR pathways are sensitive to formaldehyde exposures, it was of interest to determine if PSO2 may be involved in DPC repair. Although the pso2Δ strain in the deletion library was not sensitive to formaldehyde, additional pso2Δ strains with different backgrounds, previously shown to be sensitive to ICL-inducing agents, were obtained from Dr. Peter McHugh (University of Oxford) and Dr. Robb Moses (Oregon Health & Science University) and tested under identical conditions to those described above. The pso2Δ strains did not show sensitivity to formaldehyde under these conditions (data not shown). These data suggest that even though ICL and DPC-inducing agents may lead to stalled replication forks, there are possibly different intermediate structures that result in fundamentally different mechanisms to limit cytotoxicity in response to these agents.
3.7 DSBs do not accumulate following formaldehyde exposure
Based on the sensitivity of the HR deletion strains, one plausible model invokes a DNA break intermediate in order to perform single-strand invasion to process DPCs [18]. To test this model, pulsed field gel electrophoresis (PFGE) analyses were performed to determine if SSBs, DSBs, or both were formed during the processing of formaldehyde-induced DPCs. PFGE analyses were performed with and without the addition of S1-nuclease, which is added to convert SSBs to DSBs that can subsequently be detected by PFGE. Analyses were performed on cells that had been exposed to chronic or acute doses of formaldehyde.
To serve as controls for the induction of DSBs or SSBs, wild-type cells were treated with bleomycin or hydrogen peroxide, respectively. Following control and experimental exposures, cells were digested with lyticase, and these lysates were subsequently incubated with proteinase K, and DNAs subjected to PFGE. The bleomycin treatment resulted in DSB formation (Fig. 6A, left panel), and the hydrogen peroxide treatment produced both DSBs and SSBs (Fig. 6A, middle panel), results that are consistent with previously published data [19]. However, cells exposed to chronic formaldehyde showed no evidence of either SSB or DSB accumulation (Fig. 6A, right panel).
Fig. 6. Pulsed Field Gel Electrophoresis on Wild-Type and rad4Δ Yeast Following Formaldehyde Exposure.
(A) 20 μg/ml bleomycin for 1 hr (left panel); 10 mM H2O2 for 1 hr (middle panel); 1.5 mM formaldehyde exposure for 48 hr (right panel). The DNAs were analyzed as described in Methods, Section 2.5. (B) wild-type yeast cells. α-mating factor arrested cells exposed to 60 mM formaldehyde for 15 min. Time (hr), is the time given for the cells to recover after exposure. (C) rad4Δ yeast cells. α-mating factor arrested cells were exposed to 60 mM formaldehyde for 15 min. Time (hr), is the time given for the cells to recover after exposure. S1= S1 nuclease treatment and C = control, no treatment.
A possible interpretation of the formaldehyde exposure data was that SSBs and DSBs occur only transiently, as intermediates of the repair process, and that these breaks are not detectable under the previous conditions. To increase the number of breaks occurring at any one time, cells were arrested in G1 phase with α-mating factor prior to formaldehyde treatment. Only acute exposures could be tested on arrested cells due to the duration of a chronic exposure. Following treatment, cells were assayed for the formation of SSBs and DSBs over a 24 hr period. As shown in Fig. 6B, DSBs were not observed above background levels in PFGE after exposure at any given time point. Likewise, no DSB intermediates were observed accumulating above background levels in rad52 and mre11 deletion strains (Supplementary Fig. 3).
In contrast, SSBs were observed immediately following exposure to formaldehyde and observed in greater numbers at 4 hr after exposure. However, by 8 hr, the SSBs were almost completely repaired (Fig. 6B). Since the cells were counted at each post-exposure time point and no significant increase in cell number had occurred, we conclude that there was no dilution of the damage due to cell growth. The accumulation of SSBs following formaldehyde exposure is not evident in the rad4 deletion strain (Fig. 5 and Fig. 6), suggesting that this process is dependent on NER.
