Summary
The number of molecules with solved three-dimensional structure but unknown function is increasing rapidly. Particularly problematic are novel folds with little detectable similarity to molecules of known function. Experimental assays can determine the functions of such molecules, but are time-consuming and expensive. Computational approaches can identify potential functional sites; however, these approaches generally rely on single static structures and do not use information about dynamics. In fact, structural dynamics can enhance function prediction: we coupled molecular dynamics simulations with structure-based function prediction algorithms that identify Ca2+ binding sites. When applied to 11 challenging proteins, both methods showed substantial improvement in performance, revealing 22 more sites in one case and 12 more in the other, with a modest increase in apparent false positives. Thus, we show that treating molecules as dynamic entities improves the performance of structure-based function prediction methods.
Introduction
Understanding the function of molecules is the first step towards learning how to manipulate them. Experimental techniques for determining molecular function tend to be expensive and time consuming. Consequently, computational methods can be critical for establishing molecular function. Some techniques use similarity in the sequence of molecules: when protein sequences are at least 40% similar, they usually perform similar functions and so annotations can be transferred with some confidence. However, when sequence similarity falls below 40%, the reliability of these methods decreases (Wilson et al., 2000).
Some molecular sites of interest, such as binding pockets and enzyme active sites, may not be recognized with sequence patterns alone, as they may be comprised of loop segments that come together in three-dimensions, but are distant in the primary sequence. In these cases, similarity may exist on the structural level even when there is no detectable similarity in the corresponding sequences. In order to address this challenge, many structure-based function recognition methods use shared 3D structural environments or “3D motifs” to recognize molecular functions (Fetrow et al., 1998; Halperin et al., 2008; Wallace et al., 1997). Some methods also combine information from sequence and structure (Friedberg, 2004; Watson et al., 2005).
Since the initiation of the structural genomics (SG) efforts, the number of solved structures in the Protein Data Bank (PDB) (Berman et al., 2000) with unknown function is increasing (Chandonia and Brenner, 2006; Levitt, 2007; Terwilliger, 2004). SG efforts specifically target proteins with little sequence similarity to functionally annotated folds. Therefore, structure-based function annotation methods able to recognize distant functional relationships would be very useful (Halperin et al., 2008; Nayal and Di Cera, 1994).
The structures from the X-ray crystallographic studies in the PDB typically provide static snapshots of molecules that are averages of the structures in the crystal lattice. Crystal packing induced by crystallization conditions and experimental modifications required to facilitate experimental studies may create slight variations of the structure that do not represent the biologically functional conformations. This may frustrate attempts to annotate function based on detailed analysis of the structures.
While performing their functions, molecules undergo dynamic structural changes, which can range from subtle side-chain rearrangements to large-scale domain movements (Henzler-Wildman and Kern, 2007). Physics-based molecular dynamics (MD) simulations allow structural investigation of such motions on the femtosecond (fs), nanosecond (ns), and occasionally microsecond scales (Karplus and Kuriyan, 2005; Karplus and McCammon, 2002). These motions may reveal conformations relevant to molecular functions. At the very least, sampling near the crystal structure may help in identifying structural motifs that are not obvious in a single static structure.
The idea of using information about dynamics to improve functional analysis is not new. Alternative X-ray structures, NMR ensembles and MD-generated trajectories all provide information about dynamics useful for structure-based drug design (Damm and Carlson, 2007; Huang and Zou, 2007; Meagher and Carlson, 2004; Sivanesan et al., 2005; Wong et al., 2005). Eyrisch and co-workers (Eyrisch and Helms, 2007) used an algorithm to locate surface pockets in structures generated by the MD simulations, where those were not apparent in the original crystal structures. Among those transient surface pockets were the actual active sites of the molecules. Frembgen-Kesner and co-workers (Frembgen-Kesner and Elcock, 2006) also demonstrated that coupling a docking method with MD simulations uncovers cryptic drug binding sites.
In this manuscript we describe our work in improving the performance of structure-based function recognition methods by coupling them to molecular dynamics (see Figure 1). The MD simulations demonstrated formation of favorable, albeit transient, conformations that suggest functions not apparent in the initial PDB structures. Specifically, for eleven different molecules we used FEATURE (Wei and Altman, 2003) and a valence-based method (Nayal and Di Cera, 1994) to analyze the presence of Ca2+ binding sites in 22 structural ensembles generated by the MD simulations using GROMACS.
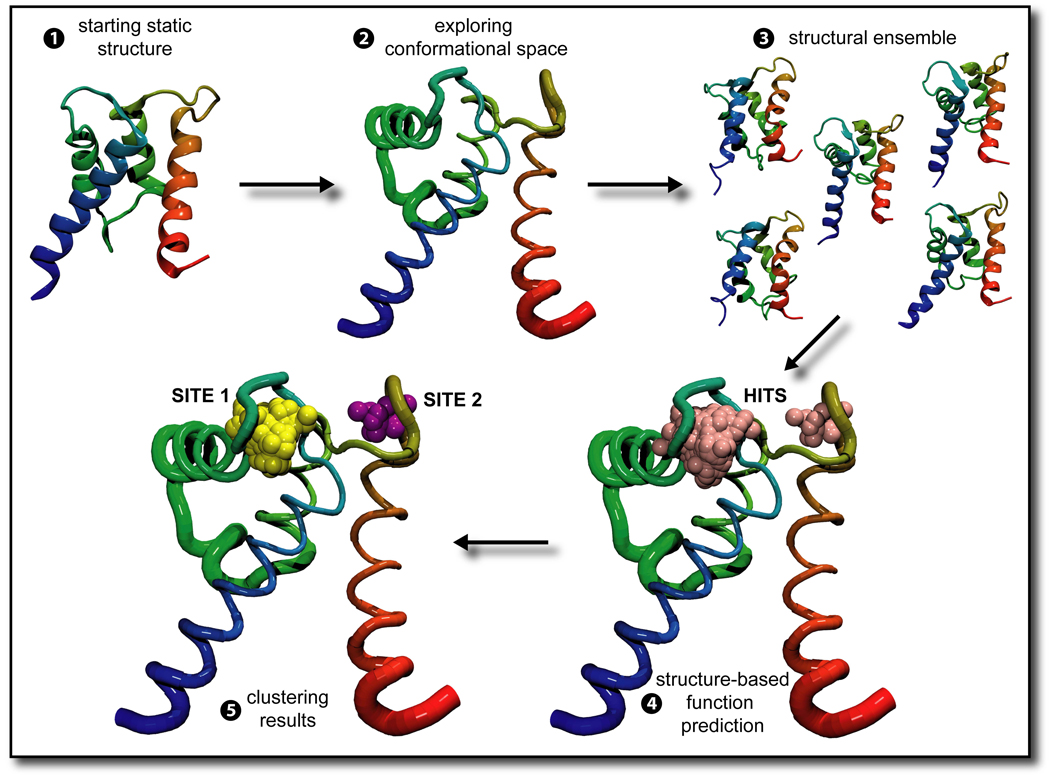
Figure 1. Overview of the scheme.
1, 2 A structure from the PDB is chosen and simulated using molecular dynamics. In our case, 11 HOLO and corresponding 11 APO structures were obtained and simulation trajectory was generated for 1 nanosecond. 3 A structural ensemble is formed with frames extracted from the simulation trajectory, in our case every 2.5 picoseconds. 4 A structure-based function prediction method, such as FEATURE or the valence method, is used to examine the structural ensemble. 5 A clustering scheme defines how many sites the structure-based function prediction methods identified, and may also assign which of those are true positive, as in 6 In 2 and 6, the thickness of the ribbon representing the backbone of the molecule represents how much of the conformational space that part of the molecule sampled over the course of the simulation. As such, in this molecule the termini and the flexible loops are thicker, because they moved more, while the α-helices are thinner, especially where the three of them form a sheet, because they did not move much.
Calcium plays a crucial role within many signaling pathways in the cell. Recognizing the ability of molecules to bind calcium may permit manipulations with direct impact on many aspects of cell life. Ca2+ binding sites tend to occur within flexible loops of the molecules and be oxygen rich, with several glutamate and aspartate residues lining the pocket. Often solvent molecules are also involved, since the binding occurs at the surface of the molecule. The straight forward coordination of calcium binding sites makes it surprising that the leading function-recognition methods fail to identify them even within structures with bound calcium. We show that allowing function recognition methods to examine the dynamic nature of the molecules dramatically improves their performance.
Results
The eleven molecules studied here have at least one documented calcium-binding site (see Table 1 and Discussion). For each of the molecules at least two structures exist in the PDB: a HOLO form, with Ca2+ bound within the structure, and an APO one, without a bound Ca2+. Existing methods for structure-based function recognition often do not identify the APO sites, and even can miss the HOLO sites. As such, these structures make an excellent set of test cases. In addition to revealing the concealed Ca2+ binding sites in the HOLO structures, can simulation identify the binding sites in the APO structures?
