Abstract
In the adult population, glioblastoma multiforme is one of the most common primary brain tumors encountered. Unfortunately, this highly malignant tumor represents over 50% of all types of primary central nervous system gliomas. The vast majority of GBMs develops quite rapidly without clinical, radiological, or morphologic evidence of a less malignant precursor lesion (primary or de novo GBMs), as compared to secondary GBMs that develop slowly by progression from diffuse low-grade astrocytomas. These GBM subtypes must be kept in mind because they may constitute distinct disease entities. Even though they look histologically quite similar, they likely involve different genetic alterations and signaling pathways. Decades of surgical therapy, radiotherapy, and chemotherapy have failed to drastically change survival. Clearly, we do not fully understand this tumor; however, the exciting genetic revolution in glioma research over the past decade is providing a promising outlook for exploring this tumor at the genetic level. Science has begun to elucidate the numerous genetic alterations and critical signaling pathways, and it has opened new exciting areas of research such as glioma stem cell biology and neoangiogenesis. This work has already begun to improve our understanding of GBM cell proliferation, migration, and invasion. Indeed, exciting novel targeted therapies are making their way to clinical trials based on this increased knowledge. This review provides the current understanding of GBM oncogenomics, signaling pathways, and glioma stem cell biology and discusses the potential new therapeutic targets on the horizon.
Keywords: glioblastoma multiforme, GBM, oncogenomics, genetics, signaling pathways
Introduction
In the adult population, glioblastoma multiforme (GBM), is a common and one of the most malignant primary brain tumors, representing up to 50% of all primary brain gliomas.1 The majority of GBMs develops rapidly without clinical, radiological, or morphologic evidence of a less malignant precursor tumor (primary or de novo GBMs) (Table 1). Secondary GBMs develop slowly by progression from low-grade or anaplastic astrocytomas. Even though they look similar histologically, primary and secondary GBMs appear to constitute distinct disease entities—they affect different age groups, clinically progress at different rates, and involve different genetic alterations and signaling pathways (Table 1).2,3
Table 1.
Major genetic alterations in primary and secondary GBM initiation and progression.*
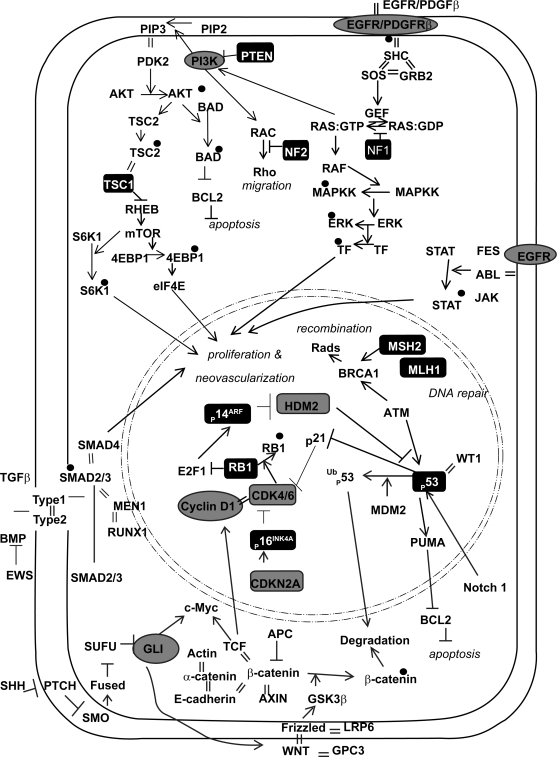
Decades of surgical therapy, radiotherapy, and chemotherapy have failed to drastically change survival for GBM. In a recent meta-analysis of 12 randomized clinical trials, the overall survival rate of patients with malignant gliomas was 40% at one year and only slightly higher (46%) after the addition of adjuvant therapies.4 We do not have a sufficient understanding of this tumor’s biology; however, the recent genetic revolution in glioma research is providing a promising outlook for exploring this tumor at a more in-depth level. Science has now begun to elucidate a protean network of genetic alterations and critical signaling pathways responsible for GBM initiation, migration, and invasion, opening the door for novel, molecular-based, targeted therapies (Fig. 1). Recent exciting work has introduced the potential for therapeutic targeting of glioma stem cells and tumor neoangiogenic pathways. We now realize that many key pathways are in play. Combination therapies directed at multiple sites have now become common in clinical trials. GBM research is making significant strides forward and exciting novel targeted therapies are making their way to clinical trials. There is real hope that the near future will bring improved survivals to patients afflicted with this universally fatal disease.
Figure 1.
Signaling pathways implicated in GBM. Oncogenes are in gray circles. Tumor suppressor genes are in black boxes. Black dots are phosphate groups. Equal signs represent protein-protein interactions.
Oncogenomics and Other Genetic Events
Oncogenomics
Comprehensive genetic screens of GBM have shown that genetic loss is scattered across the entire genome, affecting nearly all chromosomes at frequencies ranging from 2% to 80%. Particularly common regions of loss include areas on 1p, 6q, 9p, 10p, 10q, 13q, 14q, 15q, 17p, 18q, 19q, 22q, and Y.5–10 Loss of heterozygosity (LOH) on chromosome 10 is the most frequent genetic alteration in GBM, occurring in 60%–80% of cases.11 Many GBMs appear to have lost an entire copy of chromosome 10, but several LOH studies have suggested at least three distinct loci to be deleted (e.g., 10p14–p15, 10q23–24, distal to 10q25). This strongly suggests the presence of tumor suppressor genes (TSGs) at these loci.11–13 Most primary GBMs show loss of the entire chromosome, whereas secondary GBMs with LOH show partial loss of 10q.14 Allelic losses on 1p and 7q have also been seen in GBM, but at lower frequencies. Loss of 1p occurs in up to 31% of GBMs8,15,16 and in combination with 19q loss, may indicate a better prognosis and response to therapy.16–18 Loss of 7q is seen in 9% to 12% of GBMs.8,15 Loss of gene dosage from numerous genomic alterations has been documented in GBM, such as entire chromosomal loss, partial chromosomal loss, specific allelic loss, inactivating mutations, and promoter methylation. To date, few specific TSGs have been definitively implicated in GBM. It is assumed that many of these losses represent loss of specific TSGs with direct effects on gliomagenesis; however, some of these losses likely represent the inherent genomic instability that develops in tumor cells.
Gains of gene expression due to genetic alterations at the genomic level have also been demonstrated in GBM in the form of duplication of entire chromosomes, intra-chromosomal amplification of specific alleles, extra-chromosomal amplification (often in the form of double minutes [dmins]), and activating mutations.10,19,20 These forms of increased gene expression (oncogenic) occur much less frequently than losses of gene expression as noted above. Clearly, the most common oncogenic event is amplification of the epidermal growth factor receptor (EGFR) gene on chromosome 7, often in the form of dmins.20,21 A few other genes have been clearly implicated as oncogenes (Table 2 and Fig. 1). This collection of oncogenomic events is daunting and difficult to decipher. Recently developed methods for performing genome-wide screens will greatly help to better define these events with respect to tumor initiation, progression, migration, invasion, and survival in preclinical models.
Table 2.
