Summary
DNA interstrand cross-links (ICLs) are toxic DNA lesions whose repair occurs in the S phase of metazoans via an unknown mechanism. Here, we describe a novel cell-free system based on Xenopus egg extracts that supports ICL repair. During DNA replication of a plasmid containing a site-specific ICL, two replication forks converge on the cross-link. Subsequent lesion bypass involves advance of a nascent leading strand to within one nucleotide of the ICL, followed by incisions, translesion DNA synthesis, and extension of the nascent strand beyond the lesion. Immunodepletion experiments suggest that extension requires DNA polymeras ζ Ultimately, a significant portion of the input DNA is fully repaired, but not if DNA replication is blocked. Repair in this system is accompanied by activation of the Fanconi anemia and ATR checkpoint pathways. Our experiments establish a mechanism for ICL repair that reveals how this process is coupled to DNA replication.
Introduction
DNA interstand cross-links (ICLs), which covalently link the two strands of the double helix, are highly cytotoxic because they block DNA replication and transcription (Niedernhofer et al., 2005). While ICL forming agents are commonly used in cancer chemotherapy, ICLs are also formed by endogenous cellular metabolites (Scharer, 2005). The mechanism by which ICLs are repaired in eukaryotes is unknown, but important clues come from genetic studies, which have shown that various classes of proteins are required to confer cellular resistance to ICL agents (Dronkert and Kanaar, 2001; Niedernhofer et al., 2005). One such class is the structure specific endonucleases, which includes Mus81-Eme1 and the nucleotide excision repair (NER) factor, Xpf-Ercc1, both of which are thought to perform incisions near the ICL. Another class is the translesion DNA polymerases. Among these, Rev1 and DNA Polymerase ζ are particularly important (Niedzwiedz et al., 2004; Simpson and Sale, 2003). Proteins involved in homologous recombination, such as Rad54, Xrcc2, and Xrcc3, are also required (Nojima et al., 2005). Finally, thirteen Fanconi anemia gene products (FA proteins) are essential for resistance to ICLs and suppression of chromosomal instability (Niedernhofer et al., 2005; Wang, 2007). Eight FA proteins form a nuclear protein complex that is thought to mono-ubiquitylate FancD2 and FancI, an event that is crucial for cellular resistance to ICL agents. Mutations in FA proteins give rise to Fanconi anemia, a cancer predisposition syndrome.
Indirect evidence suggests that in metazoans, the principal ICL repair pathway occurs in S phase. First, regardless of when they are treated with ICL agents, mammalian cells arrest late in S phase with 4N DNA content (Akkari et al., 2000). Second, ICL agents lead to the formation of dsDNA breaks, but only after passage through S phase (Akkari et al., 2000; De Silva et al., 2000; Rothfuss and Grompe, 2004). Third, activation of the Fanconi anemia pathway occurs exclusively in S phase (Rothfuss and Grompe, 2004; Taniguchi et al., 2002), it is DNA replication-dependent (Sobeck et al., 2006), and FA mutant cells display a prolonged S phase arrest (Akkari et al., 2001). Based on these observations and the known enzymatic properties of relevant DNA repair enzymes, a model for ICL repair in S phase has been proposed [reviewed in (McHugh et al., 2001; Niedernhofer et al., 2005); Supplementary Figure 1]. Accordingly, ICL repair is initiated by collision of a DNA replication fork with the ICL. Mus81-Eme1 and/or Xpf-Ercc1 then perform dual incisions on one parental strand surrounding the ICL, causing a dsDNA break and release of one of the replicated sister chromatids. The 3' end generated in the parental strand via incision is extended by translesion DNA polymerases past the remaining mono-adduct, which is then removed, likely via nucleotide excision repair (NER). Finally, the replication fork is re-established using homologous recombination (HR). Notably, cells deficient for NER factors other than Xpf-Ercc1 are not particularly sensitive to ICL agents [(De Silva et al., 2000) and references therein]. Therefore, in the absence of NER, the adducted chromosome shown in Supplementary Figure 1D is envisioned to be the homology donor used to re-establish the replication fork. Although the above model is attractive, it is largely hypothetical.
To elucidate the molecular mechanism of ICL repair in S phase, a biochemical system that supports this process will be vital. Extracts derived from mammalian cells have been shown to promote incisions on plasmids containing ICLs [e.g. (Bessho, 2003; Li et al., 1999; Mu et al., 2000)], but the reactions are generally not performed in the context of S phase. To improve upon these approaches, we employed a soluble extract system derived from Xenopus eggs, which supports efficient replication of plasmid DNA templates using a physiological mechanism (Walter et al., 1998). Here, we show that Xenopus egg extracts support replication-dependent repair of plasmids containing a single, site-specific ICL. Using this system, we establish a detailed mechanism of ICL repair that includes several unexpected features. Thus, we observe that repair involves convergence of two DNA replication forks on the lesion. Subsequently, a multi-step lesion bypass reaction is initiated before incisions are detected near the ICL. Finally, in contrast to prevailing models, lesion bypass involves extension of a nascent leading strand, illustrating how ICL repair is coupled to DNA replication. In addition, our data suggest that ICL repair requires DNA polymerase ζ, and we identify the precise step in lesion bypass that is promoted by this translesion DNA polymerase.
Results
Convergence of DNA replication forks at a site-specific ICL
To develop a homogeneous substrate for ICL repair, we prepared plasmids containing a single, site-specific and chemically-defined ICL. This approach avoids the diverse spectrum of lesions generated when cells are treated with DNA interstrand crosslinking agents (Dronkert and Kanaar, 2001). A short duplex oligonucleotide containing a nitrogen mustard-like ICL (Figure 1A) or cisplatin ICL (Figure 1B) was ligated into a 5.6 kb plasmid, generating pICLNm and pICLPt, respectively. The sequences of pICLNm and pICLPt were identical, except for a small region of about 20 base pairs surrounding the ICL (Supplementary Figure 2A). For each construct, we also prepared a matched, undamaged plasmid of identical sequence termed pCtr. At least ~99 % of pICL plasmids contained an intact interstrand cross-link (see Experimental Procedures and below). The nitrogen mustard-like ICL was designed to fit in the major groove of DNA without distorting the double helix (Figure 1A; TA and ODS, manuscript submitted). The cisplatin ICL (Hofr and Brabec, 2001) causes a significant helical distortion [Figure 1B; (Huang et al., 1995)]. By comparing pICLNm and pICLPt, we sought to address whether the chemical structure of an ICL influences the mechanism of its repair.
Figure 1. DNA replication forks converge on an interstrand cross-link.
