Abstract
In order for cell lineages to be maintained, daughter cells must have the same patterns of gene expression as the cells from which they were divided so that they can have the same phenotypes. However, during mitosis transcription ceases, chromosomal DNA is compacted, and most sequence-specific binding factors dissociate from DNA, making it difficult to understand how the “memory” of gene expression patterns is remembered and propagated to daughter cells. The process of remembering patterns of active gene expression during mitosis for transmission to daughter cells is called gene bookmarking. Here we discuss current knowledge concerning the factors and mechanisms involved in mediating gene bookmarking, including recent results on the mechanism by which the general transcription factor TBP participates in the mitotic bookmarking of formerly active genes.
Keywords: epigenetics, active gene memory, bookmarking, TBP, TFIID, HSF2, hsp70, PP2A, histone modification, condensin, mitosis
Introduction
In order to maintain the phenotype of cell lineages, cells must “remember” which genes were active prior to cell division and propagate that pattern through mitosis to daughter cells. The problem is that during mitosis most sequence-specific DNA-binding proteins dissociate from the DNA, transcription ceases, and the chromosomes are compacted. These features of mitosis make it difficult to understand how such gene expression information could persist through this part of the cell cycle. As an explanation for this phenomenon, it is thought that before the onset of mitosis all active genes are somehow ‘marked’, so that the daughter cells know to reassemble the transcription machinery on the promoters of these genes once mitosis is completed.1-6 Since their proposed function is reminiscent of the way a bookmark marks the position of the last read page of a book, this mechanism has been given the name ‘bookmarking’, and the factors or modifications that mark the active genes have been called ‘molecular bookmarks’.1, 2 However, the precise nature of these molecular bookmarks, and the mechanism by which they function to propagate patterns of gene expression through mitosis, is only beginning to be understood.
Another proposed function of bookmarking, which appears to apply to a subset of bookmarked genes, is to endow specific genes that must be activatable in the early G1 phase of the cell cycle, such as the hsp70 gene crucial for protection from cellular stress, with the ability to quickly re-establish transcription competence to their promoters as soon as the general mitotic repression of transcription ends. According to this hypothesis, if such genes were not bookmarked the cell might not be able to transcribe from their promoters until the normal decompaction process was complete, which would perhaps delay acquisition of transcription competence until later in G1. In the case of the hsp70 gene, such a delay might leave the cell unable to induce protective heat shock proteins in response to stresses occurring in G1, potentially leading to disruption of cell function or even cell death. Similarly, bookmarking of a gene such as c-myc might be important for its expression, which is important for the proper control of cell growth.
This review describes the key characteristics of bookmarked genes and the different mechanisms that have been described thus far that mediate this bookmarking. Also discussed are recent data revealing mechanistic details involved in bookmarking of formerly active gene promoters by the general transcription factor TBP during mitosis.
Unusual structure of promoter regions in mitotic cells
Chromosomal DNA must be compacted during mitosis prior to cytokinesis in order for chromosomes to be properly segregated to daughter cells. A protein complex called condensin plays an important role in this process of mitotic chromosomal compaction.7-10 Condensin is composed of five subunits: two structural maintenance of chromosomes (SMC) protein family members named CAP-C and CAP-E, and three other subunits called CAP-G, CAP-D2, and CAP-H. This complex is converted to an active form at the beginning of mitosis by phosphorylation of the CAP-G, CAP-H, and CAP-D2 subunits by the kinase cdc2/cyclin B.7-10 For this reason, CAP-G, CAP-H, and CAP-D2 are referred to as the regulatory subunits of the condensin complex.
