Abstract
Fungus-growing ants engage in mutualistic associations with both the fungus they cultivate for food and actinobacteria (Pseudonocardia spp.) that produce selective antibiotics to defend that fungus from specialized fungal parasites. In the first system to be analyzed at the molecular level, the bacterium associated with the ant Apterostigma dentigerum produces dentigerumycin, a cyclic depsipeptide with highly modified amino acids, to selectively inhibit the parasitic fungus (Escovopsis sp.).
Mutualisms, a shorthand term for mutually beneficial interactions between different species, help shape the biological complexity and diversity of the planet. Recent investigations of several insect-fungal mutualisms have shown that bacterially produced small molecules play a key role in maintaining these complicated but widespread systems.1,2 The bacteria involved in these multipartite symbioses are typically actinobacteria, a group that has provided many biologically active small molecules, some of which have become clinically useful drugs.3 Through their mutualisms with these chemically prolific actinobacteria, insect hosts utilize bacterially produced antibiotics to combat both diseases1,4 and competitors.2 The mutualism between fungus-growing ants (Attini: Formicidae) and actinobacteria (Pseudonocardia spp.) is arguably the best studied of these systems, and three general features have emerged: (a) Evolutionary diversification of the fungus-growing ant lineage has resulted in more than 230 described ant species that provide distinct but evolutionarily related systems,5 (b) Fungus-growing ants from distinct agriculture systems use different groups of cultivated fungi (Basidiomycota Agaricales: Lepiotaceae and Pterulaceae) and actinobacteria,6 (c) The symbiotic Pseudonocardia produce selective antibiotics to inhibit the specialized parasitic fungi (Escovopsis spp.) that invade and consume the ants’ fungal garden.1,6,7 However, none of these studies has characterized any of the selective antibiotics, and their identification would permit studies on their mechanism of action on sensitive parasites, their evasion by resistant parasites, their biosynthesis, and the molecular evolution of a complex symbiotic system that is at least 50 million years old.5 Here we report the isolation, structural determination, and biological activity of one such antibiotic.
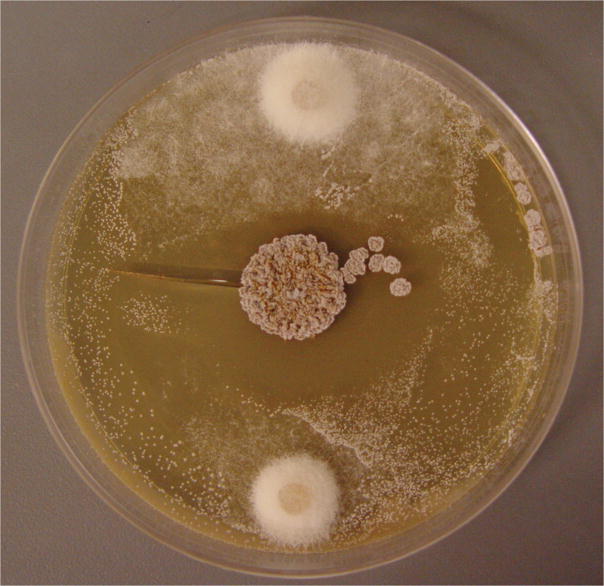
We began by isolating the bacterium (Pseudonocardia sp.), the fungal cultivar, and the parasitic fungus (Escovopsis sp.) from an ant (Apterostigma dentigerum) nest collected in Gamboa, Panama in 2001 (colony code CC011120-4, see Supplementary Methods online, Fig. 1a). The Pseudonocardia strain isolated from the cuticle of Apterostigma dentigerum strongly inhibits the Escovopsis isolate obtained from the same colony (Fig. 1b) while the fungal cultivar is relatively resistant to the bacterial strain (Supplementary Fig. 1 online). Initial LC/MS analysis of both an ethyl acetate extract of a whole liquid culture and a methanol extract of an agar plate culture of the Pseudonocardia strain revealed the presence of a single major component (Supplementary Fig. 2 online). Further investigation, involving large-scale cultivation (8 L) and assay-guided fractionation through C18 flash column and HPLC, confirmed that this major compound, which we named dentigerumycin (1, Fig. 2a), is responsible for Escovopsis inhibition.
Figure 1.
(a) Apterostigma fungus-growing ant (colony code CC011120-4), showing a worker of Apterostigma dentigerum. (b) Pairwise challenge bioassay demonstrates in vitro inhibition of the parasitic fungus Escovopsis (top and bottom) by Pseudonocardia (center) (8.5 cm diameter agar plate).
Figure 2.