(A) Structure of nitrogen mustard-like (postulated) and (B) cisplatin [based on (Huang et al., 1995)] ICLs. (C) Replication of pICLNm in Xenopus egg extracts. pCtr or pICLNm was incubated sequentially with HSS and NPE/32P-α-dATP. At the indicated times after NPE addition, replication products were analyzed on a native agarose gel. (D) The average replication efficiency of three independent experiments was plotted with error bars. (E) Model for replication of pICL. (F) pICLNm was replicated as in (C). 30 minutes after NPE addition, DNA was analyzed by electron microscopy. The predominant species, a "Figure 8" structure, is shown.
We first analyzed how pICLNm was replicated in Xenopus egg extracts (Walter et al., 1998). In this system, plasmids are first incubated in a high-speed supernatant (HSS) of egg cytoplasm, which supports the assembly of pre-replication complexes (pre-RCs) via the sequential recruitment of ORC, Cdt1, Cdc6, and MCM2–7 to DNA in a sequence non-specific manner. Subsequently, a highly concentrated nucleoplasmic egg extract (NPE) is added. NPE triggers Cdk2-dependent replication initiation from pre-RCs, and a single, complete round of DNA replication occurs which can be monitored via the incorporation of 32P-α-dATP. During replication of the undamaged control plasmid (pCtr), partially replicated plasmids and fully replicated, catenated daughter molecules appeared within 15 minutes of NPE addition (Figure 1C, lane 1)(Walter and Newport, 2000). By 30 minutes, resolved daughter molecules appeared as nicked and supercoiled circles (Figure 1C, lane 2). In contrast, during replication of pICLNm, the appearance of nicked and supercoiled products was severely inhibited (Figure 1C, lanes 5–8). Importantly, incorporation of 32P-α-dATP was identical for both plasmids (Figure 1D). These data suggest that pICLNm was fully replicated but failed to undergo decatenation (Figure 1E). In support of this interpretation, electron microscopy showed that during replication of pICLNm but not pCtr, there was a massive accumulation of "Figure 8" structures, in which each hemisphere was equivalent to the size of pICLNm (Figure 1F and Supplementary Figure 3). Together, the data demonstrate that replication initiation and elongation are highly efficient on pICL, but that forks stall after converging on the ICL.
A stepwise mechanism for bypass of an ICL lesion
To determine where replication forks arrest relative to the ICL, we mapped the ends of their leading and lagging strands. pICLNm was replicated in the presence of 32P-α-dATP and digested with Afl III, which cleaves 149 nucleotides (nts) to the left and 540 nts to the right of the ICL (Figure 2A). Nascent products were analyzed on a sequencing gel. Figure 2B shows that by 18 minutes after NPE addition, leading strand products of the leftward fork formed a prominent cluster of bands ~500–520 nts in length (lane 1, black arrow; see also red strand in Figure 2A), indicating that their 3' ends were located ~20–40 nts from the ICL. Lagging strand products of the same fork were smaller and more heterogeneous (Fig. 2B, lane 1, blue bracket), placing their 5’ ends between ~70 and ~290 nts from the cross-link (see Supplementary Figure 4 for the identification of leading and lagging strand products). Leading strand synthesis subsequently resumed, and by 45 minutes, ~50% of the ~520 nt long leading strands were extended to a length of ~540 nts (Figure 2B, lane 4, red arrowhead), indicating they were very close to the ICL. By 4 hours, the ~540 nt species disappeared and a new product of ~ 690 nts accumulated (Figure 2B, lane 8, open arrowhead). This ~690 nt "extension" product can only be generated when the Afl III restriction fragment has been completely replicated, demonstrating that the lesion was fully bypassed. No discrete replication intermediates were detected at any time using pCtr (Figure 2B, lane 9 and data not shown). By cutting out a smaller piece of replicated pICLNm surrounding the lesion and running the nascent DNA on a high resolution sequencing gel, we found that lesion bypass occurred without significant levels of deletions or insertions (Supplementary Figure 5A). Importantly, by four hours, the ratio of radioactivity in the 689 bp Afl III fragment to the 4.9 kb vector backbone was essentially the same for pICLNm as for pCtr, indicating that virtually all replicated molecules that remained in the reaction had undergone lesion bypass (Supplementary Figure 6B, compare lanes 8 and 9). Using pICLPt, virtually identical results for lesion bypass were observed (Supplementary Figures 7, 8, and 5B). Our results indicate that the leading strand of a DNA replication fork bypasses an ICL in a series of temporally resolved steps that are essentially identical for both substrates.
Figure 2. Multi-step lesion bypass of an interstrand cross-link.
(A) Structure of the replicated Afl III fragment which includes the ICL. S, primer used to generate the sequencing ladder shown in (B). (B) Mapping of nascent strands during replication of pICLNm or pCtr (final concentration 1.2 ng/µl). At the indicated times after NPE addition, reaction products were digested with Afl III and analyzed on a sequencing gel alongside a sequencing ladder derived from extension of primer S on pCtr (see A). Numbers to the left indicate the sizes of the sequencing products. Leading and lagging strands for the rightward and leftward forks are indicated. Square brackets show the positions of leading strands after the initial pausing, whereas red and green arrowheads show their location after advancing towards the ICL. Open arrowhead, extension product. (C) Enlarged and darker exposure of the bottom part of the autoradiogram shown in (B). The most prominent species observed at 45 minutes are indicated on the right, with the predicted last nucleotide denoted by a single letter. The exact sizes of species T147–A150 were confirmed in Supplementary Figure 9. (D) Cartoon-form depiction of the results in (C).
Leading strands initially stall 20–24 nts from the ICL
We next examined where exactly the leading strand stalls upon initial collision of a replisome with an ICL. Close inspection of the autoradiogram in Figure 2B showed that the earliest leading strand products to accumulate were 110 to 126 nts long (Figure 2C, lane 1), indicating that the leading strand of the rightward replication fork advanced to within 24 nts of the cross-link (see green strand in Figure 2D). Notably, the leftward fork generated a similar cluster of leading strands, with the largest prominent product also stalling 24 nts from the ICL (Supplementary Figures 4C and 4D). Since the leading strands of both replication forks stalled at precisely the same distance from the lesion on either side of the ICL, the initial pause site of DNA polymerase appears to be dictated by the inherent size of the replisome's footprint on DNA, rather than the nucleotide sequence surrounding the ICL. Interestingly, there was a range of initial pause sites on either side of the ICL. Perhaps the first fork to arrive obstructs the approach of the fork coming from the other side.
Notably, we found that on pICLPt, leading strand polymerases of both forks initially paused 4 nts closer to the ICL than on pICLNm (20 versus 24 nts; Supplementary Figures 7 and 8). This could be explained if the DNA surrounding the distorting cisplatin ICL was easier to unwind than the DNA surrounding the non-distorting nitrogen mustard-like ICL.