Table 1.
Final Results of FEATURE +/− MD and Valence +/− MD scanning.
| PDBID | Size #AA |
RMSD (Ǻ) |
Exp# | Feature(50) | Valence (1.4) | ||
|---|---|---|---|---|---|---|---|
| −MD | +MD | −MD | +MD | ||||
| 1B9A | 108 | 1.423 | 2 | 2 | 2 | 0 | 2 |
| 1B8C | 1 | 1 | 1 | 0 | 0 | ||
| 1K94 | 217 | 1.234 | 2 | 1 | l 1 | 0 | 2 |
| 1F4Q | 2 | 1 | l 1 | 0 | 0 | ||
| 3DNI | 260 | 1.750▲ | 3 | 1 | 3 | 1 | 2 |
| 1DNK | 3 | 1 | 2 | 0 | 0 | ||
| 1140 | 175 | 0.625 | 3 | 0 | 2 * | 0 | 1 |
| 1MJW | 3 | 0 | 0 | 0 | 0 | ||
| 1K96 | 90 | 4.067 | 2 | 2 | 2 | 1 | 2 |
| 1K8U | 2 | 0 | 0 | 0 | 0 | ||
| 1PSH | 119 | 0.613 | 1 | 0 | 0 1 | 0 | 0 |
| 1A3D | 1 | 0 | 1 | 0 | 0 | ||
| 1NLS | 237 | 0.371 | 1 | 1 | 1 1 | 0 | 1 |
| 1DQ0 | 1 | 0 | 1 | 0 | 0 | ||
| 1F6S | 123 | 1.194▲ | 1 | 0 | 1 1 | 0 | 1 |
| 1F6R | 1 | 1 | 1 1 | 0 | 0 | ||
| 3LHM | 130 | 0.211 | 1 | 0 | 1 1 | 1 | 1 |
| 2LHM | 1 | 0 | 1 1 | 0 | 0 | ||
| 1QMD | 370 | 0.203 | 4 | 0 | 4 1 * | 0 | 2 |
| 1QM6 | 4 | 0 | 4 4 * | 0 | 0 | ||
| 2POR | 301 | 0.939 | 4 | 1 | 4 1 | 0 | 1 |
| 3POR | 4 | 2 | 4 2 | 0 | 0 | ||
| Total H | 24 | 9 | 21 7 | 3 | 15 | ||
| Total A | 23 | 6 | 16 9 | 0 | 0 | ||
| Total | 47 | 15 | 37 16 | 3 | 15 | ||
The first column lists PDB IDs of the structures used in this work. The eleven molecules are listed in order, with HOLO structure (e.g. 1B9A) being followed by the corresponding APO structure (e.g. 1B8C) for each protein. The number of amino acids comprising each molecule is listed in column two. The next column lists the backbone RMS deviation between the HOLO and the APO structures used to start MD simulations for each molecule. The symbol ▲ denotes that the original PDB file of at least one of the structures in the pair has missing atoms or residues (see Supplemental Experimental Procedures section 1). The fourth column reports how many true Ca2+ binding sites there are in each structure. The next two columns report the global FEATURE scanning results for analysis of initial PDB structures and structural ensembles. The last two columns report the global valence-based method results for the starting PDB structures and structural ensembles. Superscript numbers inform how many false positive results were obtained. See the Results section for explanations of the results marked by *.
1. Preparation of structures
In five of the eleven pairs of structures the two partners do not have identical sequences, because one contains at least one mutation. In four pairs these mutations are far from the Ca2+ binding sites. In one HOLO – APO pair, namely 1B9A-1B8C, a mutation affecting residue 101 (GLU to ASP) appears in the coordinating loop of the 1B8C CA109 site. This mutation in the APO modifies the affinity of Ca2+ binding at the CA109 site and eliminates Ca2+ binding at the CA110 site (Cates et al., 1999). As noted below, we are able to recognize the loss of this site.
Of the 22 structures selected for this work, five structures required slight alterations, as described in the Supplemental Experimental Procedures section 1. All these were relatively modest, consisting of easily filled missing atoms or small gaps of missing residues or modified residues that could be easily replaced with the naturally occurring ones.
2. Molecular dynamics simulations
After the initial equilibration period, the energy analysis (data not shown) for each simulated system showed no significant change in potential energy over time and the RMS deviation to the starting structure generally plateaued below 6 Ǻ, suggesting that all systems were stable. In addition, the secondary structure content for each system simulated also showed no significant change over the course of the simulations (see Table 2). The trajectories were sampled every 2.5 ps and those structures were evaluated by both FEATURE and the valence method (see Experimental Procedures).
Table 2.
Details of simulation systems and analysis of MD trajectories
| Structures PDB ID | Chain Used | Ca2+ Present | Ions Other | Ions Added | % Initial Content | % Average Content | % Final Content |
|---|---|---|---|---|---|---|---|
| 1B9A | single | 2 | 1 Na+ | 50, 13 | 47, 12 | 44, 10 | |
| 1B8C | A | 1 Mg2+ | 3 Na+ | 50, 13 | 51, 10 | 56, 6 | |
| 1K94 | A | 2 | 1 CI− | 61 | 55 | 51 | |
| 1F4Q | B | 4 Na+ | 62 | 53 | 52 | ||
| 3DNI | single | 2 | 5 Na+ | 25, 26, 8 | 23, 24, 9 | 24, 25, 7 | |
| 1DNK | A | 9 Na+ | 25, 26, 9 | 25, 23, 10 | 24, 27, 12 | ||
| 1I40 | single | 5 | 1Na+, 3 CI− | None | 17, 31, 17 | 20, 27, 10 | 23, 22, 9 |
| 1MJW | A | So42− | 8 Na+ | 17, 33, 18 | 19, 28, 10 | 21, 24, 7 | |
| 1K96 | single | 2 | 1 CI− | 71 | 61 | 47 | |
| 1K8U | single | 3 Na+ | 63 | 62 | 55 | ||
| 1PSH | A | 1 | l Na+ | 45, 14 | 40, 12 | 39, 7 | |
| 1A3D | single | 1 Na+ | 2 Na+ | 45, 15 | 38, 16 | 43, 13 | |
| 1NLS | single | 1 | 1 Mn2+ | 5 Na+ | 46, 12 | 41, 9 | 41, 8 |
| 1DQ0 | single | 7 Na+ | 46, 12 | 41, 8 | 38, 8 | ||
| 1F6S | F | 1 | 5 Na+ | 32, 15 | 27, 17 | 21, 19 | |
| 1F6R | A | 6 Na+ | 31, 19 | 26, 17 | 26, 15 | ||
| 3LHM | single | 1 | 8 CI− | 30, 27 | 28, 20 | 27, 21 | |
| 2LHM | single | 6 CI− | 30, 27 | 35, 19 | 33, 17 | ||
| 1QMD | A | 3 | 2 Zn2+ | 2 Na+ | 40, 15, 10 | 35, 13, 11 | 29, 9, 13 |
| 1QM6 | A | 2 Zn2+ | 9 Na+ | 40, 15, 10 | 36, 14, 8 | 34, 15, 7 | |
| 2POR | single | 3 | 28 Na+ | 57, 12 | 49, 12 | 48, 8 | |
| 3POR | single | 34 Na+ | 57, 12 | 48, 14 | 50, 10 |
Column 1 lists PDB IDs of the structures used in this work. Column 2 shows which chain was used for the simulations. Column 3 lists how many calcium ions are present in the starting structure, which is also the number of calcium ions which were present in the system during simulation. Column 4 lists what other ions were originally present with the structure. Column 5 lists how many ions were added to each system in order to ensure neutral simulation conditions. The next three columns list the secondary structure (ss) content of the initial PDB structure, column 6, the average ss content over the course of the simulation, column 7, and the ss content of the final structure generated by the MD simulations, as calculated by the do_dssp program from the GROMACS suit. For the 1K94, 1F4Q, 1K96 and 1K8U only α-helical content is reported. For the 1B9A, 1B8C, 1PSH, 1A3D, 1F6S, 1F6R, 3LHM and 2LHM the two numbers in each column stand for α-helical and turn contents. For the 1NLS, 1DQ0, 2POR and 3POR the two numbers in each column stand for the β-sheet and turn contents. For the 1I40, 1MJW, 3DNI, 1DNK, 1QMD and 1QM6 the three numbers in each column stand for α-helical, β-sheet and turn contents.