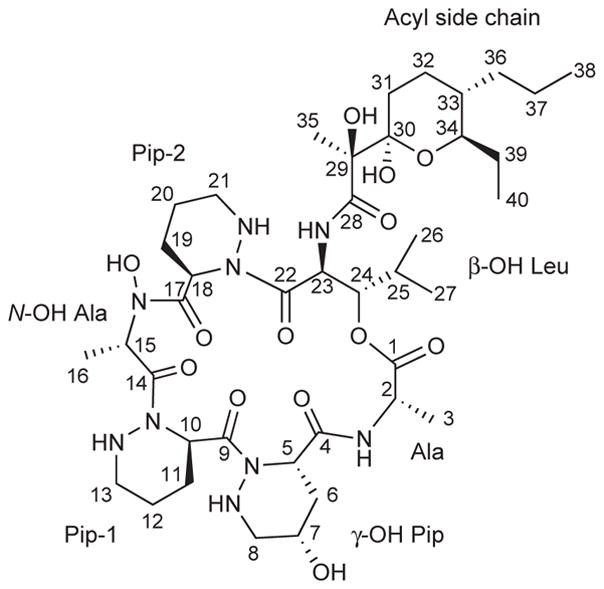
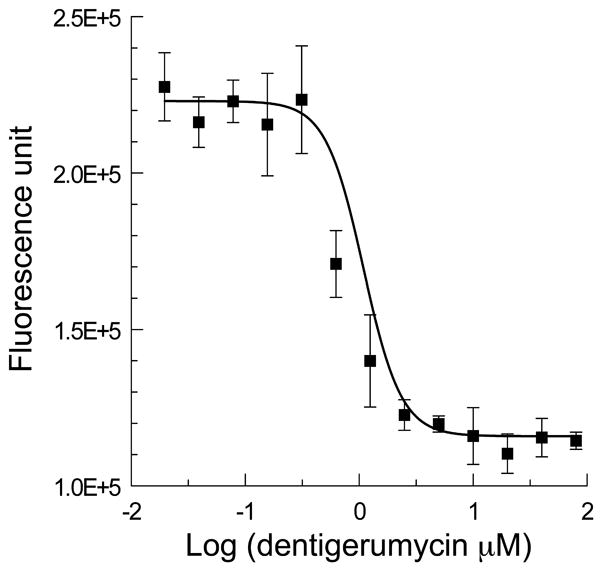
(a) The structure of dentigerumycin (1) contains highly modified amino acid units such as piperazic acids. (b) The susceptibility dose response curve plotting log concentration dentigerumycin in μM vs fungal cell density (fluorescence unit) of Escovopsis, demonstrating inhibition of the parasitic fungus by dentigerumycin with MIC of 2.8 μM.
Dentigerumycin (1) was isolated as a white powder having the molecular formula C40H67N9O13, as determined by high-resolution ESI-TOF mass spectrometry coupled with 1H and 13C NMR spectral data (Supplementary Table 1, Supplementary Fig. 3 online). The 1H NMR spectrum displayed one downfield singlet proton at 9.50 ppm, two amide doublet protons, and twelve protons between 4 and 6 ppm. The 13C NMR and gHSQC spectra showed seven amide or ester carbonyl carbons, five oxygen-bearing carbons, nine nitrogen-bearing carbons, and nineteen aliphatic carbon signals. Analysis of gHSQC spectral data allowed all one-bond carbon and proton correlations to be assigned and identified nine protons attached to hetero atoms.
Interpretation of gCOSY, TOCSY, gHSQC, and gHMBC NMR spectra assigned six amino acid-derived and one polyketide-derived sub-structures. These include two piperazic acid units (Pip-1 and Pip-2), γ-hydroxy piperazic acid (γ-OH-Pip), β-hydroxy leucine (β-OH-Leu), N-hydroxy alanine (N-OH-Ala), alanine (Ala), and a pyran-bearing acyl side chain (Fig. 2a, Supplementary Fig. 4a online). Key HMBC correlations secured the sequence of the amino acid units and the acyl side chain (Supplementary Fig. 4b online), which completed the planar structure of dentigerumycin (Fig. 2a). We determined dentigerumycin’s stereochemistry using a combination of spectroscopic analysis, degradative reactions, and derivatizations, as outlined below. The relative configuration of the pyran ring in the acyl side chain was determined by ROESY correlations (Supplementary Fig. 5a online). The relative configuration of the two consecutive stereogenic centers in β-OH-Leu was established as 23S* and 24S* by J-based configuration analysis8 (Supplementary Fig. 5b online). The relative configuration of γ-hydroxy piperazic acid was determined as 5S*,7S*-γ-OH-Pip by 1H-1H coupling constant and ROESY spectral analysis9 (Supplementary Fig. 5c online).
