Abstract
The catalytic activities of three members of the amidohydrolase superfamily were discovered using amino acid substrate libraries. Bb3285 from Bordetella bronchiseptica, Gox1177 from Gluconobacter oxydans, and Sco4986 from Streptomyces coelicolor are currently annotated as d-aminoacylases or N-acetyl-d-glutamate deacetylases. These three enzymes are 22−34% identical to one another in amino acid sequence. Substrate libraries containing nearly all combinations of N-formyl-d-Xaa, N-acetyl-d-Xaa, N-succinyl-d-Xaa, and l-Xaa-d-Xaa were used to establish the substrate profiles for these enzymes. It was demonstrated that Bb3285 is restricted to the hydrolysis of N-acyl substituted derivatives of d-glutamate. The best substrates for this enzyme are N-formyl-d-glutamate (kcat/Km = 5.8 × 106 M−1 s−1), N-acetyl-d-glutamate (kcat/Km = 5.2 × 106 M−1 s−1) and l-methionine-d-glutamate (kcat/Km = 3.4 × 105 M−1 s−1). Gox1177 and Sco4986 preferentially hydrolyze N-acyl substituted derivatives of hydrophobic d-amino acids. The best substrates for Gox1177 are N-acetyl-d-leucine (kcat/Km = 3.2 × 104 M−1 s−1), N-acetyl-d-tryptophan (kcat/Km = 4.1 × 104 M−1 s−1) and l-tyrosine-d-leucine (kcat/Km = 1.5 × 104 M−1 s−1). A fourth protein, Bb2785 from B. bronchiseptica, did not have d-aminoacylase activity. The best substrates for Sco4986 are N-acetyl-d-phenylalanine and N-acetyl-d-tryptophan. The three-dimensional structures of Bb3285 in the presence of the product acetate or a potent mimic of the tetrahedral intermediate were determined by X-ray diffraction methods. The side chain of the d-glutamate moiety of the inhibitor is ion-paired to Arg-295 while the α-carboxylate is ion-paired with Lys-250 and Arg-376. These results have revealed the chemical and structural determinants for substrate specificity in this protein. Bioinformatic analyses of an additional ∼250 sequences identified as members of this group suggest that there are no simple motifs that allow prediction of substrate specificity for most of these unknowns, highlighting the challenges for computational annotation of some groups of homologous proteins.
The functional annotation of enzymes based solely on the amino acid sequence is a difficult endeavor. This problem has become significantly more acute since the widespread application of whole organism DNA sequencing efforts. A critical assessment of the functional annotation for the more than four million genes that have been sequenced thus far suggests that approximately one third of the encoded proteins have an uncertain, unknown, or incorrect functional assignment (1). This observation suggests that a significant fraction of the metabolic diversity in microorganisms remains to be properly characterized. In the pre-genomic era most enzymes were identified and characterized based almost entirely on the ability to follow and measure a catalytic activity. In the genomic era the strategies for matching a protein/gene sequence with a definable catalytic activity must be reassessed and significantly enhanced. In many instances the amino acid sequences of these poorly characterized proteins are homologous to enzymes of known function and a clearly defined metabolic niche. The extent of the sequence identity with enzymes of known function determines, in most instances, the degree of difficulty in establishing an authentic substrate and enzymatic transformation. Our approach to this problem has been to focus on the large number of enzymes of unknown function within the amidohydrolase superfamily (2-4).
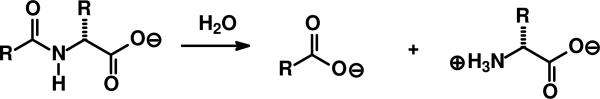
The amidohydrolase superfamily (AHS1) is a group of enzymes which has a remarkable substrate diversity embedded within active sites that are forged from a (β/α)8-barrel structural fold (5). Over 6,000 protein sequences have been identified as members of the AHS among the more than seven million gene sequences determined thus far (6). Members of this superfamily have been shown to catalyze the hydrolysis of organophosphate esters, lactones, and amides, in addition to decarboxylations, hydrations, and isomerization reactions (5, 7, 8). However, a significant fraction of the members of this broad superfamily have an ambiguous substrate and reaction specificity that remains to be unraveled (6). A group of homologous sequences that encode for enzymes of uncertain function has been provisionally annotated in public databases as N-acyl-d-amino acid deacylases. These enzymes catalyze the general reaction illustrated in Scheme 1.
Scheme 1.
The functional role of the N-acyl-d-amino acid deacylases is unclear. However, the addition of acylated d-amino acids in the growth medium of some microorganisms has been used to induce the production of these enzymes (9, 10). d-Amino acids are associated with neurotransmission and hormone synthesis in mammals and protection from proteolysis in bacteria (11). Certain organisms have the means to degrade d-amino acids (e.g. d-amino acid oxidase) and to prevent their incorporation into proteins by deacylating aminoacyl-tRNAs with a misincorporated d-amino acid (11, 12). d-Amino acids are also incorporated into key sites in some antibiotics (13). Enzymes have been identified which preferentially hydrolyze N-acylated derivatives of d-aspartate (9), d-glutamate (14, 15), d-methionine (16, 17), d-phenylalanine (18), and d-valine (19). These proteins can have high sequence identity to one another but a structural explanation for the differences in the substrate specificity is lacking.
The only structurally characterized member of this group of enzymes is d-aminoacylase (DAA) from Alcaligenes faecalis DA1 (gi| 28948588, PDB code: 1M7J). This enzyme can bind one or two divalent cations in the active site but only a single metal ion bound to the β-site is required for the expression of catalytic activity. The single divalent cation at the β-site is bound to Cys-96, His-220, and His-250 (20). It has been proposed that the nucleophilic water molecule is coordinated to the single divalent cation and hydrogen bonded to the side chain carboxylate of Asp-366 from β-strand 8. The proposed catalytic mechanism suggests that the carbonyl group of the amide bond is polarized via an interaction with the divalent cation bound to the β-site and that the water molecule utilized for hydrolysis is activated via metal coordination and a hydrogen bonding interaction with the carboxylate group of Asp-366. Once the tetrahedral adduct is formed the leaving group is activated via proton transfer from Asp-366 (21). This enzyme has been reported to preferentially hydrolyze N-acetyl-d-methionine (16).
In this paper we have determined the relative substrate specificity for three putative N-acyl-d-amino acid deacylases from the amidohydrolase superfamily. These proteins include Bb3285 from Bordetella bronchiseptica, Gox1177 from Gluconobacter oxydans, and Sco4986 from Streptomyces coelicolor. All of these proteins are currently annotated by NCBI as generic d-aminoacylases or more specifically as an N-acetyl-d-glutamate deacetylase. The protein Bb2785 from B. bronchiseptica, also investigated here, is annotated as a hypothetical protein but is similar in amino acid sequence to other proteins provisionally assigned as putative d-aminoacylases. A protein sequence alignment of Gox1177 with the structurally characterized d-aminoacylase from A. facaelis indicates that most of the residues in the active site are highly conserved although the sequence identity is only 23%. In contrast, the Bb3285 enzyme is 48% identical to the d-aminoacylase from A. facaelis. A sequence alignment of Gox1177, Bb2785, Bb3285, Sco4986 and the d-aminoacylase from A. facaelis is presented in Figure 1. Substrate libraries containing nearly all combinations of l-Xaa-d-Xaa, N-acetyl-d-Xaa, N-acetyl-l-Xaa, N-succinyl-l-Xaa, and N-succinyl-d-Xaa were utilized to establish the substrate specificities for Bb3285, Gox1177, and Sco4986. It was demonstrated that Bb3285 preferentially hydrolyzes N-acetyl-d-glutamate and N-formyl-d-glutamate whereas Gox1177 and Sco4986 hydrolyze a rather broad spectrum of N-acyl-d-amino acids, including l-Xaa-d-Xaa dipeptides by Gox1177. The three-dimensional structure of Bb3285 was determined in the presence of the product, acetate, and also with the N-methylphosphonate derivative of d-glutamate, a potent inhibitor resembling the tetrahedral intermediate formed during substrate hydrolysis.