3. Results of the HOLO – APO pairs
FEATURE examines local 3D structural environments looking at ∼80 features in six concentric spherical shells 1.25 Ǻ apart (Halperin et al., 2008). Based on a model that is built by comparing true sites and non-sites for a given property, FEATURE then assigns a score to local environments, which describes how likely they are to represent one of interest. Coupling FEATURE to the structural dynamics of molecules considerably improved its performance, revealing 22 more Ca2+ binding sites in addition to the 15 (9 in the HOLO and 6 in the APO initial PDB structures) found readily by FEATURE alone. In addition to the true Ca2+ binding sites, FEATURE found 7 false positive sites in the HOLO and 9 in the APO structural ensembles. A number of these are nonetheless interesting, as described below. The overall distributions of scores for static structures and for structures sampled during the molecular dynamics were not significantly different (see Figure 2). As expected, points that scored poorly for calcium binding were generally within the hydrophobic protein cores or at the structural periphery, where there were not enough atoms for the FEATURE algorithm to consider.
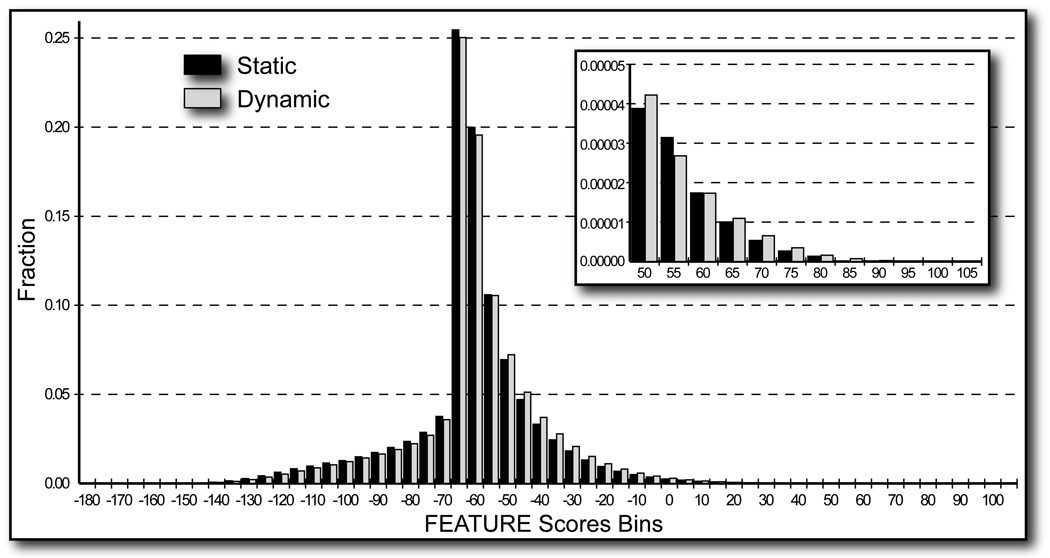
Figure 2. Global grid scanning histogram.
Global grid results for all eleven starting structures (black) and all eleven structural ensembles (light gray) are shown. The minimum and the maximum scores attained are ∼−177 and ∼105, respectively. The peak at ∼−65 is formed by points that lie on the periphery of structures such that FEATURE evaluates the empty space around the structure. The inset illustrates the magnification of the right tails of the histograms for scores 50 and above.
In order to test the generality of our approach to improve structure-based function prediction, we tested a second method which identifies Ca2+ binding sites in 3D environments. This method (Nayal and Di Cera, 1994) relies on measuring the negative charge around a potential site. Performance of the valence method also showed marked improvement with MD: it identified 15 Ca2+ binding sites in the HOLO structural ensembles, while only 3 in the initial HOLO structures.
Since FEATURE generally performed better than the valence method, the remainder of this section is focused on some results with FEATURE, which are presented below for selected HOLO – APO pairs. Results for the remaining pairs are presented in the Supplemental Results and Table 1 summarizes all results.
3DNI and 1DNK [bovine DNase I]
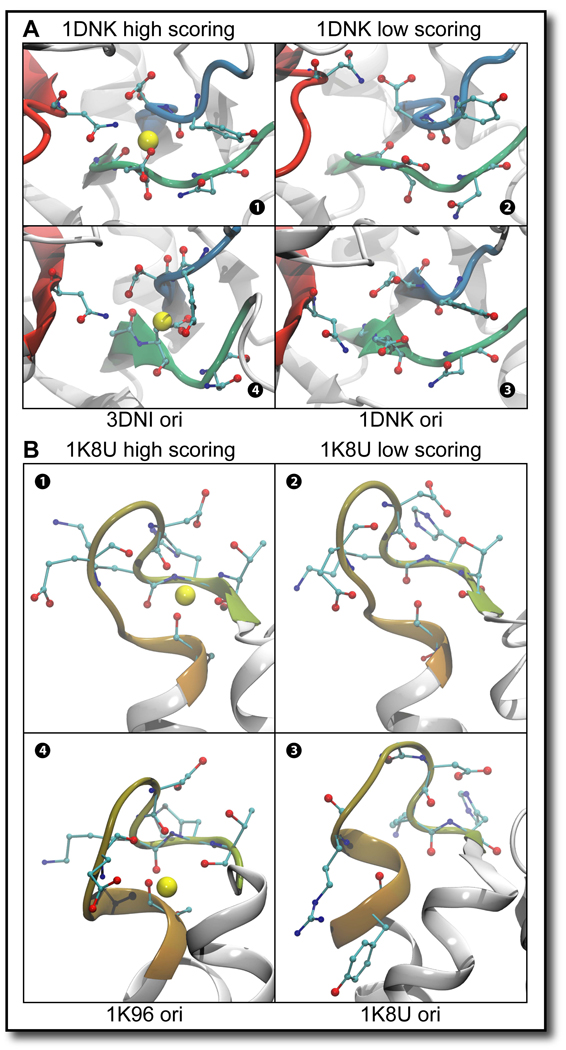
The results of FEATURE scanning for the 3DNI – 1DNK HOLO – APO pair were particularly interesting. This molecule contains only two Ca2+ binding sites, based on the simple scheme of relating our results to the Ca2+ ions bound within the HOLO structure (see Experimental Procedures). However, literature investigation revealed that this molecule contains one more characterized Ca2+ binding site at the active site (Suck et al., 1984). While only revealing a single site in the HOLO and APO initial PDB structures, FEATURE correctly identified all three Ca2+ binding sites in the HOLO structural ensemble (see Figure 4A). In the APO structural ensemble the Ca2+ binding site at the active site of the molecule and CA282 site were revealed.
Figure 4. Close-up of several Ca2+ binding sites.
The side-chains of nearby residues are shown, with cyan representing carbon, red – oxygen, and blue – nitrogen atoms. The backbone of these residues is also colored by index from red to blue. The yellow balls represent the putative site centers of the Ca2+ binding sites as identified by FEATURE, scoring over 50 on global grid FEATURE scanning. A Close-up of the Ca2+ binding site located at the active site of the HOLO – APO pair 3DNI – 1DNK The red, blue and green strands illustrate that disparate parts of the molecule come together to form this binding site. 1 1DNK highest scoring conformation is shown. 2 1DNK low scoring conformation is shown. 3 1DNK site as it appears in the original PDB file is shown. 4 3DNI conformation is shown which scores similarly to the highest scoring 1DNK conformation. B Close-up of the CA91 binding site in the HOLO – APO pair 1K96 – 1K8U and the 10 ns result of 1K8U. The single flexible loop illustrated in each panel defines this Ca2+ binding site. 1 1K8U highest scoring conformation from the 10 ns MD simulation is shown. 2 1K8U low scoring conformation is shown. 3 1K8U site as it appears in the original PDB file is shown. 4 1K96 conformation is shown which scores similarly to the highest scoring 1K8U conformation.
1I40 and 1MJW [Escherichia coli inorganic phosphatase]
In the case of the HOLO structure 1I40, our results showed a dependence on the initial structure with which MD simulation was started. In the 1I40 starting structure, CA306 is an active site cation, which usually binds in the presence of the protein substrate. However, since the substrate is absent, this Ca2+ ion is not in its native position and is instead located 5.3Ǻ away from the CA302 binding site, which influences the clustering analysis to lump the two sites into one (see Supplemental Experimental Procedures section 2). The high scoring putative Ca2+ binding site centers obtained with global grid FEATURE scanning spanned two different environments coordinated by different sets of amino acids, having in common only ASP157 (see Supplemental Movies). Since these environments correspond well to the two Ca2+ binding sites, CA302 and CA306, we conclude that our method locates both of those sites. The coordination environment of CA304 in this molecule consists of six water molecules and one oxygen atom from the protein itself. Because water molecules are not reliably present in crystal structures, FEATURE does not consider them in its models, and thus would be unlikely to recognize calcium sites coordinated mostly by water for either HOLO or APO structures. FEATURE does not identify any of the sites in the APO initial PDB structures or the APO structural ensemble.
1K96 and 1K8U [human S100A6]
While both of the expected Ca2+ binding sites present in the 1K96 HOLO structure were readily identified in the initial PDB structure and the structural ensemble, no Ca2+ binding sites were identified in its counterpart APO 1K8U system. However, the site noted as CA91 in the HOLO structure achieved scores close to the model threshold in the APO dynamic ensemble. In order to investigate whether a longer simulation might uncover this site, we continued the 1ns simulation for an additional 9 ns. The longer trajectory contained 4001 structures sampled every 2.5 ps, of which several attained a FEATURE score of more than 50, compatible with Ca2+ binding at the CA91 site, especially around the 8th nanosecond.