Genetic alterations in GBM.
| Gene | Chromosome | Mechanism | Frequency | Ref. |
|---|---|---|---|---|
| Multiple | 1p, 6q, 7q, 9p, 10p, 10q, 13q, 14q, 15q, 17p, 18q, 19q, 22q, Y | losses | 2%–75% |
5–7, 9, 123 8, 10, 15, 16 |
| Multiple | 1q, 3q, 4p, 4q, 7p, 7q, 12q, 13q, 19 | gains | up to 80% | 10, 19, 20 |
| CASP8 | 2q33 | promoter methylation | nd | 124 |
| KLF6 | 10p15 | inactivating mutation | 12% | 125 |
| MGMT | 10q26 | methylated | up to 75% | 28, 29, 126 |
| NF1 | 17q11 | inactivating mutation | nd | 38 |
| NF2 | 22q12 | deleted | nd | |
| p16 and p14 | 9p21 | homozygous deletion | up to 50% | 61, 63, 127–129 |
| p16INK4a | 9p21 | deleted, methylated | 40%–60% | 60, 71–74 |
| p14ARF | 9p21 | deleted, methylated | nd | 29 |
| p53 | 17p13 | inactivating mutation | 30%–60% | 60, 127, 130, 131 |
| PTEN | 10q23 | inactivating mutation | 5%–40% | 80, 81, 88, 89, 132 |
| RB1 | 13q14 | inactivating mutation | 30% | 61, 128, 133 |
| RUNX3 | 1p36 | methylated | nd | 134 |
| TES | 7q31 | methylated | nd | 134 |
| TMS1/ASC | 16p11 | methylated | 20% | 135 |
| CDK4 | 12q14 | amplified | nd | 19, 20 |
| CDK6 | 7q21 | amplified | nd | 20 |
| COL4A2 | 13q34 | amplified | nd | 20 |
| CSE1L | 20q13 | amplified | up to 57% | 19 |
| EGRF | 7p21 | amplified (often dmins) | 40%–60% | 19, 21, 22 |
| EGFRvIII | 7p21 | amplified | 20%–30% | 22 |
| ESR | nd | amplified | up to 36% | 19 |
| FGR | 1p35 | amplified | up to 36% | 19 |
| GLI | 12q13 | amplified | nd | 19, 136 |
| MDM2 | 12q15 | amplified | <10% | 19, 66 |
| MYC | 8q24 | amplified | nd | 19 |
| MYCN | 2p24 | amplified | nd | 19 |
| NRAS | 1p13 | amplified | nd | 19 |
| PDGFRA | 4q12 | amplified | up to 65% | 19, 20 |
| PGY1 | 7q21 | amplified | up to 36% | 19 |
| PIK3CA | 3q26 | activating mutation | nd | 136 |
| SLA/LP | 4p15 | amplified | nd | 20 |
| STIM2 | 4p15 | amplified | nd | 20 |
| TNFSF13B | 13q33 | amplified | 8% | 20 |
Many genetic alterations, such as gene amplification or deletion, represent direct glioma-inducing events, whereas others indirectly affect gliomagenesis through processes such as DNA instability. The overall mutation rate in somatic human cells is about 1.4 × 1010 nucleotides per cell division, translating into about one mutant gene per cell during an individual’s lifespan. The low spontaneous mutation rate in normal cells cannot account for the large mutation rate in GBM. In addition to many of the genetic events noted above, gliomagenesis likely involves errors in DNA replication, DNA repair, chromosomal segregation, and alteration of numerous signaling cascades not directly attributed to genomic mutations. This collection of genetic and cellular alterations gives rise to a tumor cell phenotype, previously described as a “mutator phenotype.”23 Central to this mutator phenotype are DNA repair mechanisms. There are at least four major DNA repair pathways that may go awry in GBMs, including nucleotide excision repair, base excision repair, mismatch repair, and direct reversal of lesions in recombination.23,24 As one example, elevated levels of the DNA repair enzyme O6-methylguanine-DNA methyltransferase (MGMT) have been demonstrated in GBM.25 This would confer resistance towards alkylating agents. Its expression also correlates with temozolomide (TMZ) treated GBMs.26 MGMT specifically removes promutagenic alkyl groups from the O6 position of guanine in DNA. Therefore, MGMT protects cells against carcinogenesis induced by alkylating agents. Repair of O6-alkylguanine adducts by tumor cells has been implicated in drug resistance, since it reduces the cytotoxicity of alkylating chemotherapeutic agents.27 Loss of MGMT expression may be caused by methylation of promoter CpG islands, which has been observed in gliomas.28,29 MGMT promoter methylation has been detected in 75% of secondary GBMs, much more than in primary GBMs (36%).29 Interestingly, the majority of low-grade gliomas with MGMT methylation (92%) contain a mutation in the tumor suppressor gene p53, whereas only 39% of those without MGMT methylation carry a p53 mutation.29 Furthermore, G:C to A:T transition mutations at CpG sites are significantly more frequent in low-grade gliomas with MGMT methylation (58%) than in those without (11%).29 This association with increased frequency of p53 mutations suggests that loss of MGMT expression due to promoter methylation frequently occurs at an early stage in the pathway leading to secondary GBMs.
Despite the development of a mutator phenotype and the plethora of genetic alterations it likely entails, there are a discreet number of genetic events and signaling pathways that appear to be central to GBM initiation, migration, invasion, and survival (Fig. 1). Growth factor pathways, such as epidermal and vascular epithelial growth factor receptors (EGFR, VEGFR) play a significant role glioma cell proliferation, migration, and neovascularization. Ras plays a critical intracellular crossroad between numerous pathways, allowing Ras to influence most tumor cell behaviors. P53 and RB1 pathways are often intimately connected in oncology. They both play pivotal roles in regulating cell cycling in response to stimuli for cellular repair or cellular growth. Perhaps due to its homology to proteins that interact with plasma membrane adhesions and the cytoskeletal network, PTEN-related signaling is often implicated in cellular migration in addition to its connection to the Ras cascade.
Signaling Pathways
Growth factor tyrosine kinase receptor pathway
In GBM, aberrant EGFR and other tyrosine kinase receptor autocrine signaling pathways may be the most often cited pathways. These lead to robust alterations in cellular development, proliferation, migration, and tumor-induced neovascularization. Growth factor signaling mediated by ligands like EGF activates an intricately complex network of intracellular cascades modulated by G-protein-coupled receptors and second messengers which converge at multiple sites, one of which is Ras. These clearly play a central role in brain gliomagenesis. EGFR amplification and overexpression occur in 40%–60% of primary GBMs, and rarely in secondary GBMs.2,21,30,31 Amplification of the EGFR gene is often associated with structural alterations in the gene. Seven major mutated variants of EGFR have been identified, the most common being variant III (EGFRvIII), also called de2–7EGFR or ρEGFR which is present in 20%–50% of GBMs with EGFR amplification.32–35 EGFRvIII results from a nonrandom 801-bp inframe deletion of exons 2–7 of the EGFR gene that occurs at the genomic level leading to expression of aberrant transcripts and proteins.34 This mutated protein lacks a portion of the extracellular ligand-binding domain as a result of genomic deletions, resulting in a constitutively autophosphorylated receptor, albeit at a lower level than wild type.36 In addition to enhancing growth, proliferation, migration, and tumor neovascularization, this truncated receptor also confers resistance to chemotherapies such as cisplatin through modulation of Bcl-XL and caspases in cell death pathways.37 Multiple murine glioma models have confirmed the importance of this aberrant growth factor signaling in gliomagenesis.38–40 Platelet derived growth factor receptor (PDGFR) is a similar receptor expressed in most types of gliomas,41 while EGFR is expressed mainly in GBM.42 PDGFR signals through phosphoinositide 3-kinase (PI3K) and phospholipase C gamma (PLC-γ).43
Ras signaling
In sporadic GBMs, specific mutations affecting Ras have not been detected; however, high levels of Ras guanosine triphosphate (GTP) have been documented in cell lines and primary tumors, suggesting that this signaling pathway is activated by upstream factors such as receptor tyrosine kinase activation (e.g. EGFR or PDGFR).44 Another major way of activating this pathway is via the loss of neurofibromin function, the protein product of the large neurofibromatosis 1 (NF1) gene. NF1 has been considered a mutational hot spot in the human genome.38 Germline mutations and loss-of-function mutations have been seen in the disease, which includes optic nerve gliomas, astrocytomas, and GBMs.45–47 NF1 negatively regulates Ras as an exchange factor converting Ras-GTP to Ras-GDP by its GTPase-activating (Ras-GAP) domain.38 Ras-GTP is downstream of growth factor receptors at a major signal transduction crossroad, translating extrinsic messages into the Raf-MAPKK-ERK pathway or into either the PI3K-PKB or the PI3K-Rac-Rho pathway. These influence cell survival and migration. The PI3K-Rac-Rho pathway is involved in cell motility and is negatively regulated by merlin, the protein product of the neurofibromatosis type II (NF2) tumor suppressor gene that links the cytoskeleton to the membrane.48