In light of the high degree of chromatin compaction in mitotic cells, it was therefore intriguing when studies found that the hypersensitivity of specific sites in chromatin to nucleases and chemical probes is not lost during chromosome condensation and persists through mitosis.11-14 For example, DNAse hypersensitivity sites were found to exist in the region between -300 and +1 of the promoter of the hsp70 gene in mitotic chromatin.15 This result suggests that this segment of DNA is not compacted into a structure that would be expected of mitotic DNA. This region of the hsp70 promoter contains DNA sequences that are important for stress-inducible transcription, including both of the heat shock element (HSE) arrays to which heat shock transcription factor 1 (HSF1) binds after stress exposure to increase expression of the hsp70 gene.16
Notably, several studies have demonstrated the important feature that these alterations in mitotic chromatin structure are associated with genes that are expressed, or can be expressed, in a specific cell type. For example, the promoter of the β-globin gene shows hypersensitivity to S1 nuclease, but only in cell types in which β-globin is expressed.17 Another study has shown that structural perturbations detected by KMnO4 treatment exist in the promoter regions of genes that need to be reactivated after mitosis such as the c-myc and hsp70 genes, but are not observed for the c-myc or β-globin promoters in cell types in which they are not expressed.18 The results of this study also suggest a requirement for proteins in mediating these structural perturbations. A third study, based on the chromatin immunoprecipitation technique, found that the transcription factors TFIID and TFIIB remain associated with active promoters during mitosis, but not with inactive promoters.19 Association of TFIID with DNA during mitosis was also observed by biochemical and immunochemical analyses of endogenous TFIID as well as direct fluorescence microscopy analysis of transfected TATA-binding protein (TBP)-green fluorescence protein (GFP) in mitotic cells.20, 21
Mechanisms of gene bookmarking
A number of different mechanisms for gene bookmarking have been described in the literature. The first two mechanisms that will be discussed in this review are those thought to be involved in bookmarking the bulk of active genes in a cell to transmit this activity memory through mitosis. These mechanisms are the mitotic-dependent modification of histones/binding of variant histones at the promoters of genes that had been active prior to entry into mitosis, and the prevention of the normal mitotic chromatin compaction at these formerly active promoters mediated by TFIID that remains bound at these promoters during this stage of the cell cycle.
We then discuss three mechanisms that have been described for bookmarking specific subsets of genes in cells. These are the HSF2-mediated bookmarking of the hsp70 promoter, the function of Runx protein family members, and the role of Polycomb Group (PcG) and Trithorax Group (TxG) proteins in bookmarking their target genes. In these three mechanisms specific DNA sequences present in a subset of genes recruit factors that play a key role in their bookmarking.
Mechanisms for general bookmarking of formerly active gene promoters
Histone modification/variant histones as bookmarks of active genes
The modifications that occur on histones are important regulators of chromatin structure. Based on patterns of staining of hyperacetylated histone H4 in metaphase chromosomes it was proposed that modification of histones at regions corresponding to active genes functions as a mechanism for ‘memory’ of gene expression patterns for propagation to daughter cells.22 In support of this hypothesis, it has been found that active gene regions are associated with specific patterns of histone modifications in mitotic cells, including hyperacetylation of histones H3 and H4, tri- or dimethylation of Lys4 on histone H3, and dimethylation of Lys79 on histone H3.23-25 These histone modifications have been postulated to arise from the recruitment of histone modifying enzymes by transcription factors bound to the promoters or by the RNA polymerase II complex.
The results of another study suggest that active gene regions appear to be associated with nucleosomes containing a variant of histone H3 called H3.3, in combination with acetylation and methylation of histone H3, and that these patterns persist through mitosis.26 This study also suggested that the initiation of transcription is responsible for the deposition of histone H3.3 in these regions. A recent study strengthened support for the role of H3.3 in mediating active gene memory by finding that this variant histone is associated only with promoters in embryos exhibiting memory.6, 27 This study also found that mutating the methylation target lysine 4 is associated with a loss of memory, suggesting the importance of lysine 4 for the memory mechanism.27
Mitotic TFIID function in bookmarking
The transcription factor TFIID and its associated protein complexes are central components of the basal transcription machinery in eukaryotes, where they interact with the TATA boxes of promoters via the TBP subunit of TFIID. However, using biochemical and immunocytochemical methods it was discovered that TFIID remains tightly associated with condensed chromosomes in mitotic cells.20 Identical results were obtained from direct fluorescence analysis of the localization of TBP-GFP fusion protein in cells.21 This association of TFIID with mitotic chromosomes was intriguing because it was thought that most sequence-specific DNA-binding proteins dissociate from chromatin during mitosis. Furthermore, studies using the chromatin immunoprecipitation technique demonstrated that in mitotic cells TFIID, as well as TFIIB, remain associated with promoters of genes that are active in cells but not with the promoters of genes that are not expressed.19 Consistent with this, an earlier study showed that specific perturbations are observed in the structure of TATA box regions of genes that are active in a cell but not with those of inactive genes.18
The results described above support TFIID as a good candidate for a factor that could play a role in bookmarking the promoters of active genes during mitosis. Indeed, employing TFIID as a bookmarking factor to “remember” active gene promoters during mitosis makes biological sense because 1) utilizing a transcription factor that binds to the promoters of many active genes as a bookmarking factor could explain how cells identify active genes that should be bookmarked, and 2) having TFIID remaining bound at these promoters during mitosis would also provide a straightforward mechanism for reassembling transcription complexes on them after mitosis is complete.