1F6S and 1F6R [bovine α-lactalbumin] and 3LHM and 2LHM [human lysozyme]
On the sequence and structural levels lysozyme and α-lactalbumin exhibit very high similarity, and have been postulated to be evolutionarily related: emerging from a duplication of a single ancestral gene and then diverging in their functions over time (Qasba and Kumar, 1997). As such, it is rather encouraging that FEATURE successfully identified the Ca2+ binding site present in these molecules in the HOLO and APO structural ensembles. Interesting, also, is the fact, that an identical false positive site was identified in all four structural ensembles. In the human lysozyme residues THR52, SER61, ASP67, THR70, and others nearby persistently directed available oxygen atoms towards the false positive site, and could also participate in the binding coordination of a divalent cation. The location of this false positive site was far from the locations of zinc and manganese ions binding sites. The Ca2+ binding model employed by FEATURE for this work correctly did not detect these binding sites.
Discussion
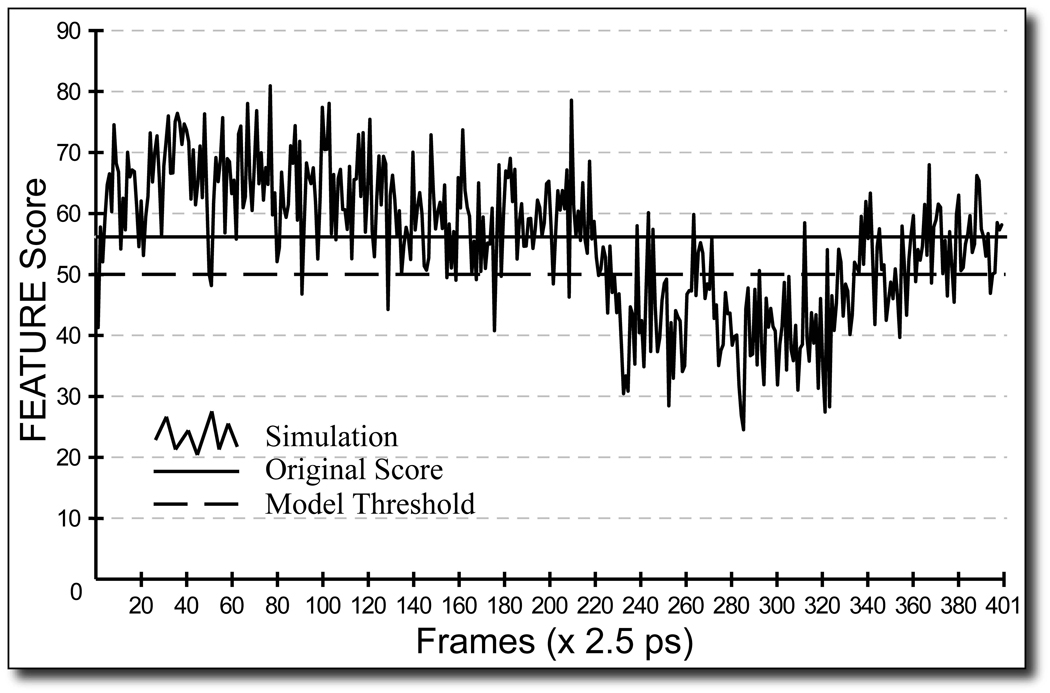
The purpose of these experiments was to test the hypothesis that short molecular dynamics simulations can improve the performance of function recognition methods. Figure 3 depicts a trace of local grid FEATURE scanning around the CA302 binding site in the 1I40 HOLO structural ensemble. This particular site is easily recognized by FEATURE as a Ca2+ binding site without the help of MD simulations, evidenced by the highest score of ∼56 attained by the initial PDB structure at this site. However, these results illustrate clearly that even HOLO crystal structures do not necessarily contain the more favorable local conformations for Ca2+ binding, while MD simulations have the potential to reveal structures which do: over the course of the simulation many structures were generated which scored much higher than the initial structure. Nearly all the sites conformed to this trend. In addition, sites exhibited dynamic behavior: the side chains within the sites changed between coordinated and uncoordinated states. On average, in order for FEATURE to identify the sites in the structural ensembles which were not obvious in the starting structures the side chains coordinating calcium binding moved by 3 Ǻ (data not shown).
Figure 3. Local grid FEATURE scanning results for a 1 ns simulation of 1I40 around the CA302 site.
A trace of the local grid FEATURE scanning for CA302 Ca2+ binding site of the HOLO 1I40 structural ensemble is shown in solid black. The horizontal black line indicates the highest score obtained for this site on a local grid in the starting structure. The model threshold is represented by a dashed line. Over the course of the simulation, several structures reveal the presence of favorable Ca2+ binding environments, by obtaining scores of 50 or above. The short dip of scores under the threshold between 0.55 ns and 0.85 ns is well explained by a local rearrangement in one of the residues coordinating this binding site. Supplemental Movies depict this change in the structure and further show how this particular residue settles towards the end of the simulation to accommodate this binding site and another one nearby.
This work demonstrates the value of examining molecular motions in the course of predicting function. In general, FEATURE performed better on the initial HOLO structures than the valence method, and the MD simulations improved the results of both methods for the HOLO structural ensembles. FEATURE outperformed the valence method in predicting Ca2+ binding sites for both the APO starting structures and the MD generated structural ensembles, which also showed marked improvement in FEATURE results. As expected, for both methods, Ca2+ sites in the APO structures were more challenging to recognize than the corresponding sites in the HOLO structures. The presence of the Ca2+ ion in the binding sites of the HOLO structures may influence the topology of the sites, with side-chains adopting favorable conformations more often. In contrast, the APO side-chain conformations are not similarly constrained, and hence may form favorable Ca2+ binding states more transiently.
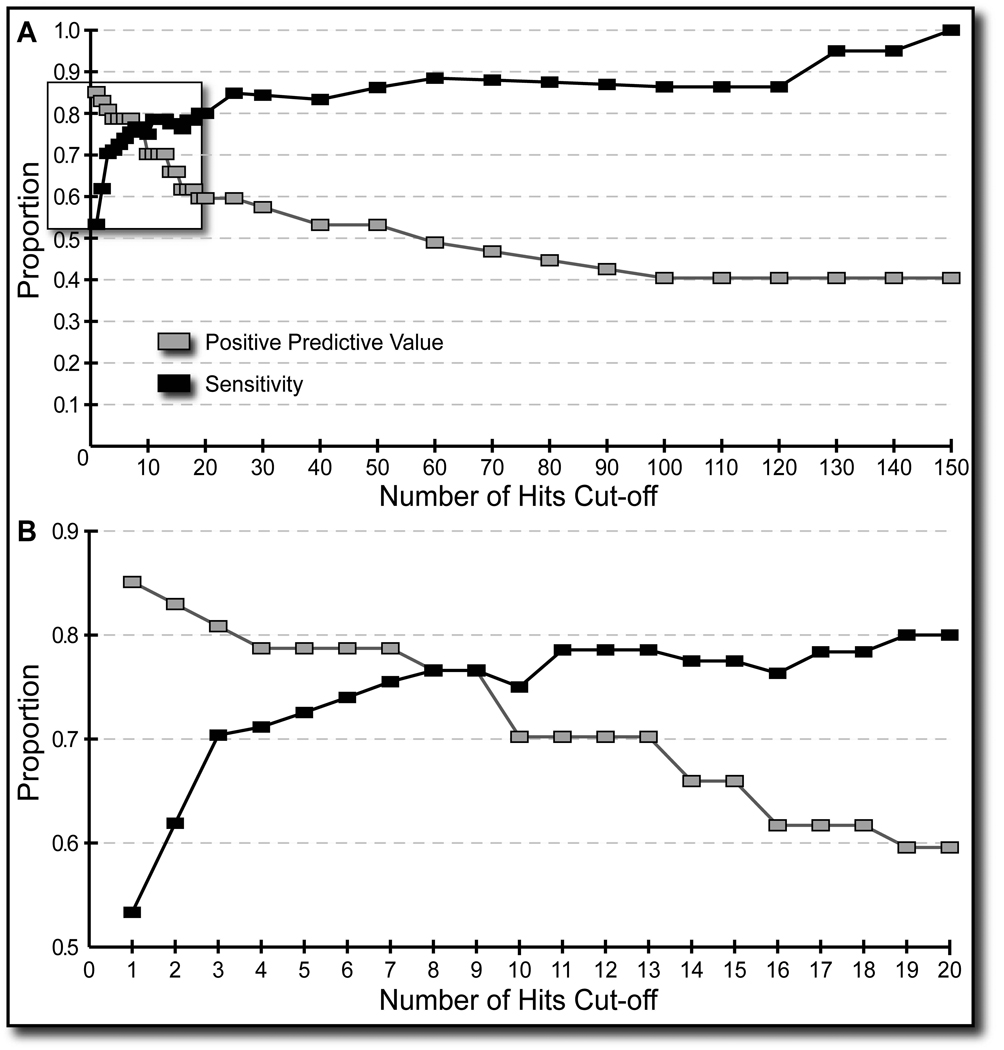
Both function prediction methods identified sites that were not documented to bind Ca2+ ions, which we label as false positive. Most of these sites differed from the true Ca2+ binding sites in two ways. Firstly, the highest score achieved in the TP clusters tended to be much higher than the highest score observed in the FP clusters. Also, FP clusters tended to consist of one or two putative site centers, while the TP clusters were mostly composed of multiple putative site centers (see Table 3 and Figure 5). Based on the Positive Predictive Value analysis, we required that at least three putative site centers should be present at the site for it to be counted as a TP or a FP result. Only 16 FP results identified by FEATURE survived this filter, out of possible 35 for FEATURE and 29 for the valence method, while only 2 and 10 TP results were lost for FEATURE and valence method, respectively. All the results reported in this work have survived this filter. Examination of the trade-off between the number of obtained TP and FP sites is also possible through varying the score cut-offs for FEATURE and the valence method. Such future investigations may be worthwhile.