In mouse models of GBM, neither activated Ras nor Akt alone induce tumors; however, the combination of both activations can.40 Similarly, mouse models deficient in NF1 and p53 develop GBM-like tumors with all the characteristic features, including invasion, neovascularity, necrosis, and atypical astrocytes.49
TGF-β signaling
Overexpression or altered signaling of growth factors and their respective pathways is a common theme in GBM. EGFR, VEGF, PDGR, and TGF have all been implicated in GBM. In response to TGF-β stimulation, two TβRI receptor-associated Smads, Smad2 and Smad3, become phosphorylated and activated. After activation, Smads form a heterodimeric complex with Smad4 and translocate to the nucleus to participate in activation of numerous target genes that contribute to proliferation and neovascularization.50 TGF-β appears to have alternative mechanisms for promoting tumorigenesis, either as a tumor suppressor gene, a mitogen, or an invasion-promoter. TGF-β can function as a tumor suppressor gene in GBM by inhibiting expression of cdks or downregulating cdk activity by inducing cdk inhibitors p15, p27, and Cip/WAF1/p21.51,52 Perhaps more interesting is that it can downregulate cell adhesion proteins, induce an epithelial to mesenchymal transition, and thereby, enhance cell migration and invasion.53–55 TGF-β can alter collagen synthesis, integran expression, cell adhesion to reconstituted basement membrane, and invasiveness in gliomas.56,57 TGF-β1 and TβRII are expressed in GBM and not in normal brain or low-grade gliomas,58 where expression levels are indirectly correlated with survival. TGF-β induces expression of PDGF-A, which may serve as the primary mediator of TGF-β growth stimulatory effects.59 Instead of the standard Smad pathway that results in growth inhibition, TGF-β can deviate upstream of Smad to activate other mitogenic pathways implicated in GBM, including MAPK (Ras-Erk) and SAPK (Rho-JNK, TAK1-p38 kinase). These pathways result in activation of different target genes leading to proliferation and transformation. Compared to the other commonly studied signaling pathways in GBM, TGF-β is a unique therapeutic target in that it may represent an important crosslink to various intracellular processes. Unfortunately, its complex and sometimes opposing actions have made it difficult to clearly understand when and how to target it.
p53-MDM2-p14ARF pathway
The majority of malignant brain tumors, including GBM, demonstrate inactivating mutations in either the p53 and/or retinoblastoma (RB) pathways. 60–63 These two pathways affect numerous cellular functions, but they are most intensely implicated in cell cycling regulation during times of cell repair or cell growth. They interact with each other via p21. P53 is a short-lived transcription factor which is upregulated in response to cellular stress such as radiation exposure, DNA strand breaks, and toxins. It facilitates DNA repair by halting the cell cycle for repair enzymes to work, or if the damage is too great, it induces cell death. As an “apoptostat” protein, it sets a cell’s apoptotic threshold in response to specific endogenous and exogenous insults. Following DNA damage, p53 is activated and induces transcription of genes such as p21Waf1/Cip1.64,65 P53 is stabilized by binding to p14ARF and degraded by MDM2.65 Secondary GBMs have a higher incidence of p53 mutations (>65%), the majority of which are present in prior lower grade biopsies. 2,31 Amplification of MDM2 is present in up to 10% of GBMs, and these all appear to be in primary GBMs that lack p53 mutations.66 Loss of p14ARF expression has often been seen in GBMs (76%), and this correlates with homozygous deletion or promoter methylation of the p14ARF gene.67 There is no difference in the overall frequency of p14ARF alterations between primary and secondary GBMs, but p14ARF promoter methylation is more frequent in secondary GBMs.67 Analysis of multiple biopsies from the same patient reveals that p14ARF methylation is present in up to a third of lower grade astrocytomas, suggesting an early event in secondary GBMs.67
The type and distribution of p53 mutations may differ between GBM subtypes. In secondary GBMs, 57% of mutations are located in two hotspot codons 248 and 273, whereas in primary GBMs, mutations are more equally distributed through all exons.2 Additionally, G:C to A:T mutations at CpG sites, especially in codons 248 and 273, appear to be an early event associated with malignant transformation in the pathway to secondary GBMs. This discrepancy between GBM subtypes is unclear. The less specific pattern of p53 mutations may simply represent increased genomic instability.
RB1-p16INK4a pathway
RB1 controls the transition from G1 into S phase of the cell cycle by inhibiting the action of elongation factor E2F1. The cyclin-dependent kinase 4 (CDK4)/cyclin D1 complex phosphorylates the RB1 protein, thereby increasing release of the E2F1 transcription factor that activates genes involved in the G1 to S transition.68 p16INK4a binds to CDK4, inhibits the CDK4/cyclin D1 complex, and thus inhibits the G1 to S transition.68 Inactivating mutations in RB1 or the upstream factor p16INK4a (also called inhibitor of CDK4a), or activating mutations in the downstream factors CDK4 or cyclin D, cause dysregulated control of E2F1. This leads to the expression of S-phase-related genes and uncontrolled cell cycling. Additionally, it leads to expression of anti-apoptotic genes like Bcl-2, causing uncontrolled cell proliferation.69
The genetic locus INK4a/ARF on chromosome 9p21 produces both p14ARF and p16INK4a by alternative splicing.64 Since p16INK4a negatively regulates CDK4 and p14ARF (p19ARF in mice) inhibits MDM2, blocking rapid ubiquitin-mediated decay of p53, simultaneous inactivation of both genes by a homozygous deletion dysregulates both the RB1 pathway and the p53 pathway. In other words, this single locus drives gliomagenesis. Up to 50% of malignant human gliomas have homozygous deletions that span the reading frame of both genes.63 This is one of the most frequent genetic changes in GBM that is acquired during tumor progression. Most GBMs that retain an intact INK4a/ARF locus display mutations in other genes of the p53 and the RB pathways, leading to unchecked cell cycling and apoptotic resistance. As alluded to above, p16INK4a is an inhibitor of CDK4, blocks CDK4-dependent phosphorylation of the RB protein70 and acts as a negative regulator of cell proliferation. Different mechanisms have been reported for inactivation of the p16INK4a gene in GBM, including homozygous deletion and hypermethylation of the promoter region.71–74 In GBM, frequent promoter hypermethylation has been noted for p14ARF and RB1.75–77
Homozygous p16INK4a deletions are more frequent in primary than in secondary GBMs.67,78 However, there is no major difference in overall frequency of any p16INK4a alteration (including homozygous deletion and promoter methylation) between these GBM subtypes.67 Promoter methylation of the RB1 gene is more frequent in secondary (43%) than in primary GBMs (14%).79 There is a correlation between loss of RB1 expression and promoter methylation of the RB1 gene in GBMs.79 RB1 promoter methylation is not detected in low grade and anaplastic astrocytomas, suggesting that this is a late event in astrocytoma progression.79
PTEN/Akt-1 pathway
Mutations of the TSG phosphatase tensin homology (PTEN) on chromosome 10q23, also called MMAC1 and TEP1, occur frequently in familial developmental and cancer syndromes such as Cowden-Bannayan syndrome and Lhermitte-Duclos disease.80,81 Many of these syndromes include GBM as part of the clinical spectrum. PTEN contains a central catalytic phosphatase core domain that negatively regulates PI3K by dephosphorylating phosphatidylinositol-3,4,5 triphosphate (PIP3) and phosphatidylinositol-3,4 diphosphate (PIP2).82 The N-terminus of PTEN is homologous to the cytoplasmic proteins tensin and auxilin, which interact with actin filaments at focal adhesions and clathrin-coated vesicles. In the case of mutant PTEN, the elevated lipid second messenger PIP3 is used by PI3K to hyperphosphorylate Akt (also known as protein kinase B [PKB]).83 This modulates the activity of proteins that play a critical role in cell survival, invasion, and proliferation.84 The catalytic activity toward phosphoinositide substrates is required for growth suppression, and PTEN-mediated growth inhibition is due to G1 cell cycle block rather than induction of apoptosis.85 The PTEN C2 domain binds phospholipid membranes and mutations in this domain reduce PTEN’s membrane affinity and ability to suppress growth and motility of GBM cells. Alternatively, tumors with an activated PTEN/Akt pathway may be sensitive to mTOR inhibitors, such as rapamycin.86