4. Discussion
In order to identify and characterize the biochemical mechanisms for repair of formaldehyde-induced DPCs, this study utilized the S. cerevisiae haploid non-essential gene deletion library to screen for genes that when deleted, confer formaldehyde sensitivity. Specific biochemical pathways were identified that protect cells from the cytotoxic effects of acute and chronic formaldehyde exposures. These investigations have shown that a large portion of the most formaldehyde-sensitive genotypes are in pathways of DNA repair, DNA damage tolerance, and chromatin remodeling (Table 1). Interestingly, S. cerevisiae cells respond in fundamentally different ways to the induction of DPCs that are dependent on whether the exposure to formaldehyde was given as a chronic, low dose or acute high dose. Following chronic exposure, cell survival is conferred by proteins of the HR pathway including, Rad50, Rad51, Rad52, Rad54, Rad55, Mre11 and Xrs2, as well as proteins associated with the resolution of stalled replication fork structures, such as Sgs1, Top3 and Rad5. These data suggest that successful tolerance and/or repair of DPCs proceed via intermediates that are substrates for HR. In comparison, only moderate sensitivity was observed in the NER deletion strains, suggesting a less critical contribution of NER to survival following chronic exposure. It is possible that after such formaldehyde exposures, DPCs encountered in the leading and lagging strands during DNA synthesis are dealt with in different manners. DPCs on the leading strand may block progression of the helicase complex that precedes the replisome and thus, replication complexes may stall well in advance of either polymerase reaching the site of the DNA adduct. This may allow for NER repair of the DPC and possibly explain the moderate sensitivity in the NER deletion strains. On the lagging strand, stalled replication complexes may temporarily uncouple synthesis in the two strands until no further lagging strand synthesis can occur. At this point, fork regression could occur, leading to a recombinogenic structure, which when processed could effectively allow for damage tolerance and replication restart. At this time, the exact nature of these intermediates remains undetermined; however, DSBs and SSBs have not been detected following chronic exposures.
In the case of acute short-term exposures, cellular survival strategies appear to switch to an NER-dependent response, with Rad1, Rad4, and Rad14 all having essential roles in conferring enhanced survival. These data suggest that the signal to transition from an HR-dependent pathway to an NER-dependent pathway is related to the rapid induction of a large number of DPCs. Our data reveal that SSBs can be readily detected under these conditions and accumulate post-exposure, indicating that the breaks are an intermediate in the processing of DPCs. The formaldehyde-induced SSBs appear to be NER-dependent as the breaks do not accumulate post-formaldehyde exposure in rad4Δ or rad1Δ strains, suggesting that these SSBs are intermediates generated during the repair process. These results are consistent with a previous report showing a Rad3-dependent accumulation of SSBs following formaldehyde exposure [20]. PFGE analyses also suggest that some SSBs may be dependent on the presence of Mre11 (Supplementary Fig. 3).
Based on our data showing similar rates of DPC removal in wild type or NER-deficient arrested cells, we suggest that under acute high-dose exposure, proteolytic degradation precedes NER recognition and that this degradation is an NER-independent active repair process, not initiated by stalled replication forks (replication-independent). This replication-independent, NER-dependent model for DPC repair is consistent with data reported for E. coli [1]. In addition, the similar rates of removal of DPCs as measured in Fig. 4 in the WT and NER-deficient strains may be indicative of a pre-NER processing event that degrades the protein crosslink to a peptide which is not detectable by this assay.
As shown in Fig. 2, strains deficient in mre11 and rad52 show less sensitivity to formaldehyde than the wild-type strain. The enhanced resistance to formaldehyde is seen consistently in the mre11 and rad52 deletion strains, suggesting that under the acute conditions, HR is not advantageous and in fact, attempted processing of formaldehyde-induced DNA adducts by HR may produce a cytotoxic intermediate or interfere with NER processing of the DPCs.