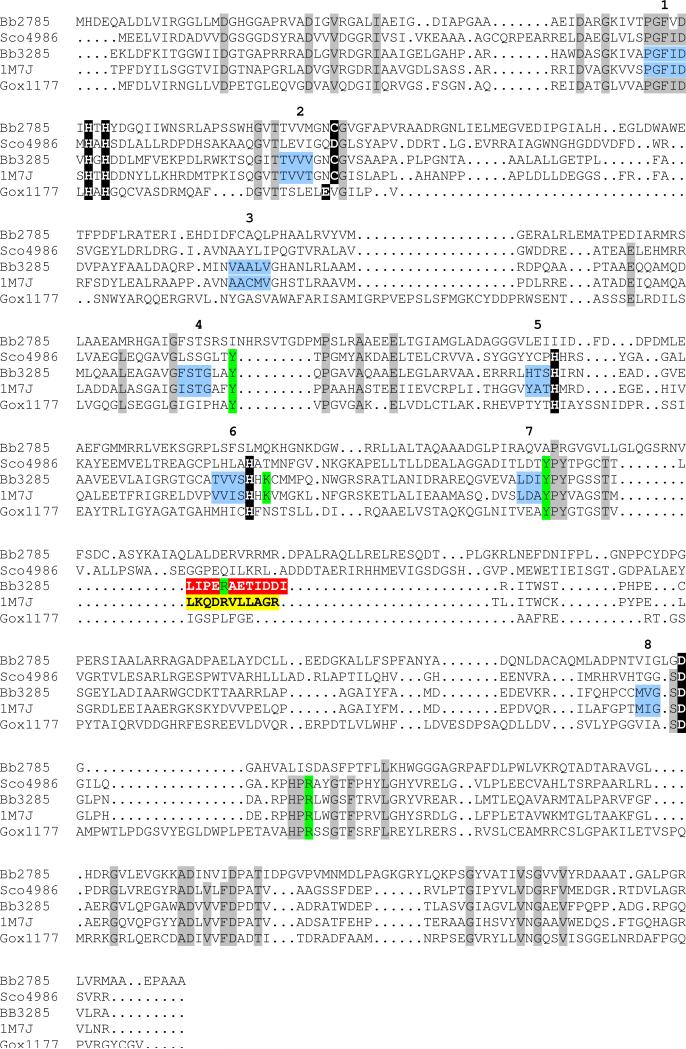
Figure 1.
Amino acid sequence alignment for the d-aminoacylase from A. facaelis (1M7J), Gox1177 from G. oxydans, Bb3285 and Bb2785 from B. bronchiseptica, and Sco4986 from S. coelicolor. Conservation patterns across these sequences with respect to the metal ligands identified in Bb3285 (see Figure 8) and the d-aminoacylase from A. facaelis are highlighted with a black background. The amino acid residues proposed to play a role in the recognition of the substrate in the active site of Bb3285 are highlighted in green. The variable substrate specificity loops in Bb3285 (291−302) and the DAA from A. facaelis (292−302) are highlighted in red and yellow, respectively. Those residues which represent the β-strands of the (β/α)8-barrel are colored light blue and the β-strands in the barrel are numbered. Those residues that are conserved in DAA (1m7j), Gox1177, Bb3285, and Sco4986 are highlighted with a grey background.
Materials and Methods
Materials
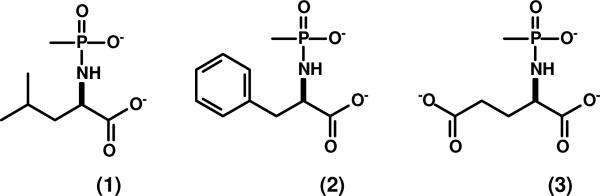
The N-acetyl derivatives of d-Cys, d-Glu, d-His, d-Lys, d-Thr, d-Gln and d-Ser were synthesized as previously described (22). The syntheses of N-succinyl-d-Glu and N-formyl-d-Glu were conducted according to the method described by Sakai et al. (23). The N-methylphosphonate derivatives of d-Leu (1), d-Phe (2) and d-Glu (3) (shown in Scheme 2) were synthesized according to the method of Xu et al. (24). The compounds, N-acetyl-d/l-Asp, N-acetyl-d-Pro, and N-acetyl-d-Asn were purchased from Sigma. All other N-acetyl-d-amino acids were purchased from Novabiochem with the exception of N-acetyl-Gly (TCI America), N-acetyl-d-Arg (Avocado), and N-acetyl-d-Tyr (Bachem). Resins and protected amino acids used for solid phase peptide synthesis were purchased from Calbiochem.
Scheme 2.
Synthesis of N-Acyl-Amino Acid Libraries
Twenty preloaded N-Fmoc-protected (or unprotected)-l-amino acid Wang resins (0.02 mmol each of N-Fmoc-Ala, N-Fmoc-Arg-(Mtr), NFmoc-Asn-(Trt), N-Fmoc-Asp-(OtBu), N-Fmoc-Cys-(Trt), N-Fmoc-Glu-(OtBu), N-Fmoc-Gln-(Trt), N-Fmoc-Gly, N-Fmoc-His-(Trt), N-Fmoc-Ile, N-Fmoc-Leu, N-Fmoc-Lys-(Boc), N-Fmoc-Met, N-Fmoc-Phe, N-Fmoc-Pro, N-Fmoc-Ser-(Trt), N-Fmoc-Thr-(Trt), N-Fmoc-Trp-(Boc), N-Fmoc-Tyr-(tBu), N-Fmoc-Val and DMF (5 mL) were shaken in a syringe for 30 minutes. The DMF was removed and then 6.0 mL of a solution containing 20% of piperidine in DMF was added and then shaken for 30 minutes to remove the Fmoc protecting group. This process was repeated. The beads were subsequently washed with DMF (4 × 5 mL) followed by the addition of acetic anhydride (82 mg, 0.8 mmol) and pyridine (63 mg, 0.8 mmol) in 6 mL DMF and then shaken overnight. After removal of the solvent, the beads were washed with DMF (4 × 5 mL) and then shaken overnight with the same reagents (acetic anhydride, pyridine and DMF). The solvent was removed and the beads were washed with DMF (4 × 5 mL), dichloromethane (4 × 5 mL), methanol (4 × 5 mL) and dried for several hours. The N-acetyl-l-amino acids were removed from the beads with 4 mL of cocktail R (TFA/thioanisole/EDT/anisole (v/v, 90/5/3/2)) and shaken for 3 hours. The library of N-acetyl-l-amino acids was obtained after the removal of cocktail R under reduced pressure at 50 °C overnight and washing with a solution of EtOAc/Et2O.
In the same fashion, a library of N-acetyl-d-amino acids was prepared using N-Fmoc-protected (or unprotected)-d-amino acid Wang resins (d-Ile, d-Pro, and glycine were omitted). The N-succinyl-d- and N-succinyl-l-amino acid libraries were prepared using the same procedure as the N-acetyl-l-amino acid libraries except that succinic anhydride was utilized instead of acetic anhydride. Analysis by mass spectrometry (ESI, positive and negative mode) was used to verify the presence of the desired compounds in each of these four libraries.
Synthesis of l-Xaa-d-Xaa Dipeptide Libraries
The dipeptide libraries were constructed in a manner similar to the preparation of the N-acetyl-l-amino acids library. Eighteen preloaded N-Fmoc-protected (or unprotected)-d-amino acid Wang resins (0.01 mmol each) were mixed and the Fmoc protecting group was removed. However, the N-Fmoc-d-Cys-(Trt) and N-Fmoc-d-Ile loaded resins were omitted. To this mixture was added N-Fmoc-l-Ala (84 mg, 0.27 mmol), N-hydroxybenzotriazole ·H2O (41 mg, 0.27 mmol) and N, N′-diisopropylcarbodiimide (34 mg, 0.27 mmol) to obtain the N-Fmoc-l-Ala-d-Xaa dipeptide library after cleavage from the beads with cocktail R. The Fmoc protecting group was removed after stirring with 20% piperidine in DMF (5 mL) for 30 minutes. The l-Ala-d-Xaa dipeptide library was obtained after removal of the solvent followed by washing with EtOAc/Et2O (v/v, 1/5) and drying overnight. The remaining l-Xaa-d-Xaa dipeptide libraries were constructed in a similar manner. Mass spectrometry (ESI, positive and negative mode) verified the formation of the 18 dipeptides in each of the 19 dipeptide libraries (the l-Cys-d-Xaa library was not synthesized).