In GBM, deletions distal to 10q25 (distal to PTEN) cover DMBT1 and FGFR2 loci,87 which suggest that acquisition of the GBM phenotype is associated with loss of other putative TSGs. The PTEN gene at 10q23 is mutated in 5%–40% of GBMs.68,80,86,88,89 PTEN mutations are almost exclusively seen in primary GBMs, but rarely in secondary GBMs.2 PTEN homozygous deletion is rare (<2%).90 Promoter methylation may be an alternative mechanism of loss of PTEN expression, but the significance of PTEN methylation in the evolution of GBMs remains to be determined.91 Nonsense mutations (12%) and deletions or insertions leading to stop codons (32%) appear to be more equally distributed throughout the exons, whereas one-third are missense mutations leading to amino acid changes, preferentially located in exons 1–6, regions homologous to tensin, auxilin, and phosphatases.2,92
Glioma stem cells
The concept of a cancer stem cell is not a recent phenomenon, but has only recently been extended to brain tumors and exploited as worthy of study for therapy. The well-characterized lineage-specific cell surface markers that define the complex hierarchical model of the hematopoietic system, including hematopoietic stem cells, provided the clues for the first elegant studies identifying cancer stem cells in acute myeloid leukemia93 and more recently in gliomas.94–97 Glioma stem cells (GSCs) represent a small fraction of the tumor cell population that are capable of asymmetric cell division into self-renewing GSCs and differentiating daughter cells that can acquire different phenotypes, subsequently losing their multipotent ability. Their capacity for limitless self-renewal and their ability to repopulate and maintain a heterogeneous tumor indicate that GSCs are a promising target for curative therapies. However, significant fundamental questions still remain about these unique GSCs. A cell capable of initiating a glioma may not necessarily be the same cell responsible for long-term glioma self-renewal; therefore, there may exist subpopulations of “GSC initiators” and “GSC propagators.” The cell of origin for GSCs is still debated with evidence supporting different theories.98 These cells may arise from developmentally arrested neural progenitors or from dedifferentiated astrocytes. Heterogeneity within a glioma may not necessarily reflect a stem cell origin of the glioma producing different phenotypes, but could be due to mutations inducing self-renewal properties in progenitor cells with more limited differentiation potential. Clearly, targeting GSCs with therapies poses new dilemmas. Theoretically, a GSC would normally be quiescent, entering the cell cycle only in response to external cues such as growth factors; therefore, quiescent GSCs would unlikely be vulnerable to classic treatments that preferentially target rapidly dividing cells. GSCs represent fundamentally unique glioma cells and as discussed below, they likely depend on genomic alterations and intracellular signaling pathways that differ from non-GSC tumor cells. Therefore, these GSC may contribute to resistance to current therapies as well as future therapies that target genes and proteins that characterize the bulk of the tumor mass. GSCs may also express high levels of drug export proteins, contributing to therapeutic resistance.
Despite the challenges, targeting GSCs is particularly attractive because of the universal pattern of recurrence in these tumors. Neurosurgeons can often remove over 90% of the visible tumor, but the cells that have migrated away from the visible tumor focus will proliferate and lead to the demise of the patient. There is circumstantial evidence that GSC may be related to these migrating glioma cells, so it is reasonable to assume that targeting GSCs will either control migrating cells from the primary focus or limit tumor growth at the secondary focus. Seen throughout all of oncology are the common genetic events and signaling pathways used in cancer cells as well as normal stem cells. Interestingly, neural stem cells have the same tremendous capacity for migration as glioma cells, supporting the idea that some of the migrating glioma cells have GSC properties. If GSCs are found to arise from neural stem cells, then targeting GSC may indeed target the migrating tumor cells and lead to a cure for patients who can tolerate the successful treatment of their primary glioma focus. EGFR overexpression is sufficient to confer migratory potential to neural progenitors supporting the theory that gliomas may originate in progenitors from the subventricular zone (SVZ).99 These cells may rapidly migrate to more favorable areas for tumor proliferation, e.g. factors that support better neoangiogenesis.
Targeting GSC will likely have to rely on genomic or proteomic characteristics that differ from other glioma tumor cells. GSCs express immature antigens specific for neural stem and progenitor cells, including the neurofilament protein nestin, the glycoprotein CD133 (a.k.a. prominin-1), Musashi-1 and BMI-1.94–96 These markers suggest the activation of similar developmentally regulated pathways.100 CD133 has been the most prevalent marker for isolating GSCs from GBM.97,101 CD133+ cells comprise 5%–30% of the tumor cell population, and as few as 100 cells can reproduce tumors in animal models.101 A marker of multipotent stem cells in blood and other tissues, much work remains to elucidate the role of CD133 in GSCs. Sonic hedgehog (SHH) and Notch (Fig. 1) are key regulators of neural progenitors in development and have also been found to be altered or overexpressed in GSCs.102 SHH has been best studied in the malignant brain tumor, medulloblastoma. However, some evidence supports its role in GBM GSCs as well.103 SHH is a critical mitogen for medulloblastoma precursor cells. Hereditary loss of function mutations in the SHH receptor Patched (PTCH) lead to constitutive activation of the SHH pathway and predisposition to medulloblastoma in Gorlin syndrome. The SHH pathway is intimately connected to cell cycling since it inactivates RB1, facilitating the over-expression of cell cycle regulators such as N-myc. Notch activation induces expression of downstream target genes, such as p53, and promotes neural stem cell growth.104 It also is better studied in medulloblastoma where activating mutations lead to stem-like cells, but it has been implicated in GSCs as well. Clearly implicated in gliomagenesis, EGFR expression also plays a role in neural progenitor development and is speculated to contribute directly to GSC maintenance.
Olig2 is expressed in myelinating oligodendroglia, but is also in mitogen-treated “transit-amplifying cells” of the SVZ, the presumed site of most neural progenitor cells and a theorized site for the origin of gliomas.105 Olig2 and Olig1 are basic-helix-loop-helix transcription factors expressed in neural and oligodendrocyte precursors. Olig2 is almost universally found in NG2-positive glia and required for development of these cells.106 NG2 is a Chondroitin sulfate proteoglycan that is thought to be another marker of oligodendrocyte progenitor cells. Olig2 sustains a replication-competent state of neural progenitors107 via suppression of p21.108 There is much work to be done, but Olig2 and NG2 may be important markers and/or targets for oligodendroglioma stem cells.
There is some interaction between genes known to regulate stem cell proliferation and genetic lesions in malignant gliomas. Some of the most common lesions in malignant gliomas are loss-of-function mutations in p16Ink4a and p19ARF negative regulators of the RB signaling pathway. Gain-of-function mutations in CDK4 are seen, in GBM, activating the RB pathway.109 Bmi1, a promoter of neural stem cell self-renewal and neural development, is expressed in most gliomas and promotes malignancy in p16Ink4a/p19ARF double-null murine gliomas. 110 As already described, Olig2, a marker of oligodendroglia precursors, is expressed in 100% adult gliomas111 and 100% of CD133 + glioma stem cells. Olig2 is required for tumor formation in p16Ink4a/p19ARF double null murine gliomas. Recent studies have confirmed the extensively studied role of members of the TGF-β superfamily in morphogenesis and specifically brain development.112 As described above, TGF-β may play a significant role in GBM development as well.