During the course of this study, two reports demonstrated a possible role for the HR pathway in the tolerance of DPCs [1, 5]. In both the E. coli study and the study using isogenic DT40 chicken cell lines, the authors proposed models for DPC processing proceeding via DSB intermediates. Our data demonstrating a prominent role for HR also led us to speculate that DSBs were a likely intermediate in a DPC processing pathway. However, PFGE analyses and neutral centrifugation analyses showed no detectable DSBs following exposure to formaldehyde, even in a HR-deficient background (Fig. 5 and Supplementary Fig. 3). These data are consistent with PFGE analyses in E. coli and comet assays performed on V79 Hamster cells following formaldehyde exposure [1, 7].
Interestingly, DSBs are not detected in cells treated with thymidine, which is known to block DNA replication. Previously, it has been shown that mammalian cells respond to hydroxyurea replication arrest differently than thymidine block [21], such that hydroxyurea induced DSBs and the repair of the strand breaks proceeded via both the NHEJ and the HR pathways. In contrast, thymidine arrest involved only HR and had no detectable DSBs. These authors concluded that the cytotoxic lesion following thymidine treatment was not persistent or transient DSBs, but a different, yet undetermined substrate for HR [21]. Additionally, an alternative non-DSB dependent model may be that HR is initiated at a DNA nick or single-strand gap generated during replication past or polymerase stalling at a DPC lesion. The plausibility of this model is supported by the observation that for most mitotic HR in S. cerevisiae, the initiating lesion is not a DSB but potentially a SSB or single-strand gap [22]. This is consistent with our data demonstrating a lack of DSBs, but an accumulation of SSBs following formaldehyde exposure. It should be noted that our observations cannot rule out the possibility that a small amount of transient DSBs may occur but which are not detectable by our methods. It is possible that the single-strand break repair pathway may have a role in preventing the cytotoxicity of DPC processing intermediates. However, the rad27 (Fen1) deletion strain was not sensitive to formaldehyde under chronic or acute conditions (Supplementary Fig 3). We cannot definitely rule out a role for this pathway since the genes for the major ligase (cdc9) and the polymerase (polδ) for this pathway are essential and were not included in our screen.
In addition to being the first genome-wide screen examining formaldehyde cytotoxicity, this study highlights that the exposure conditions (duration and concentration) can significantly affect the spectra of gene deletion strains that are identified as sensitive or resistant. These data clearly demonstrate a differential pathway response to chronic versus acute formaldehyde exposures and may have significance for risk extrapolation in human exposure studies. It is well recognized that the validity of such extrapolations can be challenged by the capacity of cells to activate or detoxify chemicals, as well as the steady-state DNA repair capacity to effectively remove damages as they occur. Consistent with dose-dependent pathway responses, rat formaldehyde inhalation studies have shown steep dose-dependent transitions in both cell proliferation assays and tumorigenesis [23]. An increased understanding of the molecular basis of transitions in the cellular response pathways may elucidate details for models of dose-dependent transitions in mechanisms of formaldehyde toxicity.
In summary, we have shown that both NER and HR are critical for survival following formaldehyde exposure. However, the relative contribution of each pathway differs depending on the dose and duration of exposure. Further, the genome-wide screen has identified other genes that contribute significantly to protection against the genotoxic effects of formaldehyde. The relationship of these other genes/pathways to NER and HR are being investigated in the context of a newly defined DPC-specific repair/tolerance pathway.
Supplementary Material
A representative plate (#13-4) from the complete library screen is shown. Replicates of the library master plate were grown in YPD media for 16 hr and ∼1 μl of each well was spotted in triplicate and grown for 2 days at 30°C. The left panel shows an unexposed plate, the right panel is a duplicate plate grown in the presence of 1.5 mM formaldehyde. An example of a strain not sensitive to this concentration of formaldehyde is shown within the grey box in the upper right corner, while a strain near the middle of the plate (black box) was identified as sensitive. Areas of no growth are intentionally designed by the supplier to allow for unambiguous plate identification.