The results of a recent study support this hypothesis.28 This study suggests that TFIID is important for maintaining the accessibility of formerly active gene promoters during mitosis. TFIID recruits the phosphatase PP2A to these promoters and also interacts with the CAP-G subunit of the condensin complex, which plays an important role in compacting chromosomal DNA during mitosis, to promote dephosphorylation and inactivation of condensin in the vicinity of these promoters and thereby prevent compaction of these specific regions of chromosomal DNA.28 A schematic model of the role of this TFIID-mediated mechanism in bookmarking formerly active gene promoters is shown in Figure 1.
Figure 1.
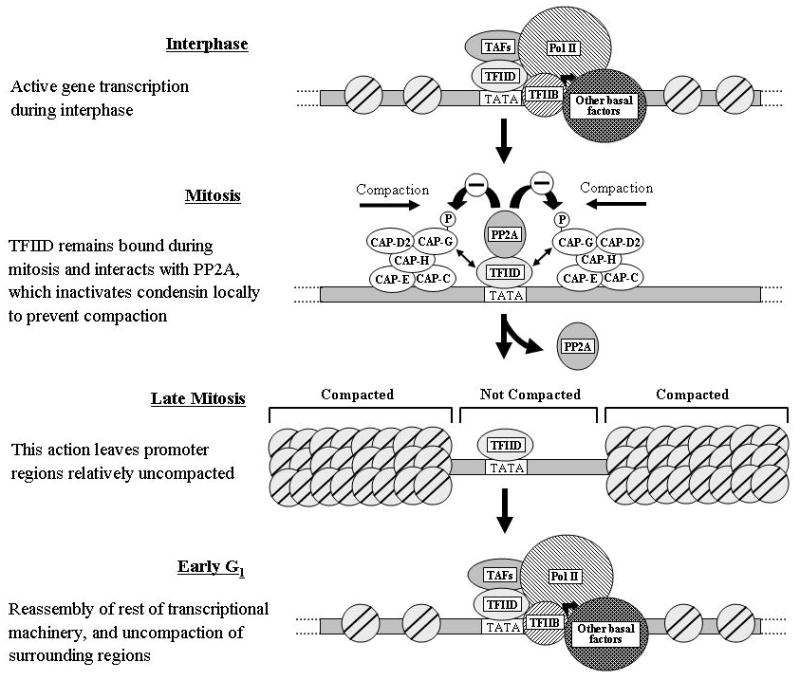
Model of TBP-mediated mechanism in “bookmarking” promoters of formerly active genes during mitosis. The model is not meant to imply that no nucleosomes are present on the promoters in mitotic cells, only that these regions are less compacted than most of the genomic DNA at this stage of the cell cycle.
Chromosome-wide “ChIP on Chip” data shows that TBP is present at a very large number of chromosomal sites in mitotic cells.28 Consistent with its suggested role in gene bookmarking, TBP binding is high in transcribed regions of the chromosomes but low in chromosomal sequences containing pseudogenes, genes whose expression is tissue-restricted, and regions that are devoid of annotated genes.
The mechanism(s) that regulate TFIID function in gene bookmarking during mitosis are not known. However, previous studies showed that TBP is a target of mitotic-dependent phosphorylation20, 29-32, which could mediate this regulation by altering the properties of the TBP protein to assist its function as a bookmarking factor.