Studies such as these strongly support the progenitor cell of origin theory for GSCs. It appears that there may be common genetic gatekeepers, such as Olig2 and SHH, for neurodevelopment and glioma formation. Developing targeted therapies may be developed to kill GSCs, but may also require sophisticated engineering in order to avoid harming normal stem cells. GSC may provide the clues to resistance of this deadly tumor to current therapies. Despite being sensitive to TMZ, the most common chemotherapy in GBM, CD133 + GSCs appear to be quite resistance to radiation therapy.113,114
Summary and future hope
Perhaps the greatest mystery in understanding the oncogenomics of GBM is deciding how and when these genetic alterations and signaling cascades interact. Even though only a few specific pathways are consistently highlighted, there are undoubtedly complex interactions among them as well as with additional unknown players. Clearly, many if not most of these genetic events described do not occur in isolation. For example, LOH 10q is not only the most frequent genetic alteration, but typically occurs in the context of other genetic alterations.2 This suggests that LOH of 10q plus other genetic events create a genomic environment that collectively contributes to the development of a majority of GBMs. Mutations of p53, p16INK4a deletion, EGFR amplification, and PTEN mutations show inverse associations with each other, except for a positive correlation between p16INK4a deletion and EGFR amplification.2,115,116 Once a thorough road map for GBM oncogenomics is designed, it should aid in the development of more rational and more effective targeted therapies. Indeed, it is highly plausible that a cocktail of therapies targeting the major intersections in the roadmap for each individual GBM will be required to effectively slow or halt this tumor.
The lack of significant progress in GBM therapy is likely multifactorial, including a lack of oncogenomic understanding, tremendous multi-drug resistance, radioresistance, insufficient preclinical models, and a tenacious blood-brain barrier. Gene expression profiles and preclinical studies have elucidated many of the oncogenomic changes, but basic questions such as the GSC origin and the hierarchy of the oncogenomic events still remain elusive.
The current standard of care for GBM patients begins with gross total resection if possible, followed by concomitant radiotherapy (typically 60 Gy in 2-Gy fractions) and TMZ chemotherapy (75 mg/m2 for 42 days). This adjuvant therapy is repeated in a 5/28-day schedule for 5 cycles (150 mg/m2) if tolerated. Surgery and radiotherapy have been mainstays in GBM treatment for decades with little modification. TMZ, an alkylating agent, was only recently established in a randomized clinical trial as the standard of care chemotherapy, demonstrating a slight increase in overall survival from 12 to 15 months.117
Our current exploration of aberrant molecular and cytogenetic pathways involved in GBM has led to numerous clinical trials testing more specific, molecular-based, therapies for this tumor. EGFR has been an attractive target, especially since it can trigger several downstream signaling pathways (PI3K/AKT/mTOR, Ras/Raf/MAPK, and PKC) and is abnormally activated in some manner in up to 60% of GBMs and 70% of all solid cancers. Targeting strategies have included monoclonal antibodies, bispecific antibodies, toxin-linked conjugates, vaccines, and small-molecule tyrosine kinase inhibitors (TKIs). Phase II clinical trials with EGFR TKIs (ZD-1839, gefitinib; OSI-774, erlotinib) demonstrated some responses, but overall minimal improvement over historical controls in unselected patients.118,119 Because of the activation of mTOR due to the frequent loss of PTEN in GBM, mTOR inhibitors have also become exciting tools. The mTOR inhibitor CCI-779 (ester of the immunosuppressive agent sirolimus [rapamycin]) did not show efficacy as monotherapy, but may be applicable to combination therapies.120 CCI-779 binds FKBP-12 to form a complex that inhibits mTOR, resulting in cell-cycle arrest. The matrix metalloproteinase inhibitor marimastat has been combined with TMZ and demonstrated increased progression-free survival in GBM patients.
These examples of molecular-based targeted therapies being tested in clinical trials represent a new era in GBM therapeutics that bring hope to those afflicted with this fatal disease. With this enthusiasm for testing therapies based on specific GBM biology, we are entering an exciting era in dealing with this deadly tumor. Numerous other targeted therapies have gone through preclinical testing.121 Unfortunately, we are still awaiting any major breakthroughs in clinical trials. Further unraveling of the critical genetic alterations that lead to the initiation and progression of GBM may be necessary for future therapies to work.
Acknowledgments
We would like to acknowledge the terrific editing skills of Janet Parsons in the Department of Pathology, Duke University.
Footnotes
Disclosure
The authors report no conflicts of interest.
References
- 1.CBTRUS. Statistical Report: Primary Brain Tumors in the United States. 1998–2002:2005. Central Brain Tumor Registry of the United States. [Google Scholar]
- 2.Ohgaki H, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64(19):6892–9. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 3.Kleihues P, Cavenee WK. WHO classification of tumors Pathology and genetics of tumors of the nervous system. Lyon: IARCpress; 2000. [DOI] [PubMed] [Google Scholar]
- 4.Stewart LA. Chemotherapy in adult high-grade glioma: a systematic review and meta-analysis of individual patient data from 12 randomised trials. Lancet. 2002;359(9311):1011–8. doi: 10.1016/s0140-6736(02)08091-1. [DOI] [PubMed] [Google Scholar]
- 5.Kim DH, et al. Chromosomal abnormalities in glioblastoma multiforme tumors and glioma cell lines detected by comparative genomic hybridization. Int J Cancer. 1995;60(6):812–9. doi: 10.1002/ijc.2910600615. [DOI] [PubMed] [Google Scholar]
- 6.Mohapatra G, et al. Genetic analysis of glioblastoma multiforme provides evidence for subgroups within the grade. Genes Chromosomes Cancer. 1998;21(3):195–206. [PubMed] [Google Scholar]
- 7.Nishizaki T, et al. Investigation of genetic alterations associated with the grade of astrocytic tumor by comparative genomic hybridization. Chromosomes Cancer. 1998;21(4):340–6. doi: 10.1002/(sici)1098-2264(199804)21:4<340::aid-gcc8>3.0.co;2-z. Genes. [DOI] [PubMed] [Google Scholar]
- 8.von Deimling A, et al. Comprehensive allelotype and genetic anaysis of 466 human nervous system tumors. J Neuropathol Exp Neurol. 2000;59(6):544–58. doi: 10.1093/jnen/59.6.544. [DOI] [PubMed] [Google Scholar]
- 9.Collins VP. Brain tumours: classification and genes. J Neurol Neurosurg Psychiatry. 2004;75(Suppl 2):ii2–11. doi: 10.1136/jnnp.2004.040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vranova V, et al. Screening of genomic imbalances in glioblastoma multiforme using high-resolution comparative genomic hybridization. Oncol Rep. 2007;17(2):457–64. [PubMed] [Google Scholar]
- 11.Rasheed BK, et al. Chromosome 10 deletion mapping in human gliomas: a common deletion region in 10q25. Oncogene. 1995;10(11):2243–6. [PubMed] [Google Scholar]
- 12.Karlbom AE, et al. Loss of heterozygosity in malignant gliomas involves at least three distinct regions on chromosome 10. Hum Genet. 1993;92(2):169–74. doi: 10.1007/BF00219686. [DOI] [PubMed] [Google Scholar]
- 13.Ichimura K, et al. Distinct patterns of deletion on 10p and 10q suggest involvement of multiple tumor suppressor genes in the development of astrocytic gliomas of different malignancy grades. Genes Chromosomes Cancer. 1998;22(1):9–15. doi: 10.1002/(sici)1098-2264(199805)22:1<9::aid-gcc2>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 14.Fujisawa H, et al. Loss of heterozygosity on chromosome 10 is more extensive in primary (de novo) than in secondary glioblastomas. Lab Invest. 2000;80(1):65–72. doi: 10.1038/labinvest.3780009. [DOI] [PubMed] [Google Scholar]