In order to establish dentigerumycin’s absolute configuration, we performed flash hydrolysis of 1 in 6 N HCl at 115°C for 1 hour.10,11 The hydrolysate was derivatized with the advanced Marfey reagents (L-FDLA (2) and D-FDLA (3), FDLA: Nα-(2,4-dinitro-5-fluoro-phenyl)-leucineamide).12 The LC/MS analysis of the derivatized alanine and β-hydroxy-leucine allowed determination of the absolute configurations of their α-carbons as 2S and 23S since L-FDLA derivatives eluted faster than D-FDLA derivatives for these amino acids. This analysis also determined a 24S configuration based on the relative stereochemistry previously determined at C-23 and C-24. The two piperazic acid units (Pip-1 and Pip-2) yielded only one product after reacting with L-FDLA, so both units have the same configuration, R, which was established by derivatizing synthetic S- and R-piperazic acids (4 and 5) with L-FDLA (see Supplementary Methods online). Although the FDLA products of theγ-hydroxy piperazic acid fragment were observed, we were not able to determine its absolute configuration because the elution sequence for S, S-γ-hydroxy piperazic acid and R, R-γ-hydroxy piperazic acid are not known. Instead, we tried the modified Mosher method on the secondary alcohol in γ-OH Pip.13 The only secondary alcohol (at C-7) was treated with R- and S-α-methoxy-α-(trifluoromethyl)phenyl acetyl chloride (MTPA-Cl) to yield S- and R-MTPA esters (6 and 7), respectively (see Supplementary Methods online). We assigned the proton chemical shifts of the γ-OH Pip by analysis of 1H, gCOSY, and TOCSY NMR spectra (Supplementary Fig. 3 online). Calculation of ΔδS-R values clearly defined the absolute configuration of C-7 as S and also established a 5S configuration from the relative stereochemistry of the γ-OH Pip (Supplementary Fig. 6 online).
Since free N-OH-Ala was not expected to react with FDLA, we converted the N-OH to N-H by treatment of dentigerumycin with TiCl3 (see Supplementary Methods online).14 We confirmed this conversion by 1H, gCOSY, TOCSY, gHSQC, and gHMBC NMR (Supplementary Fig 3 online) of the reduction product (8) and high resolution mass spectral analysis (Supplementary Table 2 online). The product (8) was then hydrolyzed, derivatized with L-FDLA, and analyzed with LC/MS as described earlier to establish a 15S configuration.
We determined the absolute configuration of the acyl side chain by means of CD (circular dichroism) spectral analysis of a Mo2(OAc)4 complex of the 1,2-diol at C-29 and C-30 (see Supplementary Methods online). Based on the empirical rule,15 the complex of dentigerumycin, which has a negative sign in the induced CD spectrum, should have 29S and 30R configurations if the 1,2-diol moiety adopts its predicted low energy conformation (Supplementary Fig. 7 online). This result also established 33S and 34R configurations based on their previously determined relative stereochemistry.
Chemical derivatizations of piperazic acids have been especially challenging,9 and their absolute configurations have been established mainly by X-ray crystallography. The combination of flash hydrolysis, advanced Marfey, and modified Mosher methods provides a generally applicable approach to characterizing these increasingly encountered amino acids.
Dentigerumycin is a previously unreported depsipeptide containing unusual amino acids piperazic acid, γ-hydroxy piperazic acid, β-hydroxy leucine, and N-hydroxy-alanine along with a polyketide-derived side chain containing a pyran ring. The natural products most similar to dentigerumycin are the polyoxypeptins16 and the aurantimycins,17 which have some amino acids in common with dentigerumycin. However, they do not have the same core structure as dentigerumycin because the other amino acids differ – most importantly through an additional piperazic acid unit in dentigerumycin, which is expected to produce a very different core conformation. Since all these were isolated from Streptomyces spp., it is likely that some biosynthetic elements were shared.18
While Petri dish assays show that dentigerumycin clearly inhibits the parasitic fungus more than the cultivated fungus (Supplementary Fig. 8 online), we evaluated its minimum inhibitory concentration (MIC = 2.8 μM) quantitatively in a liquid culture assay against the Escovopsis parasite from colony CC011120-4 (Fig. 2b). A liquid culture assay was not suitable for the cultivated fungus because it grows extremely slowly in liquid culture. Dentigerumycin also inhibited Candida albicans wild type, C. albicans ATCC10231, and amphotericin-resistant C. albicans ATCC200955 with MIC values of 1.1 μM. These results suggest that in the Apterostigma symbiosis, as in the southern pine beetle system mediated by mycangimycin,2,19 the ascomycete (Escovopsis sp.) is sensitive while the basidiomycete (cultivar) is resistant.