Amino Acid Analysis of N-Acyl-Xaa and l-Xaa-d-Xaa Dipeptide Libraries
The composition of the substituted amino acid libraries was determined by amino acid analysis after acid catalyzed hydrolysis (except for cysteine and tryptophan). The individual libraries were hydrolyzed by vapor phase 6 M HCL-2% phenol at 150 °C for 90 minutes. The amino acids norvaline and sarcosine were added to the samples before hydrolysis as internal standards. A sample of human serum albumin was hydrolyzed under the same conditions as a control to ensure that complete hydrolysis of the mixture had occurred. Amino acid analysis was conducted with a Hewlett Packard AminoQuant System (HP 1090L). Samples were suspended in borate buffer, pH 10, and the free amino acids were derivatized with o-phthalaldehyde (OPA) and 9-fluoromethylchloroformate. The derivatized amino acids were separated on a 5 μm reverse phase HPLC column (Agilent 79916AA-572) using a gradient of 100% buffer A (20 mM sodium acetate/0.018% v/v TEA/50 mM ethylenediamine/0.3% v/v THF, pH 7.2) to 60% buffer B (1:2:2 100 mM sodium acetate: acetonitrile: MeOH) over 17 minutes at a flow rate of 0.45 mL/minute. The amino acid derivatives were detected using a photodiode array (UV-DAD) spectrophotometer or by fluorescence. The excitation/emission wavelengths for the OPA- and Fmoc- derivatized amino acids were 340/450 nm and 266/305 nm, respectively. Two sets of standards containing 2 or 5 nmol of each amino acid and the internal standards were run prior to the samples of unknown amino acid composition.
Cloning of Genes for Amidohydrolase Enzymes
The genes for five putative N-acyl-d-amino acid deacylases were cloned into the NdeI/EcoRI or NdeI/HindIII restriction sites of the plasmid pET-30a+ (Novagen). The gene that encodes the protein Bb3285 (gi| 33602261) was amplified from the genomic DNA of B. bronchiseptica (ATCC BAA-588) using the primers 5′-GCAGGAGCCATATGCAGAACGCGGAAAAGCTGGATTTCAAGATTACCGG-3′ and 5′-CGCGGAATTCAGGCGCCGGCGCGCAGTACC-3′. The gene for the Bb2785 (gi| 33601761) was also cloned from the B. bronchiseptica genomic DNA with the primers 5′-GCAGGAGCCATATGCACGACGAACAGGCACTTGATCTTGTCATCCGC-3′ and 5′-CGGCAAGCTTCAGGCGGCCGCGGGCTCGGCC-3′. The gene that encodes the protein Gox1177 (gi| 58039630) was amplified from the genomic DNA of G. oxydans using the primers 5′-GCAGGAGCCATATGTTTGATCTCGTGATCCGTAATGGTCTGCTGG-3′ and 5′-CGCGGAATTCACACCACCCCGCAATACCCACGG-3′. The genomic DNA of G. oxydans was a generous gift of Dr. Armin Ehrenreich from the Georg-August University, Göttingen, Germany. The gene for Sco4986 (gi| 21223359) was cloned from the genomic DNA of S. coelicolor (ATCC BAA471-D) with the primers 5′-GCAGGAGCCATATGGAAGAGCTGGTCATCAGGGACGCCGACGTCGTGGACGG-3′ and 5′-CGCGGAATTCAGCGCGGAACCCGGCGCACCGACCGTCCGG-3′. The gene for Bll7304 (gi| 27382415) was cloned from Bradyrhizobium japonicum USDA 110 with the primers 5′-CGAGGAGCATTAATGCATGCTGCATCGGCATCCGCGGCGGTG-3′ and 5′CGCGGAATTCAGCCCTCGACGACCGGGCGGCGC-3′. The genomic DNA of B. japonicum was kindly donated by Dr. Hans-Martin Fischer from the Swiss Federal Institute of Technology, Zürich, Switzerland. The DNA polymerase chain reactions were performed using Platinum pfx polymerase from Invitrogen. The sequences of the cloned genes were confirmed by DNA sequencing using the facilities from the Gene Technologies Laboratory of Texas A&M University.
Expression and Purification
E. coli BL21(DE3) cells harboring the pET-30a+ plasmids containing the gene for the expression of Bb3285 or Bb2785 and Rosetta 2 (DE3) cells containing a pET-30a+ plasmid for the expression of Gox1177 or Sco4986 were grown in Terrific Broth with 50 μg/mL kanamycin (additionally 25 μg/mL chloramphenicol for Rosetta 2 cells) at 30 °C. When the optical density of the cells reached an OD600 of ∼0.6, 2.5 mM zinc acetate was added to the culture and expression of Bb3285 was induced with 100 μM isopropyl β-thiogalactoside (IPTG) or 1.0 mM IPTG for the expression of Bb2785, Gox1177 and Sco4986. Following induction, the cells were grown overnight at 16−19 °C. Cell lysis was achieved via sonication in 50 mM Tris, pH 7.5, and 100 μg/mL phenylmethanesulfonyl fluoride (PMSF) at 0 °C. The DNA present in the cell lysate was precipitated with protamine sulfate (2% w/w of cell mass) and removed by centrifugation. Solid ammonium sulfate was added slowly to the supernatant solution to a saturation of 60% and the protein isolated after centrifugation. The enzymes were further purified by size exclusion chromatography on a 3 L (1 L for Bb3285) column of AcA34 gel filtration medium using 50 mM Tris (Hepes for Bb3285) buffer, pH 7.5, at 4 °C. The fractions which contained the protein of the expected size were combined. The Gox1177 and Bb2785 proteins were further purified by anion exchange chromatography on a 6 mL Resource Q column from Pharmacia in 50 mM Tris, pH 7.5. The protein was eluted with a 0−1 M gradient of NaCl. The NaCl was removed in a final gel filtration step using a Superdex 200 size exclusion column in 50 mM Hepes, pH 7.5.
A total of 70 mg of Bb2785, 150 mg of Gox1177 and 142 mg of Bb3285 were isolated from 41, 36 and 66 grams of cells, respectively. The three proteins were greater than 95% pure as assessed by SDS-PAGE. The sequences of the first five amino acids for Gox1177 and Bb3285 were determined by the Protein Chemistry Laboratory at Texas A & M University. The two sequences matched those expected from the DNA sequence. The proteins Bb3285, Bb2785 and Gox1177 were found to contain 2.1, 1.8, and 1.8 equivalents of Zn as determined by ICP-MS. The protein Bll7304 was not purified since there was no expression from either BL21 (DE3) or Rosetta 2 (DE3) cells.
The Sco4986 protein was expressed in Rosetta 2 cells but the enzyme was found predominantly in the insoluble pellet after centrifugation. Rosetta 2 cells harboring the pET-30a(+) plasmid alone or the pET-30a(+)-Sco4986 plasmid were cultured under identical growth conditions and subsequently lysed via sonication in the presence of 50 mM Hepes, pH 7.5, 100 μg/mL PMSF. After centrifugation to remove insoluble proteins, the small molecules in the lysate fractions (MW <30 kDa) were separated via ultrafiltration using a PM-30 membrane (Millipore). The lysates were frozen and stored at −80 °C. The protein concentration in the crude cell lysates was determined by the Bradford assay (Biorad) with 4−28 μg/mL bovine serum albumin as a protein standard.