- 15.Wooten EC, et al. A study of loss of heterozygosity at 70 loci in anaplastic astrocytoma and glioblastoma multiforme with implications for tumor evolution. Neuro Oncol. 1999;1(3):169–76. doi: 10.1093/neuonc/1.3.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt MC, et al. Impact of genotype and morphology on the prognosis of glioblastoma. J Neuropathol Exp Neurol. 2002;61(4):321–8. doi: 10.1093/jnen/61.4.321. [DOI] [PubMed] [Google Scholar]
- 17.Ino Y, et al. Long survival and therapeutic responses in patients with histologically disparate high-grade gliomas demonstrating chromosome 1p loss. J Neurosurg. 2000;92(6):983–90. doi: 10.3171/jns.2000.92.6.0983. [DOI] [PubMed] [Google Scholar]
- 18.Hill C, Hunter SB, Brat DJ. Genetic markers in glioblastoma: prognostic significance and future therapeutic implications. Adv Anat Pathol. 2003;10(4):212–7. doi: 10.1097/00125480-200307000-00004. [DOI] [PubMed] [Google Scholar]
- 19.Hui AB, et al. Detection of multiple gene amplifications in glioblastoma multiforme using array-based comparative genomic hybridization. Lab Invest. 2001;81(5):717–23. doi: 10.1038/labinvest.3780280. [DOI] [PubMed] [Google Scholar]
- 20.Ruano Y, et al. Identification of novel candidate target genes in amplicons of Glioblastoma multiforme tumors detected by expression and CGH microarray profiling. Mol Cancer. 2006;5:39. doi: 10.1186/1476-4598-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong AJ, et al. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci U S A. 1987;84(19):6899–903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinojima N, et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63(20):6962–70. [PubMed] [Google Scholar]
- 23.Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51(12):3075–9. [PubMed] [Google Scholar]
- 24.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411(6835):366–74. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 25.Silber JR, et al. O6-methylguanine-DNA methyltransferase activity in adult gliomas: relation to patient and tumor characteristics. Cancer Res. 1998;58(5):1068–73. [PubMed] [Google Scholar]
- 26.Friedman HS, McLendon RE, Kerby T, et al. DNA mismatch repair and O6-alkylguanine-DNA alkyltransferase analysis and response to Temodal in newly diagnosed malignant glioma. J Clin Oncol. 1998;16:3851–3857. doi: 10.1200/JCO.1998.16.12.3851. [DOI] [PubMed] [Google Scholar]
- 27.Belanich M, et al. Retrospective study of the correlation between the DNA repair protein alkyltransferase and survival of brain tumor patients treated with carmustine. Cancer Res. 1996;56(4):783–8. [PubMed] [Google Scholar]
- 28.Watts GS, et al. Methylation of discrete regions of the O6-methylguanine DNA methyltransferase (MGMT) CpG island is associated with heterochromatinization of the MGMT transcription start site and silencing of the gene. Mol Cell Biol. 1997;17(9):5612–9. doi: 10.1128/mcb.17.9.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura M, et al. Promoter methylation of the DNA repair gene MGMT in astrocytomas is frequently associated with G:C→A:T mutations of the TP53 tumor suppressor gene. Carcinogenesis. 2001;22(10):1715–9. doi: 10.1093/carcin/22.10.1715. [DOI] [PubMed] [Google Scholar]
- 30.Ekstrand AJ, et al. Amplified and rearranged epidermal growth factor receptor genes in human glioblastomas reveal deletions of sequences encoding portions of the N- and/or C-terminal tails. Proc Natl Acad Sci U S A. 1992;89(10):4309–13. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe K, et al. Overexpression of the EGF receptor and p53 mutations are mutually exclusive in the evolution of primary and secondary glioblastomas. Brain Pathol. 1996;6(3):217–23. doi: 10.1111/j.1750-3639.1996.tb00848.x. discussion 23–4. [DOI] [PubMed] [Google Scholar]
- 32.Rasheed BK, et al. Molecular pathogenesis of malignant gliomas. Curr Opin Oncol. 1999;11(3):162–7. doi: 10.1097/00001622-199905000-00004. [DOI] [PubMed] [Google Scholar]
- 33.Wikstrand CJ, et al. The class III variant of the epidermal growth factor receptor (EGFRvIII): characterization and utilization as an immunotherapeutic target. J Neurovirol. 1998;4(2):148–58. doi: 10.3109/13550289809114515. [DOI] [PubMed] [Google Scholar]
- 34.Sugawa N, et al. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci U S A. 1990;87(21):8602–6. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwechheimer K, Huang S, Cavenee WK. EGFR gene amplification—rearrangement in human glioblastomas. Int J Cancer. 1995;62(2):145–8. doi: 10.1002/ijc.2910620206. [DOI] [PubMed] [Google Scholar]
- 36.Ekstrand AJ, et al. Genes for epidermal growth factor receptor, transforming growth factor alpha, and epidermal growth factor and their expression in human gliomas in vivo. Cancer Res. 1991;51(8):2164–72. [PubMed] [Google Scholar]
- 37.Nagane M, et al. Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-XL and caspase-3-like proteases. Proc Natl Acad Sci U S A. 1998;95(10):5724–9. doi: 10.1073/pnas.95.10.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cichowski K, Jacks T. NF1 tumor suppressor gene function: narrowing the GAP. Cell. 2001;104(4):593–604. doi: 10.1016/s0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- 39.Ding H, et al. Astrocyte-specific expression of activated p21-ras results in malignant astrocytoma formation in a transgenic mouse model of human gliomas. Cancer Res. 2001;61(9):3826–36. [PubMed] [Google Scholar]
- 40.Holland EC, et al. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet. 2000;25(1):55–7. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- 41.Hermanson M, et al. Platelet-derived growth factor and its receptors in human glioma tissue: expression of messenger RNA and protein suggests the presence of autocrine and paracrine loops. Cancer Res. 1992;52(11):3213–9. [PubMed] [Google Scholar]
- 42.Libermann TA, et al. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313(5998):144–7. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 43.Ronnstrand L, Heldin CH. Mechanisms of platelet-derived growth factor-induced chemotaxis. Int J Cancer. 2001;91(6):757–62. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1136>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 44.Guha A, et al. Proliferation of human malignant astrocytomas is dependent on Ras activation. Oncogene. 1997;15(23):2755–65. doi: 10.1038/sj.onc.1201455. [DOI] [PubMed] [Google Scholar]
- 45.Wallace MR, et al. Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science. 1990;249(4965):181–6. doi: 10.1126/science.2134734. [DOI] [PubMed] [Google Scholar]
- 46.Viskochil D, et al. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell. 1990;62(1):187–92. doi: 10.1016/0092-8674(90)90252-a. [DOI] [PubMed] [Google Scholar]
- 47.Cawthon RM, et al. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell. 1990;62(1):193–201. doi: 10.1016/0092-8674(90)90253-b. [DOI] [PubMed] [Google Scholar]
- 48.Shaw RJ, et al. The Nf2 tumor suppressor, merlin, functions in Racdependent signaling. Dev Cell. 2001;1(1):63–72. doi: 10.1016/s1534-5807(01)00009-0. [DOI] [PubMed] [Google Scholar]
- 49.Reilly KM, et al. Nf1; Trp53 mutant mice develop glioblastoma with evidence of strain-specific effects. Nat Genet. 2000;26(1):109–13. doi: 10.1038/79075. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, Musci T, Derynck R. The tumor suppressor Smad4/DPC 4 as a central mediator of Smad function. Curr Biol. 1997;7(4):270–6. doi: 10.1016/s0960-9822(06)00123-0. [DOI] [PubMed] [Google Scholar]
- 51.Johnson MD, et al. Transforming growth factor-beta in neural embryogenesis and neoplasia. Hum Pathol. 1993;24(5):457–62. doi: 10.1016/0046-8177(93)90156-b. [DOI] [PubMed] [Google Scholar]