Table 3.
Filtered clustered data
| FEATURE Scores | Number of Hits in Cluster | ||||
|---|---|---|---|---|---|
| PDB ID | Site | Highest Score | PDB ID | Site | Number of Hits |
| 1QMD | Zn-site | 111.24 | 1QMD | Zn-site | 24,467 |
| 1B8C | CA109 | 100.39 | 1QM6 | Zn-site | 10,456 |
| 1QMD | CA405 | 100.19 | 3DNI | CA282 | 5,824 |
| 3DNI | CA282 | 95.56 | 1B8C | CA109 | 5,342 |
| 1QM6 | Zn-site | 94.08 | 1QMD | CA405 | 4,796 |
| 1F6S | CA124 | 87.67 | 1F6S | CA124 | 3,506 |
| 1I40 | CA302 | 87.25 | 1NLS | CA239 | 2,172 |
| 1B9A | CA109 | 86.23 | 1I40 | CA302 | 2,121 |
| 2POR | CA303 | 83.90 | 1K96 | CA92 | 1,843 |
| 1K96 | CA92 | 82.76 | 1QM6 | CA405 | 1,437 |
| 1K94 | CA998 | 82.11 | 2POR | CA303 | 1,215 |
| 1QM6 | CA405 | 79.38 | 1K94 | CA998 | 660 |
| 1NLS | CA239 | 72.57 | 1B9A | CA109 | 468 |
| 1QM6 | FP1 | 71.63 | 1DQ0 | CA239 | 223 |
| 3POR | CA304 | 71.02 | 3DNI | CA281 | 169 |
| 1F6S | FP1 | 70.00 | 1QM6 | FP1 | 145 |
| 1F6R | CA124 | 69.02 | 1F6S | FP1 | 124 |
| 3DNI | CA281 | 65.41 | 1F6R | FP1 | 123 |
| 1I40 | CA306 | 65.07 | 1F6R | CA124 | 97 |
| 1DQ0 | CA239 | 64.99 | 3POR | CA304 | 85 |
| 1K94 | FP1 | 64.08 | 2POR | CA304 | 75 |
| 1K96 | CA91 | 64.06 | 1I40 | CA306 | 61 |
| 2POR | CA304 | 63.88 | 1DNK | active-site | 51 |
| 1F4Q | CA998 | 63.62 | 3POR | CA303 | 51 |
| 3POR | CA303 | 63.44 | 2LHM | FP1 | 50 |
| 1DNK | CA282 | 63.20 | 3LHM | FP1 | 46 |
| 1F6R | FP1 | 62.99 | 1A3D | CA120 | 36 |
| 1A3D | CA120 | 62.67 | 1DNK | CA282 | 34 |
| 2LHM | FP1 | 62.23 | 2POR | Ca4 | 29 |
| 1QMD | FP1 | 61.89 | 1K94 | FP1 | 23 |
| 3LHM | FP1 | 61.24 | 3POR | FP1 | 22 |
| 3LHM | CA131 | 60.15 | 1QMD | FP1 | 18 |
| 2POR | FP1 | 60.05 | 2LHM | CA131 | 18 |
| 2LHM | CA131 | 59.94 | 1NLS | FP1 | 17 |
| 1QM6 | FP2 | 59.34 | 1K96 | CA91 | 15 |
| 3DNI | active-site | 58.78 | 1K8U_8ns | CA92 | 15 |
| 1DNK | active-site | 58.33 | 1F4Q | CA998 | 13 |
| 1NLS | FP1 | 57.76 | 3POR | CA302 | 13 |
| 2POR | Ca4 | 57.71 | 1QM6 | FP2 | 10 |
| 3POR | FP1 | 57.31 | 2POR | FP1 | 10 |
| 3POR | FP2 | 56.44 | 3DNI | active-site | 9 |
| 1QM6 | FP3 | 56.41 | 3LHM | CA131 | 9 |
| 1K8U_8ns | CA92 | 56.38 | 3POR | Ca4 | 9 |
| 3POR | CA302 | 56.05 | 1QM6 | FP3 | 7 |
| 3POR | Ca4 | 55.95 | 2POR | CA302 | 7 |
| 1PSH | FP1 | 55.59 | 1F4Q | FP1 | 6 |
| 2POR | CA302 | 55.04 | 3POR | FP2 | 5 |
| 1F4Q | FP1 | 54.32 | 1PSH | FP1 | 4 |
| 1QM6 | FP4 | 53.68 | 1B9A | CA110 | 3 |
| 1B9A | CA110 | 50.62 | 1QM6 | FP4 | 3 |
The table lists the filtered clustered data in two ways: sorted by the highest FEATURE score obtained within the cluster and by the number of the identified putative Ca2+ binding sites in each super-cluster. Columns 1 and 4 show the PDB ID of the molecule in which the site is located. Columns 2 and 5 list either the Ca2+ binding sites which were identified or the name of the FP result. Column 3 lists the highest FEATURE score obtained by the specified cluster, and Column 6 lists the number of the putative Ca2+ binding sites identified in each super-cluster, starting with at least 3.
Figure 5. Analysis of trade-off between TP and FP.
The figure illustrates full analysis of our raw data with regards to the number of the identified putative Ca2+ binding site centers in each super-cluster. Panel A depicts the Positive Predictive Value and Sensitivity with respect to the number of the identified putative Ca2+ binding site centers in each super-cluster (see Table 3). Panel B is a close-up of the first 20 data points in Panel A, outlined with the rectangle. From these plots, especially panel B, we chose to filter our raw clustering results with the condition that a valid super-cluster must contain at least three putative Ca2+ binding site centers.
The accuracy of the force fields in treating Ca2+ interactions with the protein and solvent may be a limiting factor in our results. A recent study demonstrated that GROMOS force field underestimated by two-fold the attractive interactions between the Ca2+ ions and protein side-chains involved in binding coordination (Project et al., 2007). This same study implicated CHARMM and OPLS-AA force fields in underestimating by two-fold and overestimating by four-fold those same interactions. Improvements in force field parameters could further improve the efficiency of Ca2+ binding recognition when coupling structure-based function prediction methods to MD simulations. With better parameterization, it may be appealing to introduce Ca2+ atoms in the high scoring APO sites in the MD protocol in order to see if these sites continue to exhibit the high scoring conformations in the presence of the cation. In our experiments, several APO Ca2+ binding sites narrowly missed the threshold score of 50: 1DNK CA281, 1MJW CA302 and CA306, and 1K8U CA91 (1 ns simulation). It is possible that if Ca2+ were included in the APO systems in the MD simulations, the ions could enhance further the higher scoring conformations.
Our results, especially for the APO structural ensembles, conform to the preexisting equilibrium hypothesis of ligand-binding, where the molecule exists in an equilibrium of several different conformations. The binding of a ligand to the appropriate conformation shifts the equilibrium towards this particular conformation (Keskin, 2007; Tobi and Bahar, 2005; Tsai et al., 1999; Xu et al., 2008). Therefore, even without the presence of a calcium ion the APO structures may exhibit transient conformations favorable for calcium binding. In fact, with the addition of Ca2+ atoms to the APO sites in the MD simulations protocol we would not expect qualitative differences in the results, other than perhaps an increase in the frequency and persistence of high-scoring conformations.
Currently, MD simulations remain a rather expensive way of generating structural diversity. The protocol used in this work relied on the simplicity of the function of interest. The 1 ns simulations generally proved to be long enough, generating sufficient number of conformations to elicit positive identification of the existing Ca2+ binding sites (it is important to remember that our goal was simply to generate representative conformations—it was not required that the conformations were sampled with a Boltzmann probability). Several improvements can be made to this protocol, including allowing longer times for system equilibration and trying different force fields. However, such steps would be impossible to predict if the work concerned a structure with an unknown function.