Alternatively, Sco4986 was expressed in BL21 (DE3) cells harboring the pET-30a+-Sco4986 and pOFXbad-SL3 (25) plasmids. The cells were grown under the same conditions in the presence of 50 μg/mL kanamycin and ampicillin. The pOFXbad-SL3 plasmid was a generous gift from Olivier Fayet at the Laboratory for Microbiology and Molecular Genetics in Toulouse, France. The pOFXbad-SL3 plasmid contains the genes for two molecular chaperones, groES/groEL, but overexpression of these genes was not induced. The cells expressing Sco4986 (0.2 mM IPTG) in the presence of the chaperone plasmid were used in the partial purification of Sco4986. Sco4986 was partially purified, through ion exchange, in the same fashion as Gox1177, with the exception of the sonication buffer (50 mM Hepes, 10% glycerol, 100 mM NaCl, 5 mM dithiothrietol, 100 μg/mL PMSF pH 8.5), the amount of protamine sulfate (0.4 % w/w cell mass) and the saturation level of ammonium sulfate (50−70% fraction). The N-acetyl-d-phenylalanine deacetylase activity of the gel filtration fractions was used to identify the protein. The ion exchange fraction of the highest activity was submitted to the Protein Chemistry Laboratory for amino acid sequence analysis. The components of the protein sample were separated by SDS-PAGE and blotted onto a PVDF membrane. The protein band which corresponded to the expected size (58 KDa) was removed and used for amino acid sequence analysis. The amino acid sequence of this band was identical to that expected for amino acids 2−5 of Sco4986.
Crystallization and Data Collection
Crystals of two different complexes (Table 1) were grown by the hanging drop method at room temperature for Bb3285 from B. bronchiseptica : (1) wild-type Bb3285, Zn2+, acetate, and formate, and (2) wild-type Bb3285, Zn2+, and inhibitor 3. The crystallization conditions utilized the following conditions: for Bb3285, Zn2+, acetate, and formate, the protein solution contained Bb3285 (12.6 mg/mL) in 20 mM Hepes (pH 8.0), 30 mM NaCl, and 0.05 mM ZnCl2; the precipitant contained 2.0 M sodium formate and 0.1 M sodium acetate (pH 4.6). For this sample crystals appeared in 2−3 days and exhibited diffraction consistent with the space group P6122, with two molecules of Bb3285 per asymmetric unit. For Bb3285, Zn2+, and inhibitor 3, the protein solution contained Bb3285 (12.6 mg/mL) in 20 mM Hepes (pH 8.0), 30 mM NaCl, 0.05 mM ZnCl2, and 10 mM methyl phosphonate inhibitor; the precipitant contained 1.0 M ammonium phosphate, and 0.1 M sodium citrate (pH 5.6). For this sample, crystals appeared in 7 days and exhibited a diffraction pattern consistent with space group P6122, with two molecules of Bb3285 per asymmetric unit.
Table 1.
Data collection and refinement statistics for crystals of N-acyl-d-Glutamate Deacetylase from Bordetella bronchiseptica
| Bb3285·ACY·FMT | Bb3285·Inhibitor | |
|---|---|---|
| Data collection | ||
| Space group | P61 22 | P61 22 |
| No. of mol. in asym. unit | 2 | 2 |
| Cell dimensions | ||
| a, c (Å) | 90.988, 507.134 | 91.291, 507.584 |
| Resolution (Å) | 25.0−1.5 (1.55−1.50) | 25.0−1.8 (1.86−1.80) |
| No. of unique reflections | 190443 (14871) | 106991 (7244) |
| Rmerge | 0.074 (0.219) | 0.065 (0.206) |
| I / σI | 22.3 (4.9) | 23.1 (6.8) |
| Completeness (%) | 95.4 (76.1) | 91.1 (69.8) |
| Refinement | ||
| Resolution (Å) | 25.0−1.5 (1.55−1.50) | 25.0−1.8 (1.86−1.80) |
| Rcryst | 0.205 (0.343) | 0.183 (0.243) |
| Rfree | 0.215 (0.347) | 0.207 (0.258) |
| R.m.s. deviations | ||
| Bond length (Å) | 0.005 | 0.005 |
| Bond angles (°) | 1.4 | 1.5 |
| No. atoms | ||
| Protein | 7112 | 7112 |
| Waters | 684 | 664 |
| Zn ions | 2 | 4 |
| Bound ligands | 2 Acetates and 4 Formates | Methyl phosphonate inhibitor |
| Ligand atoms | 20 | 30 |
| PDB entry | 3GIP | 3GIQ |
Values in parentheses are for the highest resolution shell.
Prior to data collection, the crystals of two Bb3285 forms (Table 1) were transferred to cryoprotectant solutions composed of their mother liquids and 20% glycerol and flash-cooled in a nitrogen stream. All X-ray diffraction data sets were collected at the NSLS X4A beamline (Brookhaven National Laboratory) on an ADSC CCD detector. Diffraction intensities were integrated and scaled with programs DENZO and SCALEPACK (26). The data collection statistics are given in Table 1.
Structure Determination and Model Refinement
The two Bb3285 structures (Table 1) were solved independently by molecular replacement with fully automated molecular replacement pipeline BALBES (27), using only input diffraction and sequence data. Partially refined structures of two Bb3285 complexes (Table 1) were the outputs from BALBES without any manual intervention. Subsequently, several iterative cycles of refinement were performed for each complex including: manual model rebuilding with TOM (28), refinement with CNS (29), automatic model rebuilding with ARP (30), and solvent building with the CCP4 suite (31).
Substrate Specificity of Bb3285, Gox1177, Sco4986 and Bb2785
The substrate specificities for Bb3285 and Gox1177 were determined by mixing each protein with the N-acetyl and N-succinyl amino acid libraries. The assays were conducted in 25 mM ammonium bicarbonate, pH 8.0, at 30 °C and the concentration of each amino acid derivative in these 17−20 member libraries was ∼100 M. The N-acetyl-d-Xaa, N-acetyl-l-Xaa, N-succinyl-d-Xaa, N-succinyl-l-Xaa, Gly-d-Xaa, l-Ala-d-Xaa, and l-Asp-d-Xaa libraries were incubated for 1−24 hours in the absence and presence of variable amounts of Gox1177 or Bb3285 (1−2000 nM). The reactions were quenched by removing the enzyme with a Microcon YM-10 (Millipore) membrane. A sample equivalent to ∼10 μg of the initial N-acyl-Xaa library was dried under vacuum prior to submission to the Protein Chemistry Laboratory of Texas A&M University for a determination of the liberated amino acids after the addition of each enzyme. The chromatographic peaks from the samples of unknown amino acid composition were identified by the migration time of the derivatized amino acids from known standards. The amino acids were quantified by integration of the chromatographic peaks. The relative substrate turnover rate in each library was determined by plotting the concentration of each liberated amino acid, Q, as a function of enzyme concentration (Et) at a fixed period of time, t, from a fit to equation 1, where A is the total concentration of each substrate and k is the relative rate constant for product formation. Additionally, single 24-hour incubations of Bb3285 or Gox1177 with the Gly-d-Xaa, l-Ala-d-Xaa and l-Asp-d-Xaa libraries were prepared and submitted for amino acid analysis as described above. Nineteen l-Xaa-d-Xaa dipeptide libraries were incubated at 30 °C with 0−2000 nM Gox1177 or Bb3285 for 3 hours and 1 hour, respectively. The concentration of each dipeptide component in these libraries was ∼100 μM in 50 mM Hepes, pH 7.5. The free amino acids were quantified with ninhydrin by measuring the change in absorbance at 507 nm and the rates of hydrolysis as a function of enzyme concentration were compared for each l-Xaa-d-Xaa library.
| (1) |
For the determination of the substrate specificity of Sco4986, 18 individual N-acetyl-d-amino acids (except proline and isoleucine) at 1.0 mM were incubated with the cell lysates from the pET-30a+ and pET-30a(+)–Sco4986 transformations. The final protein concentration from the cell lysates was 0.4 mg/mL in 50 mM Hepes, pH 7.5 and the solutions were incubated for 14 hours at 30 °C. In the same manner, these 18 substrates were treated with 2 μM Bb2785 for 20 hours.