After initial fork stalling, leading strands advance to within 1 nucleotide of the ICL
Shortly after the initial stalling of the replisome, DNA synthesis resumed, and by 45 minutes ~50% of the leading strands had advanced to the site of the cross-link (Figure 2C, lane 4). Near the nitrogen mustard-like ICL, DNA synthesis came to a second stop, generating four major species of 147–150 nts (Figure 2C, lane 4). The longest product in this cluster (150 nts) ends 1 nucleotide before the template base that participates in the ICL (the "−1" position; Figure 2D). A similar result was observed for pICLPt, except that at 45 minutes, most of the products stalled at the "−1" position (see Supplementary Figure 7C lane 4, and 7D). The pausing at the “−1 position” on pICLNm and pICLPt may reflect the time required to recruit a translesion DNA polymerase such as Rev1.
The question arises whether one or both leading strands advance to the ICL from the initial −20 pause site. If both strands advanced, complete conversion of the −20 products to the −1 products should occur. However, the results for both pICLNm and pICLPt showed that a significant number of leading strand products remained paused at the initial −20 position, even after the signal at the −1 position had peaked at 45 minutes (e.g. Figures 2B and 2C, lane 4; Supplementary Figures 4 and 7). This observation suggests that on any given DNA template, only one of the two leading strands initially advances to the ICL. In summary, lesion bypass involves an initial pausing of two leading strands on either side of the cross-link ~20 nts from the lesion (24 nts for pICLNm; 20 nts for pICLPt) before one of the two forks advances to the −1 position. Subsequently, the growing leading strand is extended beyond the lesion, generating the complete 689 nt Afl III restriction fragment.
Notably, as leading strands advanced towards the ICL, there was a loss of lagging strand products, which began between 30 and 45 minutes (e.g. Figure 2B compare lanes 3 and 4; see also Supplementary Figure 4,7 and 8). Since this loss was observed before leading strands were extended past the ICL (at which time they could be ligated to downstream Okazaki fragments), it is best explained by 5' to 3' exonuclease activity, which has previously been reported in Xenopus egg extracts (Toczylowski and Yan, 2006).
Evidence for uncoupling of replicated sister chromatids
Current models for ICL repair hypothesize that dual incisions occur on either side of the ICL on one strand of the parental duplex ("unhooking"), allowing sister chromatid separation and lesion bypass [(Dronkert and Kanaar, 2001; Niedernhofer et al., 2005; Wang, 2007); see Supplementary Figure 1B–C]. To seek evidence of unhooking in our system, replicated pICLNm was digested with Hinc II, which converts the “Figure 8” structure into an 11.2 kb X-shaped species with 2.3 and 3.3 kb arms (Figure 3A). In the presence of a single incision near the ICL, Hinc II is predicted to generate an arm fragment and a Y structure, whereas dual incisions should liberate two arms and a linear 5.6 kb fragment (Figure 3A, bottom).
Figure 3. Detection of incisions near the ICL.
(A) Expected intermediates resulting from single or dual incisions near the ICL (see text). Note that the first incision could also occur to the left of the ICL, giving rise to a short arm and large Y structure. (B) pICLNm replication products were digested with Hinc II, and separated on a native agarose gel (lanes 1–8). Replicated pCtr was digested with Hinc II to generate a 5.6 kb size marker (lane 9, only 20% of the reaction loaded). (C) Advance of the leading strand to the ICL precedes incisions. At each time point in (B), the relative abundance of X-shaped molecules was quantified using a phosphorimager (blue line). At 18 and 22 minutes, before replication was complete, the level was assigned a value of 100. The relative abundance of leading strand products from both forks at the −1 position (Figure 2B and Figure 2D) (grey dashed line) is plotted. The graph shows the average of four independent experiments.
Figure 3B shows that the X shaped structure expected from Hinc II digestion was generated soon after addition of NPE (lane 1). Moreover, a very low level of all the intermediates expected from single incision events was detected. After 1 hour, there was a dramatic reduction in the abundance of the X-shaped molecules, and a concomitant increase in the 5.6 kb linear species (Figure 3B, lanes 6–8). The conversion of X-shaped molecules to linear products was not accompanied by accumulation of arm fragments (Figure 3B, lanes 6–8), as might have been expected (Figure 3A, bottom). We propose that these arms were generated by incision, but are not visible due to a highly active resection activity (see Supplementary Figure 10B,C,F). This interpretation is consistent with the 5' to 3' exonuclease activity detected in Figure 2B, which degrades lagging strand products and which should also attack newly exposed 5' ends generated by incision (as depicted in Figure 10B). It is also consistent with the fact that between ~45 and ~120 minutes after addition of NPE, we commonly observe a 40–70% loss of total replication products (e.g. Figure 4B) as well as a decline in total nascent strand products (Figure 2B, compare lanes 1 and 8). This loss of replication products is linked to repair since it is never observed with pCtr (Figure 2B, compare lanes 8 and 9 and data not shown), and we speculate it may reflect the activity of a 5' to 3' exonuclease that prepares the DNA template for homologous recombination. In summary, our results clearly show that after replication of pICLNm, the two sister chromatids that are initially connected by the ICL are separated, most likely via incisions near the ICL. This leaves a continuous daughter molecule that can serve as the DNA template for lesion bypass.
Figure 4. Replication-dependent repair of an ICL.
(A) An ICL blocks cleavage by Acc I. 15 ng of pICLNm or pCtr was digested with Acc I, separated on a native agarose gel, and stained with SYBR Gold. (B) ICL repair assay. At different times after addition of NPE/32P-α-dATP, pICLNm or pCtr was recovered and equal aliquots were digested with Hinc II (lanes 1–9) or Acc I (lanes 10–18). (Note: lanes 1–9 represent a darker exposure of panel 3B). 20% of the reaction was loaded in lanes 9 and 18. (C) At each time point shown in (B), the repair efficiency was calculated as explained in the text and graphed (Red line). Extension products from the same experiment shown in Figure 4B were plotted for comparison (grey dashed line). (D) pICLNm or pCtr was replicated using NPE lacking radioactivity and optionally supplemented with p27Kip. Plasmid was recovered, digested as indicated, and examined by Southern blotting using pCtr DNA as probe. 20% of the reaction was loaded in lane 13. Samples were supplemented with a 1.2 kb Hind III fragment of pCtr before extraction (loading control).
A leading strand advances to the ICL before there is evidence of incision
We wanted to know when the separation of sister chromatids occurs relative to the events underlying lesion bypass. We thus compared the abundance of the X-shaped species with that of the leading strand intermediates over time and plotted the results (Figure 3C). At 45 minutes, only ~15% of X-shaped molecules had disappeared, presumably due to incisions (Figure 3C, blue line), while the abundance of leading strands at the −1 to −4 position on pICLNm had already peaked (Figure 3C, grey dashed line). Assuming that only one leading strand on each replicated plasmid advances to the ICL, virtually every template in the reaction has undergone this process by 45 minutes. Similar results were observed with pICLPt (data not shown). Therefore, advance of the leading strand to within a few nucleotides of the ICL does not require prior incision.