Our experiments are not sufficient to provide statistically significant estimates of the improved performance of structure recognition. However, for many structures, we observed (as described above and in the Supplemental Results) conformational dynamics that contributed to the formation of calcium binding pockets. With improvements in molecular dynamics algorithms and implementations on special purpose hardware, it may become possible to run the dynamics for longer time period and to apply the algorithm described here on a large scale to many proteins in the PDB (Friedrichs et al., 2009). This paper reveals the potential value of such an effort at uncovering otherwise obscure binding sites.
Conclusions
The dynamic ensembles generated by the MD simulations improved the performance of two different structure-based function prediction methods. Both function recognition algorithms achieved better performance in identifying Ca2+ binding sites in structural ensembles generated by the MD simulations in 1 ns, than in the initial structures obtained from the PDB. We expect that our methodology can be easily amended to accommodate large systems or those that require significant domain motions for function determination by either generating longer simulations or employing other methods that explore conformational space more rapidly, such as normal modes or replica exchange. Furthermore, while this work employed FEATURE and the valence method, which are simple and efficient to apply in a large scale study to hundreds and thousands of structures, it should be apparent from our results that any other structure annotation algorithm can be accommodated by the methodology outlined in this work. As such, the scheme for recognizing function based on structures summarized in Figure 1 should be applicable to the novel unannotated structures generated by the Structural Genomics Consortia.
Experimental Procedures
1. Structures and definition of the HOLO and APO sets of Ca2+ binding sites
The following structures were taken from the PDB for this work: carp parvalbumin β, 1B9A (Cates et al., 1999) and 1B8C (Cates et al., 1999), human grancalcin, 1K94 (Jia et al., 2001) and 1F4Q (Han et al., 2000), bovine DNase I, 3DNI (Oefner and Suck, 1986) and 1DNK (Weston et al., 1992), Escherechia coli inorganic phosphatase, 1I40 (Samygina et al., 2001) and 1MJW (Avaeva et al., 1998), human S100A6, 1K96 (Otterbein et al., 2002) and 1K8U (Otterbein et al., 2002), Naja naja naja phospholipase A2, 1PSH (Fremont et al., 1993) and 1A3D (Segelke et al., 1998), Canavalia ensiformis concanavalin A, 1NLS (Deacon et al., 1997) and 1DQ0 (Bouckaert et al., 2000), bovine α-lactalbumin, 1F6S and 1F6R (Chrysina et al., 2000), human lysozyme, 3LHM and 2LHM (Inaka et al., 1991), Clostridium perfringens alpha-toxin, 1QMD and 1QM6 (Naylor et al., 1999), and Rhodobacter capsulatus porin, 2POR (Weiss and Schulz, 1992) and 3POR (Weiss and Schulz, 1993).
The 24 Ca2+ binging sites which formed the HOLO set were defined as the sites in which Ca2+ ion was present in the original static structures of 1B9A, 1K94, 3DNI, 1I40, 1K96, 1PSH, 1NLS, 1F6S, 3LHM, 1QMD, and 2POR with five exceptions. Only CA302, CA304, and CA304 sites were used for the 1I40 case. The active site of 3DNI and the fourth Ca2+ binding site of 2POR are characterized Ca2+ binding sites (Suck et al., 1984; Weiss and Schulz, 1992), and although no Ca2+ is present at these sites in the original PDB structures, we included them in our dataset. Similarly, the zinc-binding site of 1QMD was included as an expected site (see Supplemental Results). As such, there were 21 occupied and 3 unoccupied Ca2+ binding sites in the HOLO dataset.
By definition, the APO structures do not contain Ca2+ ions. However, since these APO structures directly correspond to the HOLO structures, we expected to observe the Ca2+ binding sites in analogous positions. With the exception of the CA110 binding site of 1B8C, discussed above, 23 out of the 24 sites forming the HOLO dataset are also present in the APO structures, which consisted of 1B8C, 1F4Q, 1DNK, 1MJW, 1K8U, 1A3D, 1DQ0, 1F6R, 2LHM, 1QM6, and 3POR.
Supplemental Experimental Procedures section 1 describes minor modifications made to some of the structures in order to ensure system completeness during the molecular dynamics simulations.
2. Molecular dynamics simulations
GROMACS is a software suit that allows generation and analysis of molecular dynamics (MD) simulations (Lindahl et al., 2001). Using GROMACS version 3.3.1, we created 1 ns simulations of the eleven pairs of structures in order to generate structural diversity for each protein set. For each of the structures, the system consisted of only one chain and appropriate ions, a 1 nanometer (nm) layer of solvent: simple point charge (SPC) (Berendsen et al., 1981) water molecules, and sodium or chloride ions for neutralization (see Table 2).
Each system, treated with a periodic boundary condition, underwent a 200-step energy minimization using the steepest descent algorithm followed by a 10 picosecond (ps) harmonic position restrained molecular dynamics simulation at 300 K, in which all the protein atoms remained motionless. Electrostatic and Van der Waals interactions and neighborlist cut-offs were set at 1 nm. Separate external temperature baths (Berendsen et al., 1984) of 300 K and coupling constant tauT = 0.1 ps were used for the protein and non-protein components of each system. The 1 ns production run simulation was carried out at constant pressure of 1 bar, kept as such by coupling weakly to pressure baths (Berendsen et al., 1984) with tauP = 0.5 ps, and constant temperature, as above. An integration time step of 2 femtoseconds (fs) was used with the LINCS (Hess et al., 1997) algorithm, constraining all covalent bonds to their equilibrium lengths. The force field used was GROMOS96 (van Gunsteren et al., 1996), 43a1. Generation of these trajectories took between 12 to 36 hours on a single 2.8 GHz processor with 2 G RAM. For each system, a structural ensemble was created by extracting structures generated by the simulation every 2.5 ps.
3. FEATURE scanning
This work employed FEATURE version 1.9 (Wei and Altman, 2003) and the Ca2+ binding site model built by Wei et al. (Wei and Altman, 1998) for analysis of static starting structures and structural ensembles generated by the MD simulations. A score of 50 or above at a given point in the structural space identified a putative Ca2+ binding site center.
FEATURE scanning was implemented in two ways: global grid and local grid. A global grid of points 1 Ǻ apart was created to encompass each starting structure and those generated by simulations. At each point, FEATURE algorithm calculated a score using the Ca2+ binding site model. For each structure examined, coordinates of the points which scored at least 50 were retained for clustering analysis described below. When looking at a novel structure, the global grid scanning would be applied as an initial analysis. Putative sites identified by the global grid scanning could then be followed up with the local grid scanning analysis, which allows examination of the behavior of the site of interest in finer detail. In our case, we were only interested in the true Ca2+ binding sites. Therefore, a local 10 × 10 × 10 Ǻ3 grid of points 1 Ǻ apart was created to encompass the Ca2+ binding sites of each starting structure and those generated by simulations. For each HOLO – APO pair, the local grids were centered on an equivalent atom, which was identified to be the closest to the Ca2+ atom in the crystallized HOLO structure. FEATURE algorithm calculated a score at each point using the Ca2+ binding site model. For each structure examined, only the information about the highest-scoring point was retained for visualizing behavior of the sites over the course of the simulations (see Figure 3).
Figure 2 illustrates the distributions of all the scores at the global grid points within 7.5 Ǻ of at least one atom in the structures, which is the radius of the FEATURE Ca2+ binding model, for the starting PDB structures and the respective structural ensembles generated by the MD simulations. We compared the two distributions using the chi-square goodness of fit test: the distribution from the starting structures was considered as theoretical values and the distribution from the structural ensembles was considered as the observed values. This analysis required counts for the calculations, so we normalized the structural ensembles distribution to the number of structures examined in each structural ensemble by reducing the number of counts for each bin by a factor of 401. The two distributions are statistically the same (calculations not shown).
As a negative control, we scanned with a local grid areas that were not near the Ca2+ binding sites in 1B9A, 1K94, and 1K96 HOLO structural ensembles. In each case, an atom was selected to be the grid center by two criteria: the atom density surrounding this atom in a sphere of radius 7.5 Ǻ and its distance to the surface were comparable to those of the central atoms of the local grids at the true Ca2+ binding sites. The local environments surrounding these points never attained conformations suitable for Ca2+ binding; the highest FEATURE scores never exceeded 30, and had a mean value of −10 for all frames in the simulations tested (data not shown). These results demonstrate that the score threshold of 50 established using static structures is a reasonably stringent cut-off to be applied to FEATURE scanning of structural ensembles generated by the MD simulations.