The fungus-growing ant system originated at least 50 million years ago,5 and the long evolutionary history of the ant-microbe symbiosis has resulted in diversification of the ants, their fungal cultivar, and the garden parasite Escovopsis.20 Morphological and physiological modification in the ants for maintaining Pseudonocardia, as well as molecular phylogenetic studies of the actinobacteria, support both an early origin and codiversification of the bacterial mutualist in the system. It is likely that over the evolutionary history of the association different Pseudonocardia bacterial symbionts evolved the production of different small molecules. Escovopsis and Pseudonocardia are predicted to undergo antagonistic coevolution, with selection for the evolution of resistance in Escovopsis and selection for bacterially produced molecules that counter this evolved resistance in Pseudonocardia. Evidence to support this prediction can be found in Petri plate bioassay experiments pairing diverse Pseudonocardia and Escovopsis strains that have revealed a variable degree of parasite inhibition between specific pairs,6 which most plausibly arises from different antibiotics being produced by different bacterial strains. Understanding both these individual interactions and their evolution at the molecular level represents an opportunity both for the discovery of new biologically active small molecules and for insights into the evolution of susceptibility, resistance, and biosynthetic pathways.
Supplementary Material
Acknowledgments
We thank the Smithsonian Tropical Research Institute (STRI) for providing facilities to work in Panama, the Autoridad Nacional del Ambiente y el Mar (ANAM) for sampling and export permit, Dr. John Heemstra and Prof. Christopher Walsh for providing synthetic piperazic acids, and A. Adams, S. Marsh, A. Pinto-Tomas, and G. Suen for comments on a manuscript draft. This work was supported by the Lundbeck and Carlsberg Foundations to M.P., NSF (MCB07020255) to C.R.C., and NIH CA24487 to J.C.
Footnotes
Note: Supplementary information and chemical compound information is available on the Nature Chemical Biology website.
AUTHOR CONTRIBUTIONS
D.C.O, M.P., C.R.C., and J.C. designed experiments and wrote manuscript. D.C.O. performed the structural determination and biological activity analysis on dentigerumycin. M.P. isolated the cultivar, the parasite, and the Pseudonocardia symbionts and performed bioassay parings.
References
- 1.Currie CR, Scott JA, Summerbell RC, Malloch D. Nature. 1999;398:701–704. [Google Scholar]
- 2.Scott JJ, et al. Science. 2008;322:63. doi: 10.1126/science.1160423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berdy J. J Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 4.Kaltenpoth M, Göttler W, Herzner G, Strohm E. Curr Biol. 2005;15:475–479. doi: 10.1016/j.cub.2004.12.084. [DOI] [PubMed] [Google Scholar]
- 5.Schultz TR, Brady SG. Proc Nat Acad Sci USA. 2008;105:5435–5440. doi: 10.1073/pnas.0711024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Currie CR, Poulsen M, Mendenhall J, Boomsma JJ, Billen J. Science. 2006;311:81–83. doi: 10.1126/science.1119744. [DOI] [PubMed] [Google Scholar]
- 7.Reynolds HT, Currie CR. Mycologia. 2004;96:955–959. [PubMed] [Google Scholar]
- 8.Matsumori N, Kaneno D, Murata M, Nakamura H, Tachibana K. J Org Chem. 1999;64:866–876. doi: 10.1021/jo981810k. [DOI] [PubMed] [Google Scholar]
- 9.Miller ED, Kauffman CA, Jensen PR, Fenical W. J Org Chem. 2007;72:323–330. doi: 10.1021/jo061064g. [DOI] [PubMed] [Google Scholar]
- 10.Fujii K, et al. Tetrahedron. 2002;56:6873–6879. [Google Scholar]
- 11.Schultz AW, et al. J Am Chem Soc. 2008;130:4507–4516. doi: 10.1021/ja711188x. [DOI] [PubMed] [Google Scholar]
- 12.Fujii K, Ikai Y, Oka H, Suzuki M, Harada K. Anal Chem. 1997;69:5146–5151. [Google Scholar]
- 13.Séco JM, Quiñoa E, Riguera R. Tetrahedron: Asymmetry. 2001;12:2915–2925. [Google Scholar]
- 14.Pennings MLM, Reinhoudt DN, Harkema S, van Hummel GJ. J Org Chem. 1983;48:486–491. [Google Scholar]
- 15.Snatzke G, Wagner U, Wolff HP. Tetrahedron. 1981;37:349–361. [Google Scholar]
- 16.Umezawa K, Nakazawa K, Ikeda Y, Naganawa H, Kondo S. J Org Chem. 1999;64:3034–3038. doi: 10.1021/jo981512n. [DOI] [PubMed] [Google Scholar]
- 17.Gräefe U, et al. J Antibiot. 1995;48:119–125. doi: 10.7164/antibiotics.48.119. [DOI] [PubMed] [Google Scholar]
- 18.Fischbach MA, Walsh CT, Clardy J. Proc Nat Acad Sci USA. 2008;105:4601–4608. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh DC, Scott JJ, Currie CR, Clardy J. Org Lett. 2009;11:633–636. doi: 10.1021/ol802709x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Currie CR, et al. Science. 2003;299:386–388. doi: 10.1126/science.1078155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.