ICLs are fully repaired in a replication-dependent manner
To determine whether any plasmids are fully repaired during incubation in Xenopus egg extract, we exploited the fact that one of the two Acc I restriction sites in pICLNm coincides with the lesion (Supplementary Figure 2A) and therefore cannot be cleaved by Acc I (Figure 4A, lane 3). However, if the ICL is repaired in an error-free manner, both Acc I sites become accessible, and Acc I digestion should yield two linear DNA restriction fragments (2.3 and 3.3 kb), as seen with pCtr (Figure 4A, lane 2). As shown in Figure 4B, lanes 10–17, incubation of pICLNm in NPE resulted in a gradual increase in the yield of 2.3 and 3.3 kb Acc I restriction fragments, indicative of repair. However, a complication is that even on unrepaired molecules, any incisions near the ICL (shown as red arrows in Figure 4B) would result in 2.3 and 3.3 kb fragments after digestion at the distal Acc I site. To identify the background level of these products, we digested an equal volume of the replicated plasmid with Hinc II, whose single restriction site coincides with the distal Acc I site. As shown in Figure 4B (lanes 1–8), 2.3 and 3.3 kb Hinc II products arose early, then declined after 90 minutes, likely due to degradation. Therefore, to quantify bona fide regeneration of the proximal Acc I site, we subtracted the Hinc II digestion products (lanes 1–8) from the Acc I digestion products (lanes 10–17), and plotted the difference (Figure 4C, red line). This analysis shows that the ICL-proximal Acc I site began to be regenerated about 90 minutes after NPE addition and reached a plateau at 120 minutes, soon after the extension step of lesion bypass (Figure 4C). By 4 hours, ~8% of the DNA molecules that had initially undergone DNA replication were cleaved at both Acc I sites. Given that there is significant degradation of the DNA template during the reaction, the repair efficiency was higher (~15%) if calculated as a percentage of molecules remaining in the reaction at each time point. Similar results were obtained for the pICLPt substrate (see below).
To address whether Acc I cleavage at the ICL is dependent on DNA replication, we inhibited replication initiation by blocking Cdk2/Cyclin E activity with p27Kip (Walter et al., 1998). pICLNm was digested with Acc I or Hinc II as in Figure 4B and visualized by Southern Blotting. Figure 4D shows that when DNA replication was inhibited, no repair products were detected (e.g. compare lanes 3 and 9), suggesting that ICL repair is replication-dependent. An alternative explanation is that the 2.3 and 3.3 kb Acc I restriction fragments reflect the replication of a contaminating, uncross-linked plasmid. However, this is unlikely for three reasons. First, Acc I cleavage was significantly delayed relative to the bulk of DNA replication, which is normally complete within 20–30 minutes of NPE addition. Second, based on quantification of the Southern blot shown in Figure 4D (lanes 7–9 versus 13), less than 0.2% of pICLNm was cleavable by Acc I at the ICL-proximal site in the absence of DNA replication, indicating that at least 99.8% of the molecules were modified at the restriction site. Third, 30 minutes after NPE addition, when DNA replication of pICLNm was essentially complete, only 0.6% of plasmids had undergone decatenation (data not shown), indicating that at least 99.4% of plasmids contained an ICL. Given that we observe between 3% to 8% regeneration of the Acc I site, we conclude that our extracts support bona fide DNA replication-dependent repair of ICLs.
Persistence of an adducted parental strand after lesion bypass
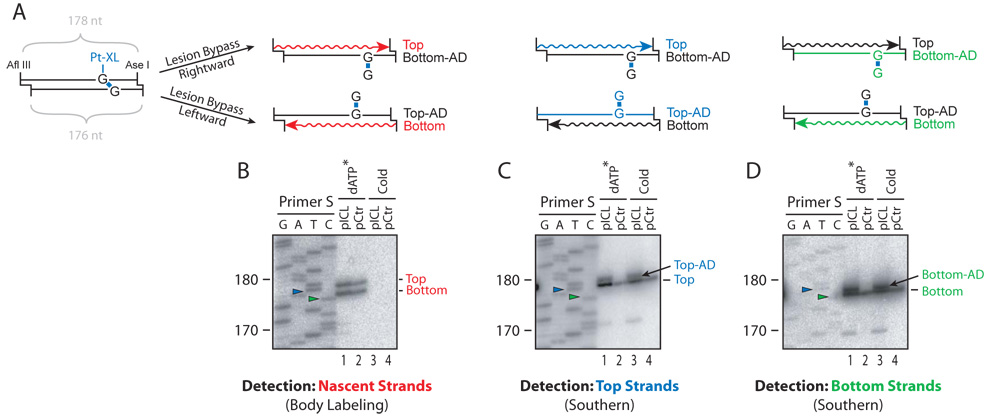
The question arises how the Acc I site is regenerated, since lesion bypass alone should yield an adducted intermediate, which is not expected to be cleavable by Acc I (Supplementary Figure 10C, bottom plasmid). Although NER is not essential for higher eukaryotic cells to survive exposure to ICL agents, it has been proposed to remove the adduct after lesion bypass (see Introduction). To examine whether this occurs in our system (as depicted in Supplementary Figure 10F–G), we used strand specific Southern blotting to monitor the presence of an adduct in the parental strands, which would retard their mobility. To generate size markers for the unadducted strands, pCtr or pICLPt was replicated in the presence of 32P-α-dATP. After 4 hours, the replicated DNA was digested with Afl III and Ase I to cut out a small fragment encompassing the ICL (Figure 5A). As expected, DNA replication generated nascent products of 178 nts (Top Strand) and 176 nts (Bottom Strand) (Figure 5B, lanes 1 and 2). To examine parental strands, the same reaction was performed in the absence of 32P-α-dATP, and Afl III / Ase I digestion products were hybridized with a strand-specific probe complementary to the Top Strand. This analysis revealed a new band migrating roughly one nucleotide above the unadducted Top Strand using pICLPt (Figure 5C, lane 3, Top-AD), but not with pCtr (Figure 5C, lane 4). Because this band was also not seen in the radioactively labeled nascent products (Figure 5B, lanes 1 and 2), it must correspond to a modified parental strand. Similarly, re-probing the stripped blot for the Bottom Strand showed a new band just above the unadducted Bottom Strand (Figure 5D, lane 3, Bottom-AD) We conclude that an adduct (AD) persisted in both parental strands. Since lesion bypass is expected to occur randomly using either the rightward or leftward fork (Figure 5A), 50% of each parental strand should be shifted if no adducts were removed, and this was in general agreement with our observations (Figure 5C and Figure 5D, lane 3). The data suggest that most of the remaining adducts were resistant to repair, although we cannot rule out that a small fraction was removed by NER, giving rise to the fully repaired products detected in Figure 4. Alternatively, fully repaired products may be generated by homologous recombination (see Discussion). The small degree of gel retardation observed in Figure 5 suggests that only a single base or nucleotide, rather than an oligonucleotide, remained attached to the parental strand via the ICL. This is either because the original incisions occurred immediately adjacent to the ICL, or because oligonucleotides generated during unhooking are trimmed by a nuclease (Hazrati et al., 2007).