4. Valence scanning
We used the valence method (Nayal and Di Cera, 1994) with default parameters to scan the starting structures and the ensembles generated by the MD simulations. Valence is the number of electrons shared by an ion during bond-formation, and is generally estimated as the overall charge of the ion (thus, Ca2+ has valence of 2). This method further assumes that atoms exert partial valence at a distance, which decreases as the distance from the ion increases. Thus, in order to locate putative ion binding sites, this method sums partial valencies provided by nearby atoms. In our tests, a point around which partial valencies summed to 1.4 or above was used for predicting Ca2+ binding. A step size of 1 Ǻ was chosen to allow comparison between valence method and FEATURE results.
5. Clustering algorithm to define true and false positive results
Both function-recognition methods applied in this study may identify multiple putative Ca2+ binding site centers in a local region that often represent the same site. Additionally, combining and analyzing global scanning results of FEATURE and the valence method posed a challenge, since the gold standard Ca2+ binding sites (defined by the HOLO structures) are difficult to define once the molecules start moving during the simulations. In general, it is challenging to compare sites in different structures, because they are separated by time and conformational motion. Accordingly, we devised a clustering algorithm that considers the nearest 50 atoms to a point, such as a known Ca2+ site, and uses a paired Wilcoxon rank sum test to assess the similarity of two points in two potentially very different structural conformations. The nearest 50 atoms were chosen because they typically filled a sphere of the volume used in scanning by FEATURE (radius = 7.5 Å).The putative site centers identified by FEATURE or the valence method were clustered in order to define predicted Ca2+ binding sites that were substantially the same across the molecular dynamics trajectories. All results reported and discussed in this manuscript are based on this clustering analysis (see details in Supplemental Experimental Procedures section 2) and a post-clustering filter (see Discussion).
6. Structure and trajectory visualization
We used Visual Molecular Dynamics (VMD) (Humphrey et al., 1996) in order to visualize structures and trajectories generated by the MD simulations, as well as the results of FEATURE and the valence method scanning. Images of structures were generated using Tachyon Ray Tracer (Frishman and Argos, 1995; Stone, 1998) from within the VMD.
Supplementary Material
The Supplemental Materials include two movies, additional results and methods.
This animation shows a close-up of the CA302 and CA306 binding sites, which are readily identified by FEATURE in the 1I40 structural ensemble generated by the MD simulations. The yellow spheres represent the putative CA302 binding site centers, while the purple spheres represent the putative CA306 binding site centers. The binding site centers here refer to the centers of the clusters generated by the first clustering step for each frame of the simulation. With the exception of one residue, namely ASP157, the two sites are defined by distinctive coordinating residues. The backbone is shown as ribbon. The coordinating residues are shown as CPK, with oxygens further represented as VDW spheres. Colors represent the following atoms: cyan – carbon; blue – nitrogen; red – oxygen. The carboxylic oxygens on ASP157 are shown somewhat bigger in the metallic red color. At the end, the movie holds for a few seconds, displaying the last structure evaluated from the MD simulations and all the putative Ca2+ binding site centers that were identified in the structural ensemble generated by the MD simulations.
This animation is identical to the previous animation, except that the putative Ca2+ binding site centers that appear either as yellow or as purple spheres leave behind a shadow when they disappear. The shadow builds up over the course of the animation, allowing visualization of the relative placement of the two sites.
Acknowledgements
This work is supported by NIH LM-05652 and the SIMBIOS National Center for Simulation of Biological Structures, GM-072970. DSG is also supported by the Stanford Genome Training Grant NIH 5 T32 HG00044. We thank Inbal Halperin, members of the Stanford Helix group and the SIMBIOS National Center for useful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avaeva SM, Rodina EV, Vorobyeva NN, Kurilova SA, Bazarova TI, Sklyankina VA, Oganessyan VY, Samygina VR, Harutyunyan EH. Three-dimensional structures of mutant forms of E.coli Inorganic Pyrophosphatase with Asp->Asn single substitution in positions 42, 65, 70, and 97. Biochemistry (Moscow) 1998;63:671–684. [PubMed] [Google Scholar]
- Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. Interaction model for water in relation to protein hydration. In: Pullman B, editor. In Intermolecular Forces. Dordrecht, The Netherlands: D. Reidel Publishing Company; 1981. pp. 331–342. [Google Scholar]
- Berendsen HJC, Postma JPM, van Gunsteren WF, Nola AD, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat T, Weissig H, Shindyalov I, Bourne P. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert J, Dewallef Y, Poortmans F, Wyns L, Loris R. The structural features of concanavalin A governing non-proline peptide isomerization. J. Biol. Chem. 2000;275:19778–19787. doi: 10.1074/jbc.M001251200. [DOI] [PubMed] [Google Scholar]
- Cates MS, Barry MB, Ho EL, Li Q, Potter JD, Phillips GN., Jr Metal-ion affinity and specificity in EF-hand proteins: coordination geometry and domain plasticity in parvalbumin. Structure. 1999;7:1269–1278. doi: 10.1016/s0969-2126(00)80060-x. [DOI] [PubMed] [Google Scholar]
- Chandonia JM, Brenner SE. The impact of structural genomics: expectations and outcomes. Science. 2006;311:347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- Chrysina ED, Brew K, Acharya KR. Crystal structures of Apo- and Holo-bovine α-Lactalbumin at 2.2-angstrom resolution reveal an effect of calcium on inter-lobe interactions. J. Biol. Chem. 2000;275:37021–37029. doi: 10.1074/jbc.M004752200. [DOI] [PubMed] [Google Scholar]
- Damm KL, Carlson HA. Exploring experimental sources of multiple protein conformations in structure-based drug design. J. Am. Chem. Soc. 2007;129:8225–8235. doi: 10.1021/ja0709728. [DOI] [PubMed] [Google Scholar]
- Deacon A, Gleichmann T, Kalb AJ, Price H, Raftery J, Bradbrook G, Yariv J, Helliwell JR. The structure of concanavalin A and its bound solvent determined with small-molecule accuracy at 0.94-angstrom resolution. J. Chem. Soc., Faraday Trans. 1997;93:4305–4312. [Google Scholar]
- Eyrisch S, Helms V. Transient pockets on protein surfaces involved in protein - protein interaction. J. Med. Chem. 2007;50:3457–3464. doi: 10.1021/jm070095g. [DOI] [PubMed] [Google Scholar]
- Fetrow JS, Godzik A, Skolnick J. Function analysis of the Escherichia coli genome using the sequence-to-structure-to-function paradigm: identification of proteins exhibiting the glutaredoxin/thioredoxin disulfide oxidoreductase activity. J. Mol. Biol. 1998;282:703–711. doi: 10.1006/jmbi.1998.2061. [DOI] [PubMed] [Google Scholar]
- Frembgen-Kesner T, Elcock AH. Computational sampling of a cryptic drug binding site in a protein receptor: explicit solvent molecular dynamics and inhibitor docking to p38 MAP kinase. J. Mol. Biol. 2006;359:202–214. doi: 10.1016/j.jmb.2006.03.021. [DOI] [PubMed] [Google Scholar]
- Fremont DH, Anderson DH, Wilson IA, Dennis EA, Xuong NH. Crystal structure of phospholipase A2 from Indian cobra reveals a trimeric association. Proc. Natl. Acad. Sci. 1993;90:342–346. doi: 10.1073/pnas.90.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg I. Automated protein function prediction - the genomic challenge. Briefings In Bioinformatics. 2004;7:225–242. doi: 10.1093/bib/bbl004. [DOI] [PubMed] [Google Scholar]
- Friedrichs S, Eastman P, Vaidyanathan V, Houston M, LeGrand S, Beberg AL, Ensign DL, Bruns CM, Pande VS. Accelerating molecular dynamic simulation on Graphics Processing Units. J. Comput. Chem. 2009 doi: 10.1002/jcc.21209. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frishman D, Argos P. Kno wledge-based secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- Halperin I, Glazer DS, Wu S, Altman RB. The FEATURE framework for protein function annotation: modelling new functions, improving performance, and extending to novel applications. BMC Genomics. 2008;16(9 Suppl 2):S2. doi: 10.1186/1471-2164-9-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q, Jia J, Li Y, Lollike K, Cygler M. Crystallization and preliminary X-ray analysis of human Grancalcin, a novel cytosolic Ca2+-binding protien present in leukocytes. Acta Crystallogr. 2000;D56:772–774. doi: 10.1107/s0907444900005096. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- Hess B, Bekker H, Berendsen HJC, Fraaije JG. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- Huang SY, Zou X. Efficient molecular docking of NMR structure: application to HIV-1 protease. Protein Sci. 2007;16:43–51. doi: 10.1110/ps.062501507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD - Visual Molecular Dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Inaka K, Kuroki R, Kikuchi M, Matsushima M. Crystal structures of the Apo- and Holomutant human lysozymes with an introduced Ca2+ binding site. J. Biol. Chem. 1991;266:20666–20671. [PubMed] [Google Scholar]