Measurement of Kinetic Constants
The deacylation of N-formyl-d-Glu, N-acetyl-d-Glu, and N-succinyl-d-Glu by Bb3285 was assayed by coupling the formation of d-glutamate with the ninhydrin reagent in a manner similar to that previously reported (32). The assays were conducted in 50 mM Hepes, pH 7.5, at 30 °C. Specifically, one volume of the enzymatic assay was quenched with four volumes (two volumes for all dipeptides) of the ninhydrin reagent. The ninhydrin reagent consisted of 0.9 % w/v ninhydrin and 1:10:80 of 1g/mL CdCl2:glacial acetic acid:EtOH. The quenched samples were heated at 85 °C (80 °C for all didpeptides) for 15 minutes (5 minutes for all dipeptides) and the reaction product was detected at 507 nm. The kinetic parameters for the hydrolysis of the N-acetyl derivatives of d-Phe, d-Tyr, d-Met and d-Trp by the Sco4986 lysate were determined using the same ninhydrin assay. The deacylation of N-formyl-d-glutamate was coupled to the formation of NADH at 340 nm using 1.2 U/mL formate dehydrogenase in the presence of 10 mM NAD+ (33).
The deacetylation of the N-acetyl amino acid derivatives by Gox1177 was monitored by coupling the formation of acetic acid to the formation of NADH with acetyl-CoA synthetase (1.1 U/mL, citrate synthase (2.3 U/mL) and malate dehydrogenase (16 U/mL) in the presence 5.0 mM NAD+, 3.8 mM l-malate, 3.1 mM ATP, 3.2 mM MgCl2, and 148 μM CoA in 130 mM triethylamine, pH 8.4. In the case of N-acetyl-d-His, N-acetyl-d-Thr, N-acetyl-d-Gln, Leu-d-Leu, Met-d-Leu and Tyr-d-Leu, the reactions were monitored by measurement of the liberated amino acid with the ninhydrin reaction. During the course of the kinetic assays of Gox1177 with dipeptide substrates, the ninhydrin reagent was kept at 0 °C prior to heating.
The kinetic constants were determined by a fit of the data to equation 2 where v is the velocity of the reaction, Et is the total enzyme concentration, kcat is the turnover number, A is the substrate concentration, and Ka is the Michaelis constant.
| (2) |
Inhibition of Gox1177 and Sco4986 by N-Methylphosphonate Modified Amino Acids
The enzymatic activity of Gox1177 was monitored by the acetic acid coupling assay in the presence of 0−90 μM of N-methyl phosphonyl-d-Leu (1) at a substrate concentration of 3.0 mM N-acetyl-d-Leu. The partially purified lysate containing Sco4986 (0.1 mg protein/mL in the final assay) was pre-incubated with 0−5 μM of N-methyl phosphonyl-d-Phe (2) for 90 minutes at 30 °C. The Sco4986 assays were initiated by the addition of 1.5 mM N-acetyl-d-Phe (5% of the total assay volume) and the rate of d-phenylalanine formation was measured with the ninhydrin assay. The competitive inhibition constants were obtained from a fit of the data to equation 3.
| (3) |
Network Analysis
BLAST (34) analysis was performed, using two experimentally characterized N-acyl-d-amino-acid deacylases as seed sequences (gi|33602261 and gi|58039630) as queries against the NCBI NR database at an E-value cutoff of 1 × 10−45. Network analysis, coupled with literature information, was used to remove sequences with likely functions other than N-acyl-d-amino-acid deacylase. Obvious fragments (sequences with lengths under 300 amino acids) were removed, leaving approximately 250 N-acyl-d-amino-acid deacylase like sequences. A protein similarity network (35) based on BLAST results was generated to explore the relationships among these sequences using Cystoscape (36). The nodes were arranged using the yFiles organic layout provided with Cytoscape version 2.4. Connections between nodes are shown as edges (depicted as lines) if the E-value of the best BLAST hit between two sequences is at least as good as 1 × 10−45. Tools used for visualization of protein networks were created by the UCSF Resource for Biocomputing, Visualization, and Informatics, supported by NIH P41 RR-01081, and are available from the Resource (http://www.rbvi.ucsf.edu).
Sequence Alignment
Sequences in each of the four clusters represented in the Cytoscape network provided in Figure 10 were aligned using MUSCLE (37). Alignments were edited by hand to remove unaligned regions at the N- and C-termini. The edited alignments for Clusters 2, 3, and 4 in Figure 10 were combined using MUSCLE's profile alignment mode. The alignment was then filtered for ease of display, removing all sequences except Bb3285, Sco4986, Gox1177, 1m7j, and Bb2785.
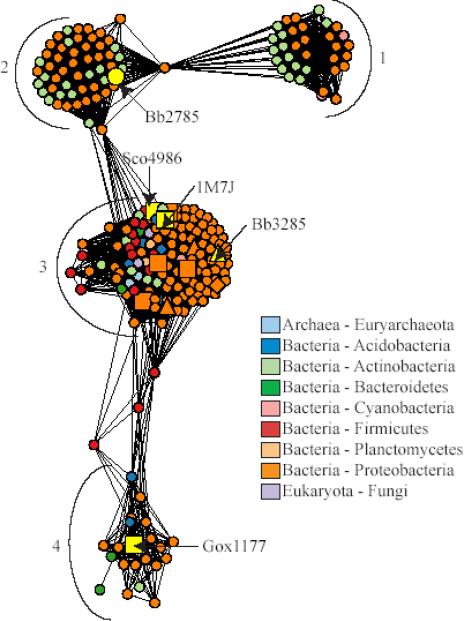
Figure 10.
Network representation of the sequence relationships in the N-acyl-d-amino-acid deacylase like sequence group. Each node in the network represents a single sequence and each edge represents the pairwise connection between two sequences with the most significant BLAST E-value (better than 1*10−45). Lengths of edges (depicted as lines) are not meaningful except that sequences in tightly clustered groups are relatively more similar to each other than sequences with few connections. Sequences that have been experimentally characterized are represented as follows: squares, N-acyl-d-amino-acid deacylases generally preferring uncharged substrates; triangles, N-acyl-d-glutamate deacylases; diamonds, N-acyl-d-aspartate deacylases. Sequences are colored according to phylum, except for 1M7J and those for the proteins whose substrate specificities were investigated in this work, which are labeled and colored yellow. The major clusters discussed in the text are indicated with numbers.
Results
Substrate Specificity of Bb3285
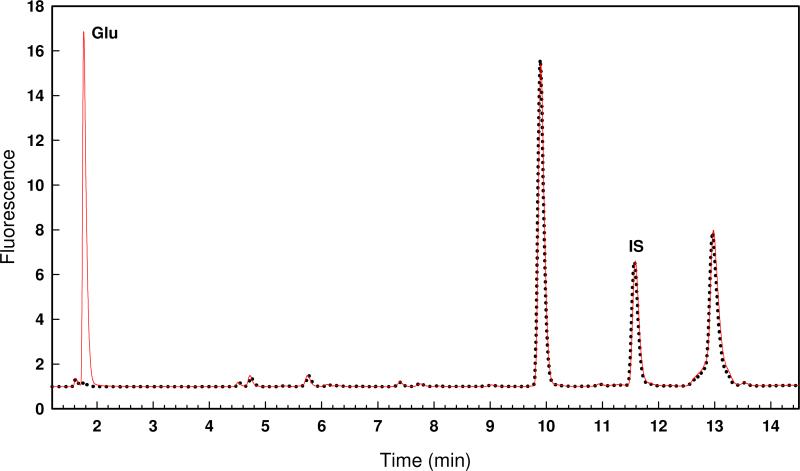
The determination of the substrate specificity of Bb3285 was initiated with the N-acetyl-d-Xaa library. Various concentrations of enzyme (0 − 0.1 μM) were mixed with a fixed concentration of the substrate library and allowed to react for 1 hour. The formation of free amino acids was measured with the ninhydrin assay and the results plotted as shown in Figure 2A. The maximum absorbance change at 507 nm was ∼0.19. If the entire substrate library had been hydrolyzed the change in absorbance would have been ∼3.5 and thus under these reaction conditions only a small fraction of the initial substrate library is hydrolyzed. A sample of the partially hydrolyzed reaction mixture was subjected to amino acid analysis to determine the identity of the N-acyl-amino acids that are functional substrates for Bb3285. The chromatogram is presented in Figure 3 as a continuous red line. A control reaction was conducted in the absence of any added Bb3285 and the chromatogram is shown in Figure 3 as a series of black dots. Relative to the control sample there is only a single amino acid, d-glutamate, that is formed from the hydrolysis of the N-acetyl-d-Xaa library after the addition of Bb3285. Therefore, the only compound to be hydrolyzed in this substrate library is N-acetyl-d-glutamate. The other members of this N-acetyl-d-amino acid library are hydrolyzed at less than 1% of the rate observed for N-acetyl-d-glutamate.