Figure 5. An adduct persists in the parental strand after lesion bypass.
(A) Cartoon illustrating replication of an Afl III / Ase I restriction fragment harboring a cisplatin ICL. Due to the different overhangs generated by these enzymes, digestion of pICLPt yields Top and Bottom Strands of 178 and 176 nts, respectively. Lesion bypass by the rightward fork yields a radioactively labeled nascent Top Strand and an adducted, parental Bottom Strand (Bottom-AD), while lesion bypass by the leftward fork results in a labeled nascent Bottom Strand and an adducted parental Top Strand (Top-AD). Strand-specific Southern blotting was used to detect either the top strands (blue lines) or the bottom strands (green lines). (B) Detection of nascent strands. pICLPt or pCtr was replicated in the presence (lanes 1, 2) or absence (lanes 3, 4) of 32P-dATP. After 4 hours, replication products were digested with Afl III and Ase I, separated on a 5% denaturing polyacrylamide gel and transferred to a Nylon membrane. Radioactive products were visualized using a phophorimager. (C) Detection of the nascent Top Strand and the adducted parental Top Strand (Top-AD) on the membrane in (B) by Southern blotting using a bottom-strand probe. (D) Detection of the nascent Bottom Strand and the adducted parental Bottom Strand by stripping and reprobing the membrane in (C) using a top-strand probe. Primer S was used to generate a sequencing ladder from pCtr that serves as a size marker (see Figure 2). Green and Blue arrowheads indicate the 176 nt and 178 nt sequencing products, respectively (see Supplementary Figure 5C for sequence information and location of primer S). The migration of the digested DNA replication products is retarded by 1 nucleotide with respect to the sequencing products (See Supplementary Figure 9 for discussion of this effect). Adducted parental strands also persisted on pICLNm (data not shown).
Rev7-depleted extracts are defective for ICL repair
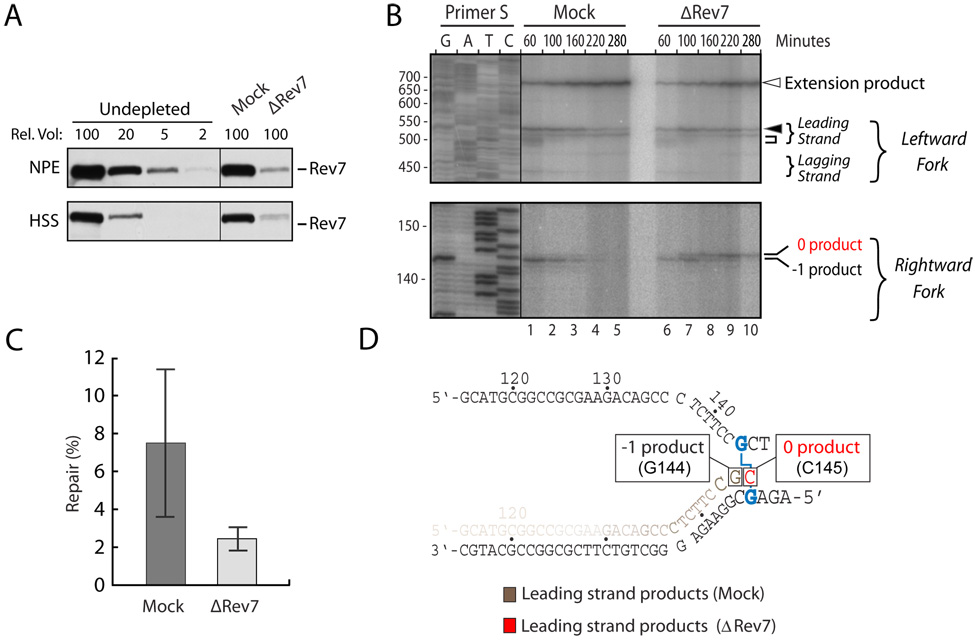
Vertebrate cells deficient for Rev7 or Rev3, the two subunits of DNA pol ζ, are extremely sensitive to cisplatin, suggesting a defect in ICL repair. To determine whether DNA pol ζ participates in ICL repair in our cell-free system, we depleted Rev7 from egg extracts. Immunodepletion with Rev7-specific antibodies removed the vast majority of Rev7 and Rev3 from HSS and NPE (Figure 6A and data not shown). In the Mock-depleted and Rev7-depleted extracts, the leading strand of the rightward fork advanced to within 1 nucleotide of the ICL in pICLPt, and this product then disappeared in both extracts with similar kinetics (Figure 6B, lower panel, "−1 product"). Strikingly, in the Rev7-depleted extract but not in the mock-depleted extract, a new species accumulated (the "0 product") which was exactly 1 nucleotide longer than the "−1 product" (Figure 6B, lower panel). The "0 product" likely reflects translesion synthesis in which a nucleotide is inserted across from the adducted template G (Figure 6D). Build-up of the "0 product" in the absence of Rev7 was observed in four independent experiments (Figure 6B and data not shown). Importantly, we also found that in Rev7-depleted extracts, regeneration of the ICL-proximal restriction site was reduced on average 3-fold relative to the mock-depleted control, demonstrating a defect in the final outcome of ICL repair (Figure 6C). The residual repair may be due to incomplete Rev7 depletion or redundant activities. The accumulation of the "0 product" in Rev7-depleted extract agrees well with the proposed role for Pol ζ in extending leading strands beyond a DNA lesion (Washington et al., 2004). It is therefore unlikely that the repair defect observed in these extracts is caused by co-depletion of unknown lesion bypass activities. Our results suggest a specific role for DNA pol ζ in ICL repair and argue that ICL repair in our cell-free system occurs via a physiological mechanism.
Figure 6. Repair of pICL is defective in Rev7-depleted extracts.