- Jia J, Borregaard N, Lollike K, Cygler M. Structure of Ca2+-loaded human Grancalcin. Acta Crystallogr. 2001;D57:1843–1849. doi: 10.1107/s0907444901016511. [DOI] [PubMed] [Google Scholar]
- Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc. Natl. Acad. Sci. U.S.A. 2005;102:6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- Keskin O. Binding induced conformational changes of proteins correlate with their intrinsic fluctuations: a case study of antibodies. BMC Struct. Biol. 2007;7:31. doi: 10.1186/1472-6807-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M. Growth of novel protein structural data. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3183–3188. doi: 10.1073/pnas.0611678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 2001;7:306–317. [Google Scholar]
- Meagher KL, Carlson HA. Incorporating protein flexibility in structure-based drug discovery: using HIV-1 protease as a test case. J. Am. Chem. Soc. 2004;126:13276–13281. doi: 10.1021/ja0469378. [DOI] [PubMed] [Google Scholar]
- Nayal M, Di Cera E. Predicting Ca2+-binding sties in proteins. Proc. Natl. Acad. Sci. U.S.A. 1994;91:817–821. doi: 10.1073/pnas.91.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor CE, Jepson M, Crane DT, Titball RW, Miller J, Basak AK, Bolgiano B. Characterisation of the calcium-binding C-terminal domain of Clostridium perfringens alpha-toxin. J. Mol. Biol. 1999;294:757–770. doi: 10.1006/jmbi.1999.3279. [DOI] [PubMed] [Google Scholar]
- Oefner C, Suck D. Crystallographic refinement and structure of DNase I at 2-angstrom resolution. J. Mol. Biol. 1986;192:605–632. doi: 10.1016/0022-2836(86)90280-9. [DOI] [PubMed] [Google Scholar]
- Otterbein LR, Kordowska J, Witte-Hoffmann C, Wang CL, Dominguez R. Crystal structures of S100A6 in the Ca2+-free and Ca2+-bound states: the calcium sensor mechanism of S100 proteins revealed at atomic resolution. Structure. 2002;10:557–567. doi: 10.1016/s0969-2126(02)00740-2. [DOI] [PubMed] [Google Scholar]
- Project E, Nachliel E, Gutman M. Parameterization of Ca2+-protein interactions for molecular dynamics simulations. J. Comput. Chem. 2007;29:1163–1169. doi: 10.1002/jcc.20876. [DOI] [PubMed] [Google Scholar]
- Qasba PK, Kumar S. Molecular divergence of lysozymes and alpha-lactalbumin. Crit. Rev. Biochem. Mol. Biol. 1997;32:255–306. doi: 10.3109/10409239709082574. [DOI] [PubMed] [Google Scholar]
- Samygina VR, Popov AN, Rodina EV, Vorobyeva NN, Lamzin VS, Polyakov KM, Kurilova SA, Nazarova TI, Avaeva SM. The structures of Escherichia coli Inorganic Pyrophosphatase complexed with Ca2+ or CaPPi, at atomic resolution and their mechanistic implications. J. Mol. Biol. 2001;314:633–645. doi: 10.1006/jmbi.2001.5149. [DOI] [PubMed] [Google Scholar]
- Segelke BW, Nguyen D, Chee R, Xuong NH, Dennis EA. Structures of two novel crystal forms of Naja naja naja phospholipase A2 lacking Ca2+ reveal trimeric packing. J. Mol. Biol. 1998;279:223–232. doi: 10.1006/jmbi.1998.1759. [DOI] [PubMed] [Google Scholar]
- Sivanesan D, Rajnarayanan RV, Doherty J, Pattabiraman N. In-silico screening using flexible ligand binding pockets: a molecular dynamics-based approach. J. Comput. Aided Mol. Des. 2005;19:213–228. doi: 10.1007/s10822-005-4788-9. [DOI] [PubMed] [Google Scholar]
- Stone J. An efficient library for parallel ray tracing and animation. Masters Thesis. Computer Science Department, University of Missouri at Rolla; 1998. [Google Scholar]
- Suck D, Oefner C, Kabsch W. Three-dimensional structure of bovine Pancreatic DNase I at 2.5 A resolution. EMBO J. 1984;3:2423–2430. doi: 10.1002/j.1460-2075.1984.tb02149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC. Structures and technology for biologists. Nat. Struct. Mol. Biol. 2004;11:296–297. doi: 10.1038/nsmb0404-296. [DOI] [PubMed] [Google Scholar]
- Tobi D, Bahar I. Structural changes involved in protein binding correlate with intrinsic motions of proteins in the unbound state. Proc. Natl. Acad. Sci. U.S.A. 2005;102:18908–18913. doi: 10.1073/pnas.0507603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Kumar S, Ma B, Nussinov R. Folding funnels, binding funnels, and protein function. Protein Sci. 1999;8:1181–1190. doi: 10.1110/ps.8.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gunsteren WF, Billeter SR, Eising AA, Hünenberger PH, Krüger P, Mark AE, Scott WRP, Tironi IG. Biomolecular Simulation: The GROMOS96 manual and user guide. (Zürich, Switzerland: Hochschulverlag AG an der ETH Zürich) 1996 [Google Scholar]
- Wallace AC, Borkakoti N, Thornton JM. TESS: a geometric hashing algorithm for deriving 3D coordinate templates for searching structural databases. Application to enzyme active sites. Protein Sci. 1997;6:2308–2323. doi: 10.1002/pro.5560061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Laskowski RA, Thornton JM. Predicting protein function from sequence and structural data. Curr. Opin. Struct. Biol. 2005;15:275–284. doi: 10.1016/j.sbi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Wei L, Altman RB. Pac. Symp. Biocomput. Maui, HI: 1998. Recognizing protein binding sites using statistical descritions of their 3D environments; pp. 497–508. [PubMed] [Google Scholar]
- Wei L, Altman RB. Recognizing complex, asymmetric functional sites in protein structures using a Bayesian scoring function. J. Bioinform. Comput. Biol. 2003;1:119–137. doi: 10.1142/s0219720003000150. [DOI] [PubMed] [Google Scholar]
- Weiss MS, Schulz GE. Structure of porin refined at 1.8 angstrom resolution. J. Mol. Biol. 1992;227:493–509. doi: 10.1016/0022-2836(92)90903-w. [DOI] [PubMed] [Google Scholar]
- Weiss MS, Schulz GE. Porin conformation in the absence of calcium. J. Mol. Biol. 1993;231:817–824. doi: 10.1006/jmbi.1993.1328. [DOI] [PubMed] [Google Scholar]
- Weston SA, Lahm A, Suck D. X-ray structure of the DNase I-d(GGTATACC)2 complex at 2-3-angstrom resolution. J. Mol. Biol. 1992;226:1237–1256. doi: 10.1016/0022-2836(92)91064-v. [DOI] [PubMed] [Google Scholar]
- Wilson C, Kreychman J, Gerstein M. Assessing annotation transfer for genomics: quantifying the relations between protein sequence, structure and function through traditional and probabilistic scores. J Mol Biol. 2000;297:233. doi: 10.1006/jmbi.2000.3550. [DOI] [PubMed] [Google Scholar]
- Wong CF, Kua J, Zhang Y, Straatsma TP, McCammon JA. Molecular docking of balanol to dynamics snapshots of Protein Kinase A. Proteins. 2005;61:850–858. doi: 10.1002/prot.20688. [DOI] [PubMed] [Google Scholar]
- Xu Y, Colletier JP, Jiang H, Silman I, Sussman JL, Weik M. Induced-fit or preexisting equilibrium dynamics? Lessons from protein crystallography and MD simulations on acetylcholinesterase and implications for structure-based drug design. Protein Sci. 2008;17:601–605. doi: 10.1110/ps.083453808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Supplemental Materials include two movies, additional results and methods.
This animation shows a close-up of the CA302 and CA306 binding sites, which are readily identified by FEATURE in the 1I40 structural ensemble generated by the MD simulations. The yellow spheres represent the putative CA302 binding site centers, while the purple spheres represent the putative CA306 binding site centers. The binding site centers here refer to the centers of the clusters generated by the first clustering step for each frame of the simulation. With the exception of one residue, namely ASP157, the two sites are defined by distinctive coordinating residues. The backbone is shown as ribbon. The coordinating residues are shown as CPK, with oxygens further represented as VDW spheres. Colors represent the following atoms: cyan – carbon; blue – nitrogen; red – oxygen. The carboxylic oxygens on ASP157 are shown somewhat bigger in the metallic red color. At the end, the movie holds for a few seconds, displaying the last structure evaluated from the MD simulations and all the putative Ca2+ binding site centers that were identified in the structural ensemble generated by the MD simulations.
This animation is identical to the previous animation, except that the putative Ca2+ binding site centers that appear either as yellow or as purple spheres leave behind a shadow when they disappear. The shadow builds up over the course of the animation, allowing visualization of the relative placement of the two sites.