Mechanisms involved in bookmarking specific sub-sets of gene promoters
Role of HSF2 in bookmarking heat shock element-containing promoters
Hsp70 is a critical cytoprotective protein whose expression is up-regulated in response to an increase in temperature and numerous other stresses. Induction of hsp70 gene expression is mediated by a protein called heat shock transcription factor 1 (HSF1), which is converted by stress to its DNA-binding form and then interacts with heat shock elements (HSEs) in the hsp70 promoter to increase its rate of transcription.33-36 These HSE elements are located in the region of the hsp70 promoter that contains site of persistent hypersensitivity to DNase in mitotic cells, suggesting it is part of the bookmarked region.15 However, the mechanism responsible for mediating the bookmarked status of the hsp70 promoter was not known.
HSF2 is another member of the HSF family of transcription factors that has a DNA-binding domain capable of recognizing the HSEs in the hsp70 gene promoter, but whose biological role in regulating hsp70 expression was not well understood.36, 37 However, results now indicate a role for HSF2 in bookmarking the hsp70 gene promoter.38 The data show that HSF2 binds to the hsp70 promoter at the beginning of mitosis, recruits the serine/threonine phosphatase PP2A to this DNA region and also interacts with the CAP-G subunit of condensin complexes that are compacting chromosomal DNA in the vicinity of the hsp70 promoter. This interaction between HSF2 and CAP-G allows the HSF2-associated PP2A to efficiently dephosphorylate the condensin complex, thereby inactivating condensin and stopping compaction of the DNA at this specific region of DNA. Thus, HSF2 functions both as a positioning factor, marking the promoter to be bookmarked by specifically binding to HSEs within it, and as a bridging factor, interacting with both PP2A and the CAP-G subunit of condensin to promote dephosphorylation of CAP-G and possibly also of the other condensin regulatory subunits, CAP-D2 and CAP-H. It is intriguing that this mechanism, namely the recruitment of PP2A to dephosphorylate/inactivate condensin, is also observed for the TBP-mediated mechanism observed for formerly active gene promoters during mitosis described above. This similarity of mechanisms employed by TBP and HSF2 on mitotic gene promoters may reflect the important role of PP2A as a negative regulator of condensin activity.39
It was previously proposed that the function of hsp70 promoter bookmarking was to allow cells to induce expression of this gene in the G1 phase of the cell cycle.1 In support of this hypothesis, blocking HSF2-mediated bookmarking by this approach was found to significantly decrease the ability of cells to induce expression of hsp70 in response to stress applied in early G1.38 This loss of inducible hsp70 expression is associated with increased cell death in response to stress, indicating the crucial biological importance of this gene bookmarking event.
Other results indicate a potential mechanism by which the bookmarking function of HSF2 may be regulated. Previous studies had demonstrated that HSF2 is covalently modified at Lys82 by the 97 amino acid protein called small ubiquitin-related modifier 1, or SUMO-1.40 As its name implies, SUMO-1 is structurally similar to ubiquitin. However, sumoylation does not target proteins for degradation but instead regulates their functional properties such as ability to interact with other proteins, subcellular localization, and transcriptional activity.41-43 Recent results have shown that sumoylation of HSF2 is up-regulated in a mitosis-dependent manner.38 Furthermore, blocking HSF2 sumoylation by mutating Lys82 to arginine decreases HSF2 association with the CAP-G subunit of condensin, indicating that this sumoylation event has a positive effect on the interaction between HSF2 and CAP-G.38 These results suggest that cell cycle-dependent sumoylation may function as a regulator of hsp70 bookmarking by modulating the ability of HSF2 to interact with condensin. The mechanism by which sumoylation of HSF2 is up-regulated in a mitosis-dependent manner remains to be determined.