Selected yeast deletion strains representing the base excision repair, single-strand break repair, and mismatch repair pathways were cultured for exponential growth. For acute exposure, cells were exposed to 60 mM formaldehyde for 15 min and washed twice prior to diluting and spotting on the plates. The cells were grown for 2 days at 30°C.
(A) rad52Δ yeast cells. α-mating factor arrested cells exposed to 60 mM formaldehyde for 15 min. Time (hr), is the time given for the cells to recover after exposure. (C) mre11Δ yeast cells. α-mating factor arrested cells exposed to 60 mM formaldehyde for 15 min. Time (hr), is the time given for the cells to recover after exposure. S1= S1 nuclease treatment and C = control, no treatment.
Acknowledgments
We thank Robb Moses (OHSU), Peter McHugh (University of Oxford) and Leona Samson (MIT) for the generous gifts of yeast strains. We would also like to thank Christian Dan and Erik Bom for technical assistance. We are grateful to R. Stephen Lloyd, Irina Minko, and Anuradha Kumari for valuable comments and critically reading the manuscript. This work was supported in part by NIH RO1 CA106858 and NIH PO1 ES05355.
Abbreviations
- BER
base excision repair
- DPC
DNA-protein crosslink
- DSB
double strand break
- NER
nucleotide excision repair
- HR
homologous recombination
- PFGE
pulsed field gel electrophoresis
- SSB
single-strand break
Footnotes
Conflict of Interest Statement: The authors declare that there are no conflicts of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nakano T, et al. Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein crosslinks. Mol Cell. 2007;28(1):147–58. doi: 10.1016/j.molcel.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 2.Baker DJ, et al. Nucleotide excision repair eliminates unique DNA-protein cross-links from mammalian cells. J Biol Chem. 2007;282(31):22592–604. doi: 10.1074/jbc.M702856200. [DOI] [PubMed] [Google Scholar]
- 3.Fornace AJ, Jr, Seres DS. Repair of trans-Pt(II) diamminedichloride DNA-protein crosslinks in normal and excision-deficient human cells. Mutat Res. 1982;94(2):277–84. doi: 10.1016/0027-5107(82)90291-3. [DOI] [PubMed] [Google Scholar]
- 4.Quievryn G, Zhitkovich A. Loss of DNA-protein crosslinks from formaldehyde-exposed cells occurs through spontaneous hydrolysis and an active repair process linked to proteosome function. Carcinogenesis. 2000;21(8):1573–80. [PubMed] [Google Scholar]
- 5.Ridpath JR, et al. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 2007;67(23):11117–22. doi: 10.1158/0008-5472.CAN-07-3028. [DOI] [PubMed] [Google Scholar]
- 6.Gantt R. A cell cycle-associated pathway for repair of DNA-protein crosslinks in mammalian cells. Mutat Res. 1987;183(1):75–87. doi: 10.1016/0167-8817(87)90048-4. [DOI] [PubMed] [Google Scholar]
- 7.Speit G, Schutz P, Merk O. Induction and repair of formaldehyde-induced DNA-protein crosslinks in repair-deficient human cell lines. Mutagenesis. 2000;15(1):85–90. doi: 10.1093/mutage/15.1.85. [DOI] [PubMed] [Google Scholar]
- 8.Minko IG, Zou Y, Lloyd RS. Incision of DNA-protein crosslinks by UvrABC nuclease suggests a potential repair pathway involving nucleotide excision repair. Proc Natl Acad Sci U S A. 2002;99(4):1905–9. doi: 10.1073/pnas.042700399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minko IG, et al. Initiation of repair of DNA-polypeptide cross-links by the UvrABC nuclease. Biochemistry. 2005;44(8):3000–9. doi: 10.1021/bi0478805. [DOI] [PubMed] [Google Scholar]
- 10.Reardon JT, Sancar A. Repair of DNA-polypeptide crosslinks by human excision nuclease. Proc Natl Acad Sci U S A. 2006;103(11):4056–61. doi: 10.1073/pnas.0600538103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barber LJ, et al. DNA interstrand cross-link repair in the Saccharomyces cerevisiae cell cycle: overlapping roles for PSO2 (SNM1) with MutS factors and EXO1 during S phase. Mol Cell Biol. 2005;25(6):2297–309. doi: 10.1128/MCB.25.6.2297-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grossmann KF, et al. S. cerevisiae has three pathways for DNA interstrand crosslink repair. Mutat Res. 2001;487(34):73–83. doi: 10.1016/s0921-8777(01)00106-9. [DOI] [PubMed] [Google Scholar]