Figure 2.
Enzyme/time courses for the hydrolysis of various substrate libraries by Bb3285. (A) N-acetyl-d-Xaa (black circles), N-acetyl-l-Xaa (open circles), and N-succinyl-d-Xaa (red circles). (B) N-succinyl-d-Xaa (red circles) and N-succinyl-l-Xaa (open circles). (C) l-Ala-d-Xaa and (D) l-Asp-d-Xaa. The enzyme concentration is shown along the abscissa. In panels A, B, and C the enzyme was incubated with the substrate library for 1 hour whereas in panel D the substrate library was incubated with the enzyme for 24 hours at 30 °C. Additional details are provided in the text.
Figure 3.
HPLC chromatogram of the N-acetyl-d-Xaa library treated with no enzyme (black dots) and 20 nM Bb3285 (red line) for 1 hour at 30 °C. The OPA-derivatized d-glutamate was detected at a retention time of 1.7 minutes in the sample treated with Bb3285. The internal standard is labeled as IS. Additional details are provided in the text.
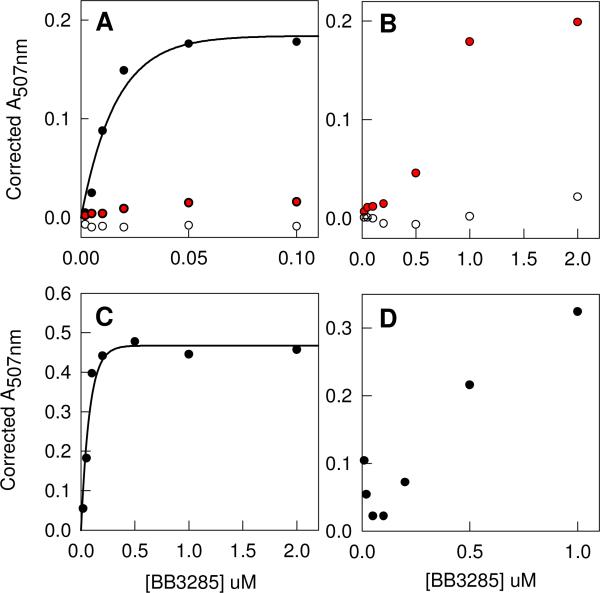
Bb3285 was also utilized as a catalyst for the hydrolysis of the N-succinyl-d-Xaa library and the results are presented in Figure 2A. Relative to the N-acetyl-d-Xaa library, the rate of hydrolysis is measurably slower. When the concentration of Bb3285 is increased 20-fold to 2.0 μM, the fraction of the N-succinyl-d-Xaa library that is hydrolyzed is approximately equal to that of the N-acetyl-d-Xaa library as illustrated in Figure 2B. Under these reaction conditions there is no evidence for the hydrolysis of the N-acetyl-l-Xaa (Figures 2A and 2B) or N-succinyl-l-Xaa substrate libraries (data not shown). Bb3285 was tested with two l-Xaa-d-Xaa dipeptide libraries and the results are shown in Figures 2C and 2D. With the l-Ala-d-Xaa dipeptide library, 1−2 dipeptides are hydrolyzed in 1 hour at an enzyme concentration of 0.2 μM. The hydrolysis of the l-Asp-d-Xaa library is considerably slower. Amino acid analyses of these substrate libraries indicate that only those compounds with a d-glutamate at the carboxy terminus are substrates for Bb3285. Bb3285 was subsequently screened for dipeptidase activity towards eighteen l-Xaa-d-Xaa libraries. The relative rates of hydrolysis for these libraries are summarized in Table 2.
Table 2.
Relative rates of hydrolysis for l-Xaa-d-Xaa dipeptide libraries.
| Library | Bb3285 | Gox1177 |
|---|---|---|
| l-Ala-d-Xaa | 16 | 48 |
| l-Arg-d-Xaa | 2 | 3 |
| l-Asn-d-Xaa | 4 | 26 |
| l-Asp-d-Xaa | 1 | 0.1 |
| l-Gln-d-Xaa | 2 | 5 |
| l-Glu-d-Xaa | 1 | 1 |
| l-Gly-d-Xaa | 64 | 5 |
| l-His-d-Xaa | 2.6 | 22 |
| l-Ile-d-Xaa | 5 | 7 |
| l-Leu-d-Xaa | 86 | 92 |
| l-Lys-d-Xaa | 5 | 1 |
| l-Met-d-Xaa | 100 | 76 |
| l-Phe-d-Xaa | 10 | 71 |
| l-Pro-d-Xaa | 37 | 52 |
| l-Ser-d-Xaa | 4 | 27 |
| l-Thr-d-Xaa | 3 | 37 |
| l-Trp-d-Xaa | 6 | 80 |
| l-Tyr-d-Xaa | 7 | 100 |
| l-Val-d-Xaa | 7 | 6 |
The kinetic constants were determined for the hydrolysis of N-formyl-, N-acetyl-, N-succinyl-, N-Met-, and N-Leu-derivatives of d-glutamate by Bb3285 at pH 7.5. The kinetic constants from fits of the data to equation 2 are presented in Table 3. Of the compounds tested, N-formyl-d-glutamate exhibited the highest values for kcat (2200 s−1) and kcat/Km (5.6 × 106 M−1 s−1). Somewhat lower values were obtained for N-acetyl-d-glutamate.
Table 3.
Kinetic parameters for Bb3285 with selected substrates at pH 7.5a
| Substrate | kcat, s−1 | KM, μM | kcat/KM, M−1s−1 |
|---|---|---|---|
| N-formyl-d-Glu | 2200 ± 72 | 380 ± 35 | (5.8 ± 0.6) × 106 |
| N-acetyl-d-Glu | 460 ± 9 | 88 ± 8 | (5.2 ± 0.5) × 106 |
| N-succinyl-d-Glu | 5.8 ± 0.4 | 60 ± 22 | (9.6 ± 3.5) × 104 |
| l-Leu-d-Glu | 3.6 ± 0.1 | 12 ± 1.4 | (3.0 ± 0.4) × 105 |
| l-Met-d-Glu | 2.9 ± 0.1 | 8.4 ± 1.4 | (3.4 ± 0.6) × 105 |
These data were obtained from a fit of the data to equation 2.