More recent studies have added further insight into HSF2 bookmarking function. For example, results indicate that HSF2 also binds to the HSE-containing promoters of other genes, including those of the hsp90, hsp27, and c-fos genes, indicating that HSF2 is likely involved in bookmarking other genes beside the hsp70 gene.44 Other work has shown that HSF2, along with HSF1, is bound to the hsp70 gene promoter even in mature spermatozoa.45 This may provide an explanation for previous observations that the hsp70 gene is one of the first to be expressed after fertilization, and that this expression is higher in the male pronucleus of the fertilized embryo.46, 47
Involvment of Runx2 in transmitting gene expression patterns
Runx2 is a transcription factor in development of osteogenic cell lineages.48-50 It was found that this factor remains bound to promoters of rDNA genes during mitosis, and that these chromosomal loci exhibit an open chromatin state.51 The results of another study revealed that Runx2 also binds a number of other target gene promoters in mitotic cells, that this factor is important for transmission of gene expression patterns of specific loci from parent to daughter cells, and that this could be mediated by Runx2-mediated patterns of histone modifications at the target gene promoters.52 These results suggest that one of the functions of Runx2 is to epigenetically maintain phenotype through mitosis, in order to to ensure that the identity of cell lineages is retained in progeny cells. A recent paper showed that another member of the Runx family of proteins, Runx3, is bound to nucleolar organizing regions during mitosis, suggesting that this factor may also be involved in epigenetic maintenance of gene expression patterns.53
Polycomb/trithorax and switchable epigenetic memory
Groups of proteins called Polycomb group (PcG) and Trithorax group (TrxG) complexes bind to specific DNA sequences in target genes and are thought to have opposite effects on these genes. PcG complexes function as repressors of genes, whereas TrxG proteins are recruited in response to activating signals and act positively to maintain gene activity.54-57
PcG and TrxG complexes both contain histone methyltransferase activities and appear to mediate their antagonistic effects on these DNA regions by promoting patterns of histone modification that are characteristic of each complex.58-65 The histone methylation pattern conferred by PcG complexes is associated with a more closed chromatin configuration while those mediated by TrxG complexes appear to be more open.66-69 These effects persist through cell divisions, indicating that PcG and TrxG both function as bookmarking factors for their common target genes. Thus, modification by PcG and TrxG represents a mechanism for regulated bookmarking in which the outcome with respect to transcription competence depends on which complex's activity predominates at a specific gene region.
Concluding remarks and future perspectives
The studies described above revealed a number of important features of gene bookmarking. First, it appears that there are at least two biological functions of bookmarking: one is the bookmarking of active genes to insure the faithful transmission of phenotype through mitosis to daughter cells; the other is the bookmarking of genes that require the ability to be expressed in early G1, such as in the case of the hsp70 gene whose ability to regain inducibility in early G1 allows the cell to be protected against stress should it occur during this part of the cell cycle. Second, it appears that more than one type of molecular bookmark and bookmarking mechanism exists in our cells. This is not surprising in light of the importance of bookmarking for cell function. Bookmarking at some gene loci in mitotic cells might be mediated by selective histone modifications, the presence of histone variants, or the retention of TFIID to genes that are active prior to mitosis. Other genes or subsets of genes, such as the hsp70 gene family, might depend on sequence-specific DNA-binding proteins such as HSF2.
Having at least two general mechanisms for marking active genes would make biological sense because it would provide functional redundancy to ensure that the bookmarking of these genes does indeed take place. Another possible reason is that since some promoters lack TATA boxes, these promoters would presumably need to be bookmarked by a non-TBP-dependent mechanism such as histone modification, the presence of nucleosomes containing a histone variant such H3.3, or some as-yet-undiscovered mechanism. The existence of bookmarking mechanisms specific to a gene family could reflect a need for potential regulation, such as in the bookmarking of homeotic genes by PcG or TrxG, or the need to express inducible genes in early G1, or perhaps in some cases to ensure the ability to express critical genes that reside in chromosomal regions that are prone to adopt repressive structures during mitotic compaction and/or that may be difficult to de-compact.