(A) Rev7 immunodepletion. Undepleted, mock-depleted, and Rev7-depleted HSS and NPE were analyzed by Western blotting using Rev7 antibody. A relative volume of 100 corresponds to 0.3 µl extract. (B) Accumulation of a new lesion bypass intermediate in Rev7-depleted extracts. pICLPt was replicated in mock-depleted or Rev7-depleted HSS and NPE (4 ng/µl final DNA concentration). At the indicated times, products were digested with Afl III and analyzed on a sequencing gel (as in Supplementary Figure 7B). Numbers to the left indicate the size of the sequencing products. The new replication intermediate is indicated ("0 product"). (C) The average repair efficiency in Mock- and Rev7-depleted extracts in 4 independent experiments is plotted with error bars. (D) Cartoon depicting the intermediate that accumulates in Rev7- depleted extracts. We infer that a C residue is inserted at position 145, since the translesion step is likely performed by the cytidyl transferase Rev1, and some of the products are digestible with Sap I.
Collision of a DNA replication fork with an ICL leads to checkpoint activation and triggers the Fanconi Anemia pathway
When mammalian cells are treated with agents such as mitomycin C that induce ICLs, FancD2 is ubiquitylated and the ATR checkpoint kinase is activated (Wang, 2007). We therefore examined whether these damage-response pathways are also activated upon replication of purified ICL-containing plasmids. As shown in Supplementary Figure 11A, replication of pICLPt caused a massive induction of Chk1 phosphorylation at serine 344 (Chk1-P; lower panel, lanes 2–6) that was comparable to the level seen in the presence of aphidicolin, which induces helicase uncoupling (lanes 12–13) (Byun et al., 2005). Replication of undamaged plasmid did not induce Chk1 phosphorylation. pICLPt-induced Chk1 phosphorylation was completely dependent on replication initiation (Supplementary Figure 11B, lower panel, lanes 1 and 2). The data indicate that collision of a DNA replication fork with an interstrand cross-link activates the ATR signaling pathway. Replication of pICLPt also converted FancD2 to a slower migrating form (Supplementary Figure 11A, top panel, lanes 2–6), which represents an ubiquitylated species (data not shown). Like ICL repair, FancD2 ubiquitylation was critically dependent on DNA replication (Supplementary Figure 11B). The replication-dependent activation of the Fanconi anemia and ATR pathways by pICL further suggests that repair of these DNA templates in Xenopus egg extracts involves a physiological mechanism.
Discussion
Using Xenopus egg extracts and model DNA templates containing a single, site-specific ICL, we have established an in vitro system that supports ICL repair in S phase. Unlike many studies that use cellular sensitivity to ICL agents to make inferences about the process of ICL repair, our approach directly examines the process of repair using a DNA template that contains no DNA lesions other than ICLs. Using this system, we show that ICL repair is directly coupled to DNA replication. Examination of repair intermediates suggests the following mechanism of ICL repair (Figure 7). Initially, two DNA replication forks converge on the ICL, generating a structure in which the leading strands of each fork stall ~20–40 nts from the ICL, and the 5' ends of the lagging strands are located at a greater and more variable distance from the lesion (Figure 7B). After a ~20 minute delay, lesion bypass is initiated when the leading strand of one fork is extended to within a few nucleotides of the ICL (Figure 7C). After a further ~30 minutes, the two sister chromatids that are joined via the ICL are uncoupled, likely via dual incisions surrounding the ICL (Figure 7D). Concurrent with sister chromatid uncoupling, a nucleotide is incorporated across from the adducted base that formed part of the ICL (Figure 7E), an event termed translesion synthesis. At present, we cannot determine whether plasmid uncoupling or translesion synthesis occurs first. Soon after incision/translesion synthesis, the leading strand is extended beyond the ICL and ligated to the first downstream Okazaki fragment (Figure 7F). Immunodepletion of Rev7 suggests that this extension step is dependent on DNA polymerase ζ. Finally, fully repaired daughter duplexes are formed, as evidenced by Acc I digestion of the DNA sequence underlying the ICL. The Acc I site is likely regenerated by homologous recombination (HR) and/or nucleotide excision repair (NER) (Figure 7G; see also Supplementary Figure 10), although an involvement of base excision repair cannot be ruled out.
Figure 7. Model for ICL repair in Xenopus egg extracts.
See Discussion for details.
The question arises why not more of the input DNA was repaired, since lesion bypass is highly efficient. One likely explanation is that most molecules retain an adduct in the parental strand that precludes digestion by Acc I (Figure 5). This observation suggests that NER is not highly active in our system. The remaining adduct is unlikely to be lethal, since it will not disrupt chromosome segregation. Another potential explanation is that the repair is error-prone, although we disfavor this idea, as explained below. Finally, recombinational repair may be inefficient or aberrant. Indeed, although we observe X-shaped molecules during 2-D gel electrophoresis that are consistent with Holliday junction intermediates (data not shown) and 5' to 3' resection (Figure 2B), the latter process may be so rapid as to destroy many daughter molecules before they can engage in HR (Supplementary Figure 10F, top plasmid).
The mechanism of ICL repair characterized here differs in important ways from current models for this process, which have been inferred largely from genetic data and the biochemical properties of relevant DNA repair enzymes (Dronkert and Kanaar, 2001; Niedernhofer et al., 2005; Niedernhofer et al., 2004; Niedzwiedz et al., 2004; Wang, 2007). Thus, prevailing models of ICL repair envision that an incised parental strand is extended past the DNA lesion (Supplementary Figure 1D), whereas our results clearly show that this reaction involves one of the two nascent strands (Figure 7). This observation reveals how ICL repair is coupled to DNA replication. A potential benefit of using a nascent strand is that replicative polymerases, and possibly translesion DNA polymerases, need not be recruited de novo. Moreover, in current models, lesion bypass can begin only after incision has occurred, since the latter process generates a 3' end within the parental strand that will be extended past the lesion. In contrast, by extending a nascent strand, lesion bypass can begin before incisions. Indeed, our data indicate that after the initial pause at −20, the leading strand advances to within a single nucleotide of the before any incisions occur. We inferred the absence of incisions at early time points from the persistence of the X-shaped molecule generated by Hinc II digestion (Figure 3B), since any incisions near the ICL should cause this structure to collapse (Figure 3A). A potential exception is an incision that occurs close to the ICL, internal to a significant region of duplex DNA, since in this scenario the arm fragment may not be released (Supplementary Figure 12A). However, such an incision would be revealed as soon as the replication fork advances to the incision (Supplementary Figure 12B). We therefore believe that incisions are not made until after the leading strand moves to the −1 position. A beneficial consequence of initiating lesion bypass before incisions have occurred is that repair of one sister chromatid is already underway at the moment of incision. This reduces the lag period between the time that dsDNA breaks are generated and when a fully duplex sister chromatid is available for homology-directed repair of the break.