- 52.Matsuura I, et al. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430(6996):226–31. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- 53.Cui W, et al. TGFbeta1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell. 1996;86(4):531–42. doi: 10.1016/s0092-8674(00)80127-0. [DOI] [PubMed] [Google Scholar]
- 54.Oft M, et al. TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 1996;10(19):2462–77. doi: 10.1101/gad.10.19.2462. [DOI] [PubMed] [Google Scholar]
- 55.Yin JJ, et al. TGF-beta signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. J Clin Invest. 1999;103(2):197–206. doi: 10.1172/JCI3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Merzak A, et al. Control of human glioma cell growth, migration and invasion in vitro by transforming growth factor beta 1. Br J Cancer. 1994;70(2):199–203. doi: 10.1038/bjc.1994.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paulus W, et al. Effects of transforming growth factor-beta 1 on collagen synthesis, integrin expression, adhesion and invasion of glioma cells. J Neuropathol Exp Neurol. 1995;54(2):236–44. doi: 10.1097/00005072-199503000-00010. [DOI] [PubMed] [Google Scholar]
- 58.Yamada N, et al. Enhanced expression of transforming growth factorbeta and its type-I and type-II receptors in human glioblastoma. Int J Cancer. 1995;62(4):386–92. doi: 10.1002/ijc.2910620405. [DOI] [PubMed] [Google Scholar]
- 59.Jennings MT, et al. Transforming growth factor beta as a potential tumor progression factor among hyperdiploid glioblastoma cultures: evidence for the role of platelet-derived growth factor. J Neurooncol. 1997;31(3):233–54. doi: 10.1023/a:1005767616500. [DOI] [PubMed] [Google Scholar]
- 60.Fulci G, et al. p53 gene mutation and ink4a-arf deletion appear to be two mutually exclusive events in human glioblastoma. Oncogene. 2000;19(33):3816–22. doi: 10.1038/sj.onc.1203700. [DOI] [PubMed] [Google Scholar]
- 61.He J, Olson JJ, James CD. Lack of p16INK4 or retinoblastoma protein (pRb), or amplification-associated overexpression of cdk4 is observed in distinct subsets of malignant glial tumors and cell lines. Cancer Res. 1995;55(21):4833–6. [PubMed] [Google Scholar]
- 62.Ishii N, et al. Frequent co-alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathol. 1999;9(3):469–79. doi: 10.1111/j.1750-3639.1999.tb00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Labuhn M, et al. Quantitative real-time PCR does not show selective targeting of p14(ARF) but concomitant inactivation of both p16 (INK4A) and p14(ARF) in 105 human primary gliomas. Oncogene. 2001;20(9):1103–9. doi: 10.1038/sj.onc.1204197. [DOI] [PubMed] [Google Scholar]
- 64.Sherr CJ, Weber JD. The ARF/p53 pathway. Curr Opin Genet Dev. 2000;10(1):94–9. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 65.Stott FJ, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. Embo J. 1998;17(17):5001–14. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reifenberger G, et al. Amplification and overexpression of the MDM2 gene in a subset of human malignant gliomas without p53 mutations. Cancer Res. 1993;53(12):2736–9. [PubMed] [Google Scholar]
- 67.Nakamura M, et al. p14ARF deletion and methylation in genetic pathways to glioblastomas. Brain Pathol. 2001;11(2):159–68. doi: 10.1111/j.1750-3639.2001.tb00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13(12):1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 69.Gomez-Manzano C, et al. Transfer of E2F-1 to human glioma cells results in transcriptional up-regulation of Bcl-2. Cancer Res. 2001;61(18):6693–7. [PubMed] [Google Scholar]
- 70.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366(6456):704–7. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 71.Park SH, et al. 5’CpG island methylation of p16 is associated with absence of p16 expression in glioblastomas. J Korean Med Sci. 2000;15(5):555–9. doi: 10.3346/jkms.2000.15.5.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lang FF, et al. Pathways leading to glioblastoma multiforme: a molecular analysis of genetic alterations in 65 astrocytic tumors. J Neurosurg. 1994;81(3):427–36. doi: 10.3171/jns.1994.81.3.0427. [DOI] [PubMed] [Google Scholar]
- 73.Ohgaki H, et al. Genetic alterations associated with the evolution and progression of astrocytic brain tumours. Virchows Arch. 1995;427(2):113–8. doi: 10.1007/BF00196514. [DOI] [PubMed] [Google Scholar]
- 74.Cheng Y, et al. Molecular analysis of microdissected de novo glioblastomas and paired astrocytic tumors. J Neuropathol Exp Neurol. 1999;58(2):120–8. doi: 10.1097/00005072-199902000-00002. [DOI] [PubMed] [Google Scholar]
- 75.Costello JF, et al. Silencing of p16/CDKN2 expression in human gliomas by methylation and chromatin condensation. Cancer Res. 1996;56(10):2405–10. [PubMed] [Google Scholar]
- 76.Watanabe T, et al. Methylation of the p73 gene in gliomas. Acta Neuropathol (Berl) 2002;104(4):357–62. doi: 10.1007/s00401-002-0549-1. [DOI] [PubMed] [Google Scholar]
- 77.Gonzalez-Gomez P, et al. CpG island methylation status and mutation analysis of the RB1 gene essential promoter region and protein-binding pocket domain in nervous system tumours. Br J Cancer. 2003;88(1):109–14. doi: 10.1038/sj.bjc.6600737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Biernat W, et al. Alterations of cell cycle regulatory genes in primary (de novo) and secondary glioblastomas. Acta Neuropathol (Berl) 1997;94(4):303–9. doi: 10.1007/s004010050711. [DOI] [PubMed] [Google Scholar]
- 79.Nakamura M, et al. Promoter hypermethylation of the RB1 gene in glioblastomas. Lab Invest. 2001;81(1):77–82. doi: 10.1038/labinvest.3780213. [DOI] [PubMed] [Google Scholar]
- 80.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 81.Steck PA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15(4):356–62. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 82.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273(22):13375–8. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 83.Maehama T, Dixon JE. PTEN: a tumour suppressor that functions as a phospholipid phosphatase. Trends Cell Biol. 1999;9(4):125–8. doi: 10.1016/s0962-8924(99)01519-6. [DOI] [PubMed] [Google Scholar]
- 84.Maier D, et al. The PTEN lipid phosphatase domain is not required to inhibit invasion of glioma cells. Cancer Res. 1999;59(21):5479–82. [PubMed] [Google Scholar]
- 85.Furnari FB, Huang HJ, Cavenee WK. The phosphoinositol phosphatase activity of PTEN mediates a serum-sensitive G1 growth arrest in glioma cells. Cancer Res. 1998;58(22):5002–8. [PubMed] [Google Scholar]
- 86.Vogt PK. PI 3-kinase, mTOR, protein synthesis and cancer. Trends Mol Med. 2001;7(11):482–4. doi: 10.1016/s1471-4914(01)02161-x. [DOI] [PubMed] [Google Scholar]
- 87.Fujisawa H, et al. Acquisition of the glioblastoma phenotype during astrocytoma progression is associated with loss of heterozygosity on 10q25-qter. Am J Pathol. 1999;155(2):387–94. doi: 10.1016/S0002-9440(10)65135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dahia PL. PTEN, a unique tumor suppressor gene. Endocr Relat Cancer. 2000;7(2):115–29. doi: 10.1677/erc.0.0070115. [DOI] [PubMed] [Google Scholar]
- 89.Knobbe CB, Merlo A, Reifenberger G. Pten signaling in gliomas. Neuro-oncol. 2002;4(3):196–211. [PMC free article] [PubMed] [Google Scholar]
- 90.Knobbe CB, Reifenberger G. Genetic alterations and aberrant expression of genes related to the phosphatidyl-inositol-3′-kinase/protein kinase B (Akt) signal transduction pathway in glioblastomas. Brain Pathol. 2003;13(4):507–18. doi: 10.1111/j.1750-3639.2003.tb00481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baeza N, et al. PTEN methylation and expression in glioblastomas. Acta Neuropathol (Berl) 2003;106(5):479–85. doi: 10.1007/s00401-003-0748-4. [DOI] [PubMed] [Google Scholar]