It should be noted that these postulated bookmarking mechanisms are not necessarily mutually exclusive, and in fact more than one mechanism could be involved in the bookmarking of a given gene. Indeed, the finding that binding of Polycomb and Trithorax complexes to DNA sequences leads to specific patterns of histone modification mediated by the action of the histone methyltransferases associated with these complexes illustrates this point clearly.58-65 In addition, the finding that acetylation of Lys9 and Lys14 residues of histone H3 increases binding of TFIID to a promoter region could explain at the biochemical level how histone modifications characteristic of active genes contribute to maintaining transcriptional memory, namely by stabilizing the binding of TFIID to ensure that it remains associated with the promoters of these genes during mitosis.26, 70
The results described above reveal intriguing aspects of the bookmarking mechanisms present in eukaryotic cells. However, they also raise many interesting new questions. For example, do events such as histone modifications or the association of histone variants with active genes contribute to transmission of the transcription-competent gene pattern to daughter cells by stabilizing the binding of factors such as TFIID, or by some other mechanism(s)? Are there other factors, like HSF2, that bookmark the promoter of a specific gene or families of genes that have common promoter DNA elements with which the factors(s) interact. It seems likely that there are indeed other bookmarking factors of this type awaiting discovery. Further, if there are other bookmarking factors, do they operate by a mechanism similar to that used by TFIID and HSF2, namely, recruiting a phosphatase and interacting with condensin subunits to promote efficient dephosphorylation and inactivation of condensin near these chromosomal regions, or do they mediate bookmarking of these regions by a different mechanism? Answers to these and other questions will provide valuable and interesting insights into the molecular mechanisms, regulation, and biological functions of gene bookmarking.
Acknowledgments
We apologize to colleagues whose work we could not cite because of space considerations. We would like to thank members of the laboratory and department for insightful discussions concerning gene bookmarking, and acknowledge the support of NIH grant GM64606 to K.D.S.
References
- 1.John S, Workman JL. Bookmarking genes for activation in condensed mitotic chromosomes. Bioessays. 1998;20:275–9. doi: 10.1002/(SICI)1521-1878(199804)20:4<275::AID-BIES1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 2.Sarge KD, Park-Sarge OK. Gene bookmarking: keeping the pages open. Trends Biochem Sci. 2005;30:605–10. doi: 10.1016/j.tibs.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Zhou GL, Liu DP, Liang CC. Memory mechanisms of active transcription during cell division. Bioessays. 2005;27:1239–45. doi: 10.1002/bies.20327. [DOI] [PubMed] [Google Scholar]
- 4.Zhou GL, Xin L, Liu DP, Liang CC. Remembering the cell fate during cellular differentiation. J Cell Biochem. 2005;96:962–70. doi: 10.1002/jcb.20572. [DOI] [PubMed] [Google Scholar]
- 5.Hirose S. Crucial roles for chromatin dynamics in cellular memory. J Biochem. 2007;141:615–9. doi: 10.1093/jb/mvm092. [DOI] [PubMed] [Google Scholar]
- 6.Ng RK, Gurdon JB. Epigenetic inheritance of cell differentiation status. Cell Cycle. 2008;7:1173–7. doi: 10.4161/cc.7.9.5791. [DOI] [PubMed] [Google Scholar]
- 7.Belmont AS. Mitotic chromosome structure and condensation. Curr Opin Cell Biol. 2006;18:632–8. doi: 10.1016/j.ceb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–22. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 9.Uhlmann F, Hopfner KP. Chromosome biology: the crux of the ring. Curr Biol. 2006;16:R102–5. doi: 10.1016/j.cub.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 10.Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 11.Weintraub H. Assembly and propagation of repressed and depressed chromosomal states. Cell. 1985;42:705–11. doi: 10.1016/0092-8674(85)90267-3. [DOI] [PubMed] [Google Scholar]
- 12.Kerem BS, Goitein R, Diamond G, Cedar H, Marcus M. Mapping of DNAase I sensitive regions on mitotic chromosomes. Cell. 1984;38:493–9. doi: 10.1016/0092-8674(84)90504-x. [DOI] [PubMed] [Google Scholar]
- 13.Groudine M, Weintraub H. Propagation of globin DNAase I-hypersensitive sites in absence of factors required for induction: a possible mechanism for determination. Cell. 1982;30:131–9. doi: 10.1016/0092-8674(82)90019-8. [DOI] [PubMed] [Google Scholar]