A second difference is that while current models of ICL repair propose collision of a single replication fork with the lesion, we observe convergence of two forks on the ICL. Based on the following considerations, the latter situation may occur frequently, even in somatic cells. First, since initiations at neighboring origins generally take place concurrently (Berezney et al., 2000), two converging forks will arrive at an ICL contemporaneously if the lesion is located near the midpoint between two origins. Second, if the ICL is located immediately adjacent to one origin, one of the two converging forks will reach the ICL at most 40 minutes before the other [assuming an average inter-origin distance of ~50 kb and a fork speed of ~1.25 kb/min; (Berezney et al. 2000)]. Importantly, repair is quite slow, taking several hours, both in vitro (this paper) and in mammalian cells (Akkari et al., 2000). Moreover, it has been proposed that when one replication fork stalls, new replication forks are rapidly established from nearby, surplus pre-RCs (Ge et al., 2007). Therefore, even when an ICL is located adjacent to an origin, two DNA replication forks may still arrive at the lesion almost contemporaneously. A potential advantage of the dual fork collision mechanism is that there is no need to re-establish a processive replication fork after lesion bypass is complete. This consideration is particularly relevant given the absence of a known replication restart pathway in higher eukaryotes. In some instances, only one fork can collide with the ICL, for example when two such lesions occur between neighboring pre-RCs, but this situation should be rare. In summary, it seems safe to conclude that cells must be able to cope with single and dual collisions of replication forks with ICLs, and future work will be required to determine whether the mechanisms of repair differ in the two situations.
Our results raise important questions about the mechanism of lesion bypass. First, what happens during the 20 minutes between the arrival of the leading strand at position −20 and its further extension towards the ICL? It is possible that parts of the replication fork must be disassembled before the leading strand can be extended. For example, we speculate that the replicative DNA helicase, the MCM2–7 complex (Takahashi et al., 2005), must be removed to allow the leading strand to advance to the ICL. Notably, many nascent products were observed between the initial pause site and the site of the ICL (Figure 2C and supplementary Figure 7C), suggesting that the leading strand was extended in a non-processive fashion. These results indicate that the mode of DNA synthesis changes substantially after the initial pause, possibly due to a reliance on strand-displacement synthesis.
On pICLNm, the leading strand pauses again at the −4 to −1 positions (Figure 2C). We propose that the pauses seen at the −4 to −2 positions result from the difficulty of unwinding DNA around the ICL. The pause at −1, which is also observed on pICLPt, probably reflects the time required to recruit a translesion DNA polymerase, a step that may involve PCNA ubiquitylation (Lehmann et al., 2007). Based on genetic experiments, the dCMP transferase Rev1 is a prime candidate for this polymerase (Niedzwiedz et al., 2004; Simpson and Sale, 2003). Since the nitrogen mustard-like and cisplatin ICLs are formed between G residues, the involvement of Rev1 suggests that the translesion step itself should be largely error-free. Indeed, we infer that some products must be error-free given that they can be digested with restriction enzymes. Genetic experiments suggest that the Rev1 product is subsequently extended by DNA polymerase ζ, a heterodimer of Rev3 and Rev7 (Lehmann et al., 2007). Our results provide strong support for this idea, since we see accumulation of the translesion product when pICLPt is replicated in Rev7-depleted extracts. Importantly, there is generally a gap of several hundred nucleotides between the ICL and the 5' end of the first lagging strand product located downstream. Since DNA polymerase ζ is error-prone, we speculate that there might be a switch back to a replicative DNA polymerase to avoid high mutation rates. Intriguingly, we observed no accumulation of the translesion product when pICLNm was replicated in Rev7-depleted extracts (data not shown). This observation suggests that a nitrogen mustard-like ICL can be handled by other translesion DNA polymerases. It also shows that removing DNA polymerase ζ from our extracts results in highly specific defects.
We have shown that replication of pICL in Xenopus egg extracts leads to activation of the ATR checkpoint pathway. This observation is interesting because replication-dependent ATR activation is normally thought to occur as a result of helicase uncoupling (Byun et al., 2005), which should not be possible in the presence of an ICL. We speculate that the checkpoint is launched on single-stranded DNA that is exposed on the lagging strand template after a fork collides with the ICL. We also found that pICL replication promotes FancD2 ubiquitylation, suggesting that the FA pathway participates in ICL repair. The recently reported replication-independent ubiquitylation of FancD2 by undamaged plasmids in egg extracts likely resulted from the use of very high DNA concentrations (Sobeck et al., 2007). The cell-free system we have developed is ideally suited to elucidate the roles of these DNA damage response pathways, as well as known repair enzymes, in the process of interstrand cross-link repair.
Experimental Procedures
Preparation of pICL
A cisplatin-ICL duplex was prepared (Hofr and Brabec, 2001), purified on a Mono-Q column, and ligated into pSVRLuc (See Supplementary Figure 2). The analysis of the cross-link-containing substrate (restriction digest, real time PCR) confirmed the presence of ICL in ≥ 99% of the plasmids. For pICLNm, Oligos containing a single modified guanosine base, 7-deaza-7- (2,3-dihydroxy-propyl)-guanine (G*), were annealed and processed to generate the ICL depicted in Fig. 1A (TA and ODS, submitted). The cross-linked DNA duplex was ligated into pSVRLuc. About 99% of pICLNm contained an ICL, as judged by AccI restriction digest and quantitative PCR. Both the nitrogen mustard-like and cisplatin ICLs are stable under the neutral reaction conditions employed in this study.
Electron Microscopy
DNA was purified from replication reactions and EM was performed using the cytochrome C drop spreading method (Thresher and Griffith, 1992). The contour length of all DNA molecules was determined using image J software and converted to basepairs using the 200 nm scale bar in each micrograph, assuming 3.4 A° / basepair.
Xenopus Egg Extracts and Replication
DNA replication and preparation of Xenopus egg extracts (NPE and HSS) were as described (Walter et al., 1998). Briefly, plasmid was incubated with HSS for 20 minutes, followed by addition of two volumes of NPE containing α32P-dATP (0.25 µCi/µl, 3000Ci/mmol final).
Unless otherwise indicated, the final plasmid concentration in the reaction was 1.8 ng/µl. To measure DNA replication efficiency (Figure 1C and D), 2 µl aliquots of the reaction were analyzed on native agarose gels (Takahashi et al., 2004). For other applications, 10 µl aliquots of replication reactions were stopped with 90 µl Stop Solution A (1% SDS, 1 mM EDTA, 100 mM Tris pH 8), treated with RNAse A followed by 0.5 µg/µl Proteinase K at 37 °C, followed by phenol/chloroform extraction. DNA was ethanol precipitated in the presence of glycogen (30 µg/µl final) and resuspended in 5–10 µl of 10 mM Tris Buffer pH 7.5.