Substrate Specificity of Gox1177
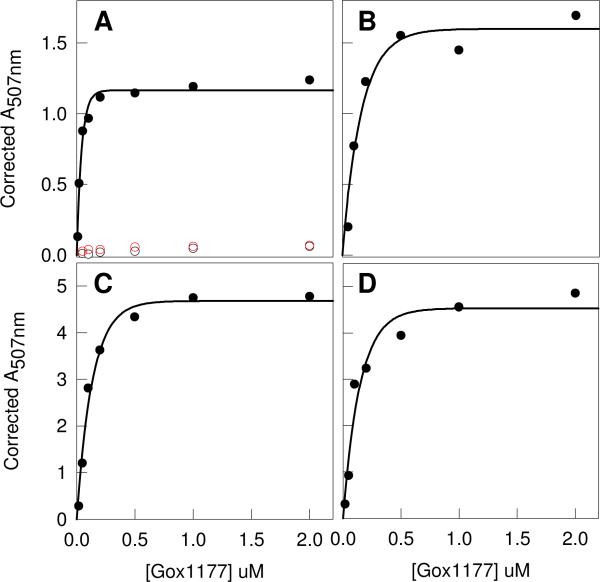
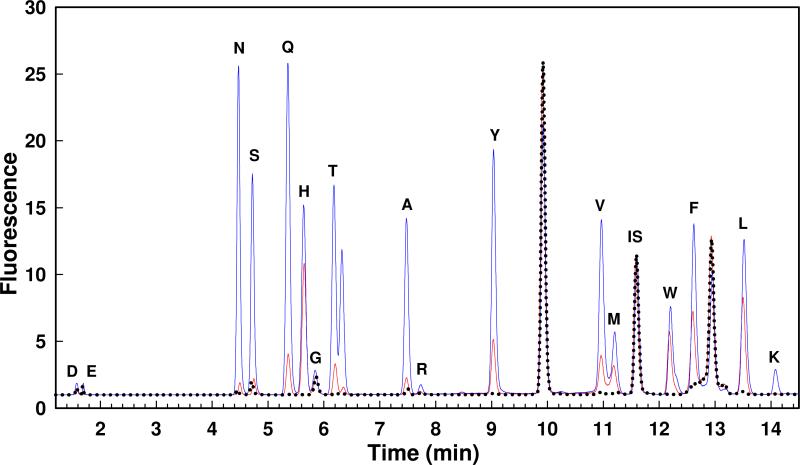
The substrate specificity for Gox1177 was initially interrogated with the N-acetyl-d-Xaa library. The enzyme (0−2 μM) was incubated for three hours with the N-acetyl-d-Xaa library and then the reaction was quenched by the addition of the ninhydrin reagent to determine the concentration of the free amino acids that were released following hydrolysis of the amide bond. The plot for the change in absorbance at 507 nm as a function of enzyme concentration is presented in Figure 4A. The maximum absorbance change at the highest enzyme concentrations is ∼1.1. This result indicates that approximately one third of the N-acyl-d-amino acids in this library were hydrolyzed under these reaction conditions. A fraction of the hydrolyzed N-acetyl-d-Xaa library was subjected to amino acid analysis to determine the specific compounds that are substrates for Gox1177. The HPLC chromatogram is presented in Figure 5 for samples that were incubated for 90 minutes with either 20 nM (red) or 200 nM (blue) Gox1177. This enzyme has substantially broader substrate specificity than does Bb3285 and shows a kinetic preference for the hydrolysis of N-acyl derivatives of the hydrophobic amino acids leucine, methionine, phenylalanine, tryptophan, tyrosine and valine, in addition to alanine, asparagine, glutamine, histidine, serine and threonine. The enzyme was unable to hydrolyze compounds contained within the N-acetyl-l-Xaa or N-succinyl-d-Xaa substrate libraries (Figure 4A).
Figure 4.
Enzyme/time courses for the hydrolysis of various substrate libraries by Gox1177. (A) N-acetyl-d-Xaa (black circles), N-acetyl-l-Xaa (open black circles) and N-succinyl-d-Xaa (red circles). (B) Gly-d-Xaa. (C) l-Ala-d-Xaa. (D) l-Asp-d-Xaa. In panels A, B, and C the enzyme was incubated with the substrate library for 3 hours whereas in panel D the substrate library was incubated with Gox1177 for 24 hours.
Figure 5.
HPLC chromatograms for the hydrolysis of the N-acetyl-d-Xaa library treated with no enzyme (black dots), 20 nM Gox1177 (red line) and 200 nM (blue line) Gox1177 for 90 minutes at 30 °C. The OPA-derivatized amino acids are indicated with their single letter code. The OPA-derivatized norvaline internal standard is indicated as IS. The unlabelled peak near threonine is likely oxidized methionine. Additional details are provided in the text.
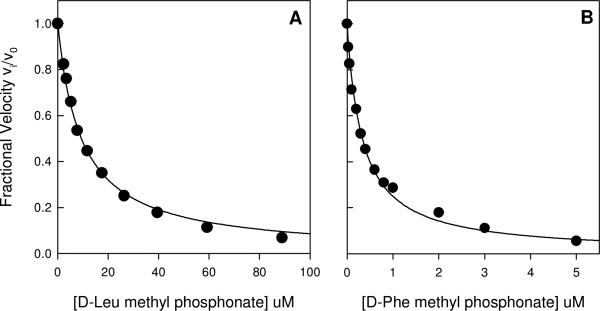
The breadth of the substrate profile for Gox1177 was further examined by utilizing the dipeptide libraries Gly-d-Xaa (Figure 4B), l-Ala-d-Xaa (Figure 4C) and l-Asp-d-Xaa (Figure 4D). A significant fraction of the compounds found in these libraries were hydrolyzed as substrates by Gox1177. To determine the relative ability of the dipeptide libraries to be hydrolyzed by Gox1177, the Gly-d-Xaa and 18 l-Xaa-d-Xaa (except l-cysteine) dipeptide libraries were mixed with 0−2 μM Gox1177 and the free amino acids produced after 3 hours were determined with the ninhydrin reagent. The relative rate constants are listed in Table 2. For the dipeptide libraries the enzyme has a preference for Tyr, Leu, Trp, Met, and Phe at the N-terminus. The kinetic parameters for the hydrolysis of l-Met-d-Leu, l-Leu-d-Leu, l-Tyr-d-Leu, and eleven N-acetyl-d-amino acid substrates by Gox1177 are provided in Table 4. At the C-terminal end of these N-acyl-d-amino acids, the preferred amino acids are leucine, tryptophan, phenylalanine, and tyrosine. The N-methylphosphonate derivative of d-leucine (1) was tested as a competitive inhibitor of Gox1177. At pH 8.4 this compound was found to be a competitive inhibitor versus N-acetyl-d-leucine with a Ki value of 4.9 ± 0.1 μM from a fit of the data to equation 3.
Table 4.
Kinetic parameters for Gox1177 with selected substratesa
| Compound | kcat, s−1 | Km, mM | kcat/Km, M−1s−1 |
|---|---|---|---|
| N-acetyl-d-Ala | 50 ± 4 | 18 ± 2 | (2.9 ± 0.4) × 103 |
| N-acetyl-d-Val | 16 ± 1 | 4.1 ± 0.2 | (3.9 ± 0.2) × 103 |
| N-acetyl-d-Leu | 101 ± 3 | 3.2 ± 0.2 | (3.2 ± 0.2) × 104 |
| N-acetyl-d-Met | 40 ± 2 | 1.8 ± 0.3 | (2.2 ± 0.4) × 104 |
| N-acetyl-d-His | nd | nd | (8.5 ± 0.2) × 103 |
| N-acetyl-d-Trp | 89 ± 2 | 2.2 ± 0.2 | (4.1 ± 0.3) × 104 |
| N-acetyl-d-Phe | 59 ± 2 | 2.5 ± 0.3 | (2.4 ± 0.3) × 104 |
| N-acetyl-d-Tyr | 66 ± 2 | 7.1 ± 0.5 | (9.3 ± 0.7) × 103 |
| N-acetyl-d-Thr | nd | nd | (6.0 ± 0.1) × 103 |
| N-acetyl-d-Asn | nd | nd | 480 ± 20 |
| N-acetyl-d-Gln | nd | nd | (2.7 ± 0.1) × 103 |
| N-formyl-d-Leu | 13 ± 1 | 5.9 ± 0.5 | (2.2 ± 0.2) × 103 |
| N-succinyl-d-Leu | nd | nd | 30 ± 1.4 |
| N-propionyl-d-Leu | 23 ± 1 | 1.2 ± 0.1 | (2.0 ± 0.1) × 104 |
| l-Leu-d-Leu | 31 ± 1 | 3.3 ± 0.2 | (9.2 ± 0.7) × 103 |
| l-Met-d-Leu | 20 ± 1 | 1.6 ± 0.2 | (1.3 ± 0.2) × 104 |
| l-Tyr-d-Leu | 19 ± 1 | 1.3 ± 0.2 | (1.5 ± 0.2) × 104 |
From fits of the data to equation 2.