- 13.Kelly DE, Lamb DC, Kelly SL. Genome-wide generation of yeast gene deletion strains. Comp Funct Genomics. 2001;2(4):236–42. doi: 10.1002/cfg.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455(7214):770–4. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bjergbaek L, et al. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J. 2005;24(2):405–17. doi: 10.1038/sj.emboj.7600511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trask DK, DiDonato JA, Muller MT. Rapid detection and isolation of covalent DNA/protein complexes: application to topoisomerase I and II. EMBO J. 1984;3(3):671–6. doi: 10.1002/j.1460-2075.1984.tb01865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergstralh DT, Sekelsky J. Interstrand crosslink repair: can XPF-ERCC1 be let off the hook? Trends Genet. 2008;24(2):70–6. doi: 10.1016/j.tig.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Baldwin EL, et al. Mms22p protects Saccharomyces cerevisiae from DNA damage induced by topoisomerase II. Nucleic Acids Res. 2005;33(3):1021–30. doi: 10.1093/nar/gki246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ribeiro GF, Corte-Real M, Johansson B. Characterization of DNA damage in yeast apoptosis induced by hydrogen peroxide, acetic acid, and hyperosmotic shock. Mol Biol Cell. 2006;17(10):4584–91. doi: 10.1091/mbc.E06-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magana-Schwencke N, Ekert B, Moustacchi E. Biochemical analysis of damage induced in yeast by formaldehyde. I. Induction of single-strand breaks in DNA and their repair. Mutat Res. 1978;50(2):181–93. doi: 10.1016/0027-5107(78)90023-4. [DOI] [PubMed] [Google Scholar]
- 21.Lundin C, et al. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol Cell Biol. 2002;22(16):5869–78. doi: 10.1128/MCB.22.16.5869-5878.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lettier G, et al. The role of DNA double-strand breaks in spontaneous homologous recombination in S. cerevisiae. PLoS Genet. 2006;2(11):e194. doi: 10.1371/journal.pgen.0020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monticello TM, et al. Correlation of regional and nonlinear formaldehyde-induced nasal cancer with proliferating populations of cells. Cancer Res. 1996;56(5):1012–22. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A representative plate (#13-4) from the complete library screen is shown. Replicates of the library master plate were grown in YPD media for 16 hr and ∼1 μl of each well was spotted in triplicate and grown for 2 days at 30°C. The left panel shows an unexposed plate, the right panel is a duplicate plate grown in the presence of 1.5 mM formaldehyde. An example of a strain not sensitive to this concentration of formaldehyde is shown within the grey box in the upper right corner, while a strain near the middle of the plate (black box) was identified as sensitive. Areas of no growth are intentionally designed by the supplier to allow for unambiguous plate identification.
Selected yeast deletion strains representing the base excision repair, single-strand break repair, and mismatch repair pathways were cultured for exponential growth. For acute exposure, cells were exposed to 60 mM formaldehyde for 15 min and washed twice prior to diluting and spotting on the plates. The cells were grown for 2 days at 30°C.
(A) rad52Δ yeast cells. α-mating factor arrested cells exposed to 60 mM formaldehyde for 15 min. Time (hr), is the time given for the cells to recover after exposure. (C) mre11Δ yeast cells. α-mating factor arrested cells exposed to 60 mM formaldehyde for 15 min. Time (hr), is the time given for the cells to recover after exposure. S1= S1 nuclease treatment and C = control, no treatment.