- 14.Struhl G. A gene product required for correct initiation of segmental determination in Drosophila. Nature. 1981;293:36–41. doi: 10.1038/293036a0. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Balbas MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 16.Wu BJ, Kingston RE, Morimoto RI. Human HSP70 promoter contains at least two distinct regulatory domains. Proc Natl Acad Sci U S A. 1986;83:629–33. doi: 10.1073/pnas.83.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsen A, Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982;29:609–22. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- 18.Michelotti EF, Sanford S, Levens D. Marking of active genes on mitotic chromosomes. Nature. 1997;388:895–9. doi: 10.1038/42282. [DOI] [PubMed] [Google Scholar]
- 19.Christova R, Oelgeschlager T. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat Cell Biol. 2002;4:79–82. doi: 10.1038/ncb733. [DOI] [PubMed] [Google Scholar]
- 20.Segil N, Guermah M, Hoffmann A, Roeder RG, Heintz N. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996;10:2389–400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- 21.Chen D, Hinkley CS, Henry RW, Huang S. TBP Dynamics in Living Human Cells: Constitutive Association of TBP with Mitotic Chromosomes. Mol Biol Cell. 2002;13:276–84. doi: 10.1091/mbc.01-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeppesen P. Histone acetylation: a possible mechanism for the inheritance of cell memory at mitosis. Bioessays. 1997;19:67–74. doi: 10.1002/bies.950190111. [DOI] [PubMed] [Google Scholar]
- 23.Kouskouti A, Talianidis I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 2005;24:347–57. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valls E, Sanchez-Molina S, Martinez-Balbas MA. Role of histone modifications in marking and activating genes through mitosis. J Biol Chem. 2005;280:42592–600. doi: 10.1074/jbc.M507407200. [DOI] [PubMed] [Google Scholar]
- 25.Xin L, Zhou GL, Song W, Wu XS, Wei GH, Hao DL, et al. Exploring cellular memory molecules marking competent and active transcriptions. BMC Mol Biol. 2007;8:31–9. doi: 10.1186/1471-2199-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chow CM, Georgiou A, Szutorisz H, Maia e Silva A, Pombo A, Barahona I, et al. Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division. EMBO Rep. 2005;6:354–60. doi: 10.1038/sj.embor.7400366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–9. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- 28.Xing H, Vanderford NL, Sarge KD. The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat Cell Biol. 2008;10:1318–23. doi: 10.1038/ncb1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottesfeld JM, Wolf VJ, Dang T, Forbes DJ, Hartl P. Mitotic repression of RNA polymerase III transcription in vitro mediated by phosphorylation of a TFIIIB component. Science. 1994;263:81–4. doi: 10.1126/science.8272869. [DOI] [PubMed] [Google Scholar]
- 30.White RJ, Gottlieb TM, Downes CS, Jackson SP. Mitotic regulation of a TATA-binding-protein-containing complex. Mol Cell Biol. 1995;15:1983–92. doi: 10.1128/mcb.15.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leresche A, Wolf VJ, Gottesfeld JM. Repression of RNA polymerase II and III transcription during M phase of the cell cycle. Exp Cell Res. 1996;229:282–8. doi: 10.1006/excr.1996.0373. [DOI] [PubMed] [Google Scholar]
- 32.Heix J, Vente A, Voit R, Budde A, Michaelidis TM, Grummt I. Mitotic silencing of human rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO J. 1998;17:7373–81. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akerfelt M, Trouillet D, Mezger V, Sistonen L. Heat shock factors at a crossroad between stress and development. Ann N Y Acad Sci. 2007;1113:15–27. doi: 10.1196/annals.1391.005. [DOI] [PubMed] [Google Scholar]
- 34.Voellmy R. On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones. 2004;9:122–33. doi: 10.1379/CSC-14R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabindran SK, Giorgi G, Clos J, Wu C. Molecular cloning and expression of a human heat shock factor, HSF1. Proc Natl Acad Sci U S A. 1991;88:6906–10. doi: 10.1073/pnas.88.16.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarge KD, Zimarino V, Holm K, Wu C, Morimoto RI. Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding ability. Genes Dev. 1991;5:1902–11. doi: 10.1101/gad.5.10.1902. [DOI] [PubMed] [Google Scholar]
- 37.Schuetz TJ, Gallo GJ, Sheldon L, Tempst P, Kingston RE. Isolation of a cDNA for HSF2: evidence for two heat shock factor genes in humans. Proc Natl Acad Sci U S A. 1991;88:6911–5. doi: 10.1073/pnas.88.16.6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xing H, Wilkerson DC, Mayhew CN, Lubert EJ, Skaggs HS, Goodson ML, et al. Mechanism of hsp70i gene bookmarking. Science. 2005;307:421–3. doi: 10.1126/science.1106478. [DOI] [PubMed] [Google Scholar]