Substrate Specificity of Sco4986 and Bb2785
The protein Sco4986 was unable to be purified to homogeneity but the substrate specificity was measured using clarified cell lysates. Control experiments were conducted using cells that contained the pET-30a(+) plasmid lacking the gene for expression of Sco4986. Incubation of the nineteen N-acetyl-d-amino acids with the lysates of Rosetta 2 BL21 (DE3) cells harboring the pET-30a(+) or pET-30a(+)-Sco4986 plasmids was used to determine if any background d-aminoacylase activity was present in E. coli and if the presence of the Sco4986 gene in the plasmid resulted in the lysate having d-aminoacylase activity. There was a significant increase in the rate of formation of free amino acids when the substrates and the Sco4986 lysate were together in solution vs. either of these components alone. It was determined that all of the N-acetyl-d-amino acids were substrates for Sco4986, with the exception of the N-acetyl derivatives of d-Asp, d-Glu, d-His, d-Lys, and d-Arg. For the N-acetyl-derivatives of d-Ala, d/l-Cys, d-Phe, d-Leu, d-Met, d-Gln, d-Val, d-Trp and d-Tyr, the rate of hydrolysis of the substrates by the pET-30a(+) control lysate ranged from 1% to 4% of that observed for the lysate containing Sco4986. The hydrolysis of the N-acetyl-Gly, N-acetyl-d-Asn, N-acetyl-d-Ser and N-acetyl-d-Thr substrates by the control lysate was 11−12% of the rate observed for the lysate containing Sco4986. The relative rates of hydrolysis are summarized in Table 5. The best of these substrates is N-acetyl-d-Phe (Km = 0.53 ± 0.03 mM) followed by N-acetyl-d-Trp (Km = 0.28 ± 0.03 mM), N-acetyl-d-Tyr (Km = 9.1 ± 0.5 mM), and N-acetyl-d-Met (Km = 6.2 ± 1.0 mM). The N-methylphosphonate derivative of d-Phe (2) was found to be an inhibitor of Sco4986 with a Ki of 87 ± 4 nM. The inhibition plots for Gox1177 and Sco4986 with compounds 1 and 2 are presented in Figure 6. No hydrolysis could be detected upon the incubation of Bb2785 with the 18 N-acetyl-d-amino acid compounds.
Table 5.
Relative rates of hydrolysis by Sco4986a.
| Substrate | Relative Rate |
|---|---|
| N-acetyl-d-Ala | 7 |
| N-acetyl-d-Asn | 5 |
| N-acetyl-d-Gln | 3 |
| N-acetyl-d-Gly | 1 |
| N-acetyl-d-Leu | 7 |
| N-acetyl-d-Met | 30 |
| N-acetyl-d-Phe | 100 |
| N-acetyl-d-Ser | 7 |
| N-acetyl-d-Thr | 3 |
| N-acetyl-d-Trp | 48 |
| N-acetyl-d-Tyr | 25 |
| N-acetyl-d-Val | 8 |
pH 7.5 at 1.0 mM substrate.
Figure 6.
Inhibition of Gox1177 and Sco4986 by the N-methylphosphonate derivative of d-leucine (1) and d-phenylalanine (2), respectively. (A) A competitive inhibition constant of 4.9 μM was obtained from a fit of the data to equation 3 at a substrate concentration of 3.0 mM N-acetyl-d-Leu. (B) A competitive inhibition constant of 87 nM was obtained from a fit of the data to equation 3 at a substrate concentration of 1.5 mM N-acetyl-d-Phe. Additional details are provided in the text.
Three-Dimensional Structure of Bb3285
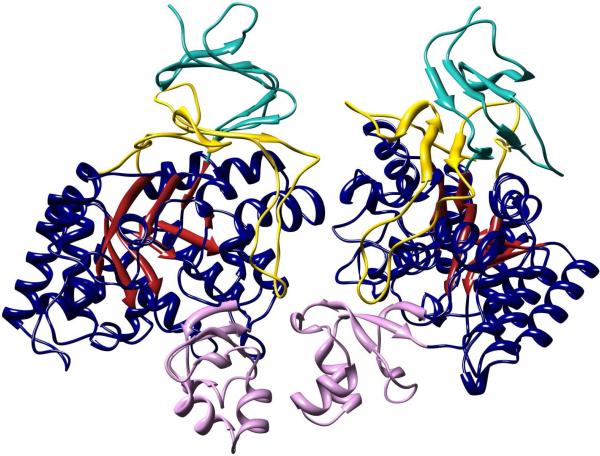
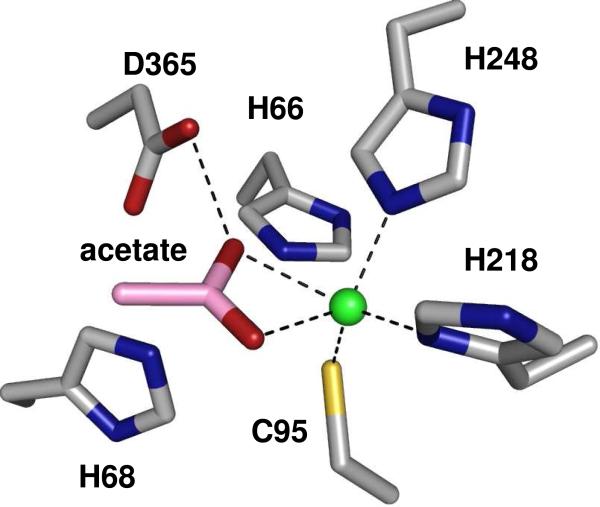
The crystal structure of Bb3285 was determined to a resolution of 1.5 Å as a homodimer with one zinc, one acetate and two formate molecules bound in the active site (Figure 7). The N-terminal residues 1−4 and the C-terminal residues 479−480 are disordered in both structures and are not included in the final model. Only one residue, Met-253, lies in the disallowed region of the Ramachandran plot. This residue is located in the loop L7 (after β-strand 7 of the barrel) and participates in hydrogen bond interactions with adjacent loop L6. In addition to the signature (β/α)8-barrel (shown in dark blue and red) two additional domains are present. The first of these domains is an insertion colored in pink (residues 287−344) between β-strand 7 and α-helix 7 of the (β/α)8-barrel. The second domain is a nine stranded β-barrel encompassing residues 5−61 (colored in teal) and residues 413−478 (colored in yellow) contributed from the N- and C-termini of the polypeptide. A long loop (residues 432−451) is inserted between the seventh and eighth β-strands of the β-barrel. The single zinc is bound in the β-metal site and is ligated by Cys-95, His-218, and His-248 and the acetate product as shown in Figure 8. In this complex one of the carboxylate oxygens from acetate is positioned 2.3 Å from the zinc and 2.6 Å away from one of the oxygens of the catalytic aspartate residue from the end of β-strand 8 (Asp-365). The other carboxylate oxygen in the acetate is 2.6 Å away from the phenolic oxygen of Tyr-190 and 2.3 Å from the zinc. The two histidines at the end of β-strand 1 (His-66 and His-68) do not ligate a second zinc in the α-metal binding site. One of the formate molecules is interacting with Lys-250, Tyr-282, and Arg-376 at distances of 2.7, 2.5, and 2.8 Å, respectively. The other formate forms a polar interaction with the side chain of Arg-295.
Figure 7.
Homodimeric structure of Bb3285. The (β/α)8-barrel is colored with red β-strands and dark blue α-helices and adjoining loops. The first insertion domain (from residue 287 to 344) containing the substrate specificity loop is colored pink. The second insertion domain consisting of residues 5−61 and 413−478 are colored cyan and yellow, respectively. Molecular graphics images in Figures 7 and 9 were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, supported by NIH P41 RR-01081 (38).
Figure 8.
Metal center of Bb3285 with one Zn ion (green) and acetate (pink carbons) bound in the active site. This image was created using the Pymol for Windows version 1.1r1 (DeLano, W.L. The PyMOL Molecular Graphics System (2002) on World Wide Web http://www.pymol.org).