- 92.Broderick DK, et al. Mutations of PIK3CA in anaplastic oligodendrogliomas, high-grade astrocytomas, and medulloblastomas. Cancer Res. 2004;64(15):5048–50. doi: 10.1158/0008-5472.CAN-04-1170. [DOI] [PubMed] [Google Scholar]
- 93.Baum CM, et al. Isolation of a candidate human hematopoietic stemcell population. Proc Natl Acad Sci U S A. 1992;89(7):2804–8. doi: 10.1073/pnas.89.7.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Galli R, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64(19):7011–21. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 95.Hemmati HD, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100(25):15178–83. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ignatova TN, et al. Human cortical glial tumors contain neural stemlike cells expressing astroglial and neuronal markers in vitro. Glia. 2002;39(3):193–206. doi: 10.1002/glia.10094. [DOI] [PubMed] [Google Scholar]
- 97.Singh SK, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–8. [PubMed] [Google Scholar]
- 98.Stiles CD, Rowitch DH. Glioma stem cells: a midterm exam. Neuron. 2008;58(6):832–46. doi: 10.1016/j.neuron.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 99.Aguirre A, et al. Overexpression of the epidermal growth factor receptor confers migratory properties to nonmigratory postnatal neural progenitors. J Neurosci. 2005;25(48):11092–106. doi: 10.1523/JNEUROSCI.2981-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3(12):895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 101.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 102.Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353(8):811–22. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 103.Clement V, et al. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17(2):165–72. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Androutsellis-Theotokis A, et al. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442(7104):823–6. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- 105.Jackson EL, et al. PDGFR alpha-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron. 2006;51(2):187–99. doi: 10.1016/j.neuron.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 106.Ligon KL, et al. Development of NG2 neural progenitor cells requires Olig gene function. Proc Natl Acad Sci U S A. 2006;103(20):7853–8. doi: 10.1073/pnas.0511001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee SK, et al. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19(2):282–94. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ligon KL, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53(4):503–17. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sauvageot CM, Kesari S, Stiles CD. Molecular pathogenesis of adult brain tumors and the role of stem cells. Neurol Clin. 2007;25(4):891–924. vii. doi: 10.1016/j.ncl.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 110.Bruggeman SW, et al. Bmi1 controls tumor development in an Ink4a/Arf-independent manner in a mouse model for glioma. Cancer Cell. 2007;12(4):328–41. doi: 10.1016/j.ccr.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 111.Ligon KL, et al. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol. 2004;63(5):499–509. doi: 10.1093/jnen/63.5.499. [DOI] [PubMed] [Google Scholar]
- 112.Golestaneh N, Mishra B. TGF-beta, neuronal stem cells and glioblastoma. Oncogene. 2005;24(37):5722–30. doi: 10.1038/sj.onc.1208925. [DOI] [PubMed] [Google Scholar]
- 113.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 114.Beier D, et al. Temozolomide preferentially depletes cancer stem cells in glioblastoma. Cancer Res. 2008;68(14):5706–15. doi: 10.1158/0008-5472.CAN-07-6878. [DOI] [PubMed] [Google Scholar]
- 115.Hegi ME, et al. Hemizygous or homozygous deletion of the chromosomal region containing the p16INK4a gene is associated with amplification of the EGF receptor gene in glioblastomas. Int J Cancer. 1997;73(1):57–63. doi: 10.1002/(sici)1097-0215(19970926)73:1<57::aid-ijc10>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 116.Hayashi Y, et al. Association of EGFR gene amplification and CDKN2 (p16/MTS1) gene deletion in glioblastoma multiforme. Brain Pathol. 1997;7(3):871–5. doi: 10.1111/j.1750-3639.1997.tb00890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 118.Rich JN, et al. Phase II trial of gefitinib in recurrent glioblastoma. J Clin Oncol. 2004;22(1):133–42. doi: 10.1200/JCO.2004.08.110. [DOI] [PubMed] [Google Scholar]
- 119.Vogelbaum MAPDM, Stevens G, Barnett G, Brewer C. Phase II study of erlotinib single agent therapy in recurrent glioblastoma multiforme. Eur j Cancer Suppl. 2005;3:135. [Google Scholar]
- 120.Chang SM, et al. Phase II study of CCI-779 in patients with recurrent glioblastoma multiforme. Invest New Drugs. 2005;23(4):357–61. doi: 10.1007/s10637-005-1444-0. [DOI] [PubMed] [Google Scholar]
- 121.Omuro AM, Faivre S, Raymond E. Lessons learned in the development of targeted therapy for malignant gliomas. Mol Cancer Ther. 2007;6(7):1909–19. doi: 10.1158/1535-7163.MCT-07-0047. [DOI] [PubMed] [Google Scholar]
- 122.Ohgaki H. Genetic pathways to glioblastomas. Neuropathology. 2005;25(1):1–7. doi: 10.1111/j.1440-1789.2004.00600.x. [DOI] [PubMed] [Google Scholar]
- 123.Rasheed A, et al. Molecular markers of prognosis in astrocytic tumors. Cancer. 2002;94(10):2688–97. doi: 10.1002/cncr.10544. [DOI] [PubMed] [Google Scholar]
- 124.Martinez R, et al. CpG island promoter hypermethylation of the proapoptotic gene caspase-8 is a common hallmark of relapsed glioblastoma multiforme. Carcinogenesis. 2007 doi: 10.1093/carcin/bgm014. [DOI] [PubMed] [Google Scholar]
- 125.Jeng YM, Hsu HC. KLF6, a putative tumor suppressor gene, is mutated in astrocytic gliomas. Int J Cancer. 2003;105(5):625–9. doi: 10.1002/ijc.11123. [DOI] [PubMed] [Google Scholar]
- 126.Martinez R, et al. Frequent hypermethylation of the DNA repair gene MGMT in long-term survivors of glioblastoma multiforme. J Neurooncol. 2006 doi: 10.1007/s11060-006-9292-0. [DOI] [PubMed] [Google Scholar]
- 127.Ichimura K, et al. Deregulation of the p14ARF/MDM2/p53 pathway is a prerequisite for human astrocytic gliomas with G1-S transition control gene abnormalities. Cancer Res. 2000;60(2):417–24. [PubMed] [Google Scholar]
- 128.Ichimura K, et al. Human glioblastomas with no alterations of the CDKN2A (p16INK4A MTS1) and CDK4 genes have frequent mutations of the retinoblastoma gene. Oncogene. 1996;13(5):1065–72. [PubMed] [Google Scholar]
- 129.Ueki K, et al. CDKN2/p16 or RB alterations occur in the majority of glioblastomas and are inversely correlated. Cancer Res. 1996;56(1):150–3. [PubMed] [Google Scholar]
- 130.Chung R, et al. TP53 gene mutations and 17p deletions in human astrocytomas. Genes Chromosomes Cancer. 1991;3(5):323–31. doi: 10.1002/gcc.2870030502. [DOI] [PubMed] [Google Scholar]
- 131.von Deimling A, et al. Astrocytic gliomas: characterization on a molecular genetic basis. Recent Results Cancer Res. 1994;135:33–42. doi: 10.1007/978-3-642-85039-4_5. [DOI] [PubMed] [Google Scholar]
- 132.Wang SI, et al. Somatic mutations of PTEN in glioblastoma multiforme. Cancer Res. 1997;57(19):4183–6. [PubMed] [Google Scholar]
- 133.Henson JW, et al. The retinoblastoma gene is involved in malignant progression of astrocytomas. Ann Neurol. 1994;36(5):714–21. doi: 10.1002/ana.410360505. [DOI] [PubMed] [Google Scholar]
- 134.Mueller W, et al. Downregulation of RUNX3 and TES by hypermethylation in glioblastoma. Oncogene. 2007;26(4):583–93. doi: 10.1038/sj.onc.1209805. [DOI] [PubMed] [Google Scholar]
- 135.Martinez R, Schackert G, Esteller M. Hypermethylation of the proapoptotic gene TMS1/ASC: prognostic importance in glioblastoma multiforme. J Neurooncol. 2006 doi: 10.1007/s11060-006-9264-4. [DOI] [PubMed] [Google Scholar]
- 136.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10(8):789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]