Nascent Strand Analysis
Extracted replication products were digested with the indicated enzymes followed by addition of 0.5 volumes of Stop Solution B (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol FF). Restriction fragments (3 µl) were separated on 42 cm long, 5% polyacrylamide sequencing gels prepared with Rapidgel-XL in 0.8X GTG Buffer (USB Corporation, Cleveland). Gels were transferred to filter paper, dried, and visualized with a phosphorimager. Sequencing ladders were generated using the Cycle Sequencing kit from USB.
Antibodies
FancD2 antiserum was prepared against residues 1–172 of Xenopus FancD2 and its specificity confirmed using Western blotting. Antibody against full length Xenopus Rev7 protein was prepared using the same procedure. Rev7 was removed from HSS and NPE using three rounds of depletion using the Rev7 antiserum.
Supplementary Material
Supplemental Data for this article include figures, tables, and Supplemental Experimental procedures and can be found with this article online at
Acknowledgements
We are grateful to Alan D'Andrea for advice and encouragement during early stages of this project. This work was supported by NIH grants GM62267 (JCW), GM55390 (TE), and GM31819 (JDG), a Leukemia and Lymphoma Scholar Award (JCW), a Dutch Cancer Society Fellowship (PK), and by grants from the New York State Office of Science and Technology and Academic Research NYSTAR, C040069 (ODS) and the Swiss Cancer League OCS-01413-080-2003 (ODS). We thank Agata Smogorzewska, Ralph Scully, and Alan D'Andrea for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akkari YM, Bateman RL, Reifsteck CA, D'Andrea AD, Olson SB, Grompe M. The 4N cell cycle delay in Fanconi anemia reflects growth arrest in late S phase. Mol Genet Metab. 2001;74:403–412. doi: 10.1006/mgme.2001.3259. [DOI] [PubMed] [Google Scholar]
- Akkari YM, Bateman RL, Reifsteck CA, Olson SB, Grompe M. DNA replication is required To elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol Cell Biol. 2000;20:8283–8289. doi: 10.1128/mcb.20.21.8283-8289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R, Dubey DD, Huberman JA. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–484. doi: 10.1007/s004120050399. [DOI] [PubMed] [Google Scholar]
- Bessho T. Induction of DNA replication-mediated double strand breaks by psoralen DNA interstrand cross-links. J Biol Chem. 2003;278:5250–5254. doi: 10.1074/jbc.M212323200. [DOI] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol. 2000;20:7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat Res. 2001;486:217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazrati A, Ramis-Castelltort M, Sarkar S, Barber LJ, Schofield CJ, Hartley JA, McHugh PJ. Human SNM1A suppresses the DNA repair defects of yeast pso2 mutants. DNA Repair (Amst) 2007 doi: 10.1016/j.dnarep.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Hofr C, Brabec V. Thermal and thermodynamic properties of duplex DNA containing site-specific interstrand cross-link of antitumor cisplatin or its clinically ineffective trans isomer. J Biol Chem. 2001;276:9655–9661. doi: 10.1074/jbc.M010205200. [DOI] [PubMed] [Google Scholar]
- Huang H, Zhu L, Reid BR, Drobny GP, Hopkins PB. Solution structure of a cisplatin-induced DNA interstrand cross-link. Science. 1995;270:1842–1845. doi: 10.1126/science.270.5243.1842. [DOI] [PubMed] [Google Scholar]
- Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Li L, Peterson CA, Lu X, Wei P, Legerski RJ. Interstrand cross-links induce DNA synthesis in damaged and undamaged plasmids in mammalian cell extracts. Mol Cell Biol. 1999;19:5619–5630. doi: 10.1128/mcb.19.8.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2:483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- Mu D, Bessho T, Nechev LV, Chen DJ, Harris TM, Hearst JE, Sancar A. DNA interstrand cross-links induce futile repair synthesis in mammalian cell extracts. Mol Cell Biol. 2000;20:2446–2454. doi: 10.1128/mcb.20.7.2446-2454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Nojima K, Hochegger H, Saberi A, Fukushima T, Kikuchi K, Yoshimura M, Orelli BJ, Bishop DK, Hirano S, Ohzeki M, et al. Multiple repair pathways mediate tolerance to chemotherapeutic cross-linking agents in vertebrate cells. Cancer Res. 2005;65:11704–11711. doi: 10.1158/0008-5472.CAN-05-1214. [DOI] [PubMed] [Google Scholar]
- Rothfuss A, Grompe M. Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol Cell Biol. 2004;24:123–134. doi: 10.1128/MCB.24.1.123-134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharer OD. DNA interstrand crosslinks: natural and drug-induced DNA adducts that induce unique cellular responses. Chembiochem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]
- Simpson LJ, Sale JE. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. Embo J. 2003;22:1654–1664. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobeck A, Stone S, Costanzo V, de Graaf B, Reuter T, de Winter J, Wallisch M, Akkari Y, Olson S, Wang W, et al. Fanconi anemia proteins are required to prevent accumulation of replication-associated DNA double-strand breaks. Mol Cell Biol. 2006;26:425–437. doi: 10.1128/MCB.26.2.425-437.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobeck A, Stone S, Hoatlin ME. DNA structure-induced recruitment and activation of the Fanconi anemia pathway protein FANCD2. Mol Cell Biol. 2007;27:4283–4292. doi: 10.1128/MCB.02196-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi TS, Wigley DB, Walter JC. Pumps, paradoxes and ploughshares: mechanism of the MCM2-7 DNA helicase. Trends Biochem Sci. 2005;30:437–444. doi: 10.1016/j.tibs.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol. 2004;6:991–996. doi: 10.1038/ncb1177. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D'Andrea AD. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- Thresher R, Griffith J. Electron microscopic visualization of DNA and DNA-protein complexes as adjunct to biochemical studies. Methods Enzymol. 1992;211:481–490. doi: 10.1016/0076-6879(92)11026-f. [DOI] [PubMed] [Google Scholar]
- Toczylowski T, Yan H. Mechanistic analysis of a DNA end processing pathway mediated by the Xenopus Werner syndrome protein. J Biol Chem. 2006;281:33198–33205. doi: 10.1074/jbc.M605044200. [DOI] [PubMed] [Google Scholar]
- Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Walter J, Sun L, Newport J. Regulated chromosomal DNA replication in the absence of a nucleus. Mol Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- Washington MT, Minko IG, Johnson RE, Haracska L, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minor-groove N2-guanine adduct by the sequential action of yeast Rev1 and DNA polymerase zeta. Mol Cell Biol. 2004;24:6900–6906. doi: 10.1128/MCB.24.16.6900-6906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data for this article include figures, tables, and Supplemental Experimental procedures and can be found with this article online at