- 39.Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–21. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- 40.Goodson ML, Hong Y, Rogers R, Matunis MJ, Park-Sarge OK, Sarge KD. Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J Biol Chem. 2001;276:18513–8. doi: 10.1074/jbc.M008066200. [DOI] [PubMed] [Google Scholar]
- 41.Bossis G, Melchior F. SUMO: regulating the regulator. Cell Div. 2006;1:13. doi: 10.1186/1747-1028-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–80. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 43.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 44.Wilkerson DC, Skaggs HS, Sarge KD. HSF2 binds to the Hsp90, Hsp27, and c-Fos promoters constitutively and modulates their expression. Cell Stress Chaperones. 2007;12:283–90. doi: 10.1379/CSC-250.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilkerson DC, Murphy LA, Sarge KD. Interaction of HSF1 and HSF2 with the Hspa1b promoter in mouse epididymal spermatozoa. Biol Reprod. 2008;79:283–8. doi: 10.1095/biolreprod.107.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christians E, Campion E, Thompson EM, Renard JP. Expression of the HSP 70.1 gene, a landmark of early zygotic activity in the mouse embryo, is restricted to the first burst of transcription. Development. 1995;121:113–22. doi: 10.1242/dev.121.1.113. [DOI] [PubMed] [Google Scholar]
- 47.Fiorenza MT, Bevilacqua A, Canterini S, Torcia S, Pontecorvi M, Mangia F. Early transcriptional activation of the hsp70.1 gene by osmotic stress in one-cell embryos of the mouse. Biol Reprod. 2004;70:1606–13. doi: 10.1095/biolreprod.103.024877. [DOI] [PubMed] [Google Scholar]
- 48.Marie PJ. Transcription factors controlling osteoblastogenesis. Arch Biochem Biophys. 2008;473:98–105. doi: 10.1016/j.abb.2008.02.030. [DOI] [PubMed] [Google Scholar]
- 49.Deng ZL, Sharff KA, Tang N, Song WX, Luo J, Luo X, et al. Regulation of osteogenic differentiation during skeletal development. Front Biosci. 2008;13:2001–21. doi: 10.2741/2819. [DOI] [PubMed] [Google Scholar]
- 50.Komori T. Regulation of bone development and maintenance by Runx2. Front Biosci. 2008;13:898–903. doi: 10.2741/2730. [DOI] [PubMed] [Google Scholar]
- 51.Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, et al. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007;445:442–6. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- 52.Young DW, Hassan MQ, Yang XQ, Galindo M, Javed A, Zaidi SK, et al. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc Natl Acad Sci U S A. 2007;104:3189–94. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pande S, Ali SA, Dowdy C, Zaidi SK, Ito K, Ito Y, et al. Subnuclear targeting of the Runx3 tumor suppressor and its epigenetic association with mitotic chromosomes. J Cell Physiol. 2009;218:473–9. doi: 10.1002/jcp.21630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Breiling A, Sessa L, Orlando V. Biology of polycomb and trithorax group proteins. Int Rev Cytol. 2007;258:83–136. doi: 10.1016/S0074-7696(07)58002-2. [DOI] [PubMed] [Google Scholar]
- 55.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–45. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 56.Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–32. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 57.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 58.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 60.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–96. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 61.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 62.Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–28. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 63.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–17. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 64.Byrd KN, Shearn A. ASH1, a Drosophila trithorax group protein, is required for methylation of lysine 4 residues on histone H3. Proc Natl Acad Sci U S A. 2003;100:11535–40. doi: 10.1073/pnas.1933593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klymenko T, Muller J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 2004;5:373–7. doi: 10.1038/sj.embor.7400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grimaud C, Negre N, Cavalli G. From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res. 2006;14:363–75. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- 67.Cernilogar FM, Orlando V. Epigenome programming by Polycomb and Trithorax proteins. Biochem Cell Biol. 2005;83:322–31. doi: 10.1139/o05-040. [DOI] [PubMed] [Google Scholar]
- 68.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–43. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 69.Lund AH, van Lohuizen M. Polycomb complexes and silencing mechanisms. Curr Opin Cell Biol. 2004;16:239–46. doi: 10.1016/j.ceb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 70.Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–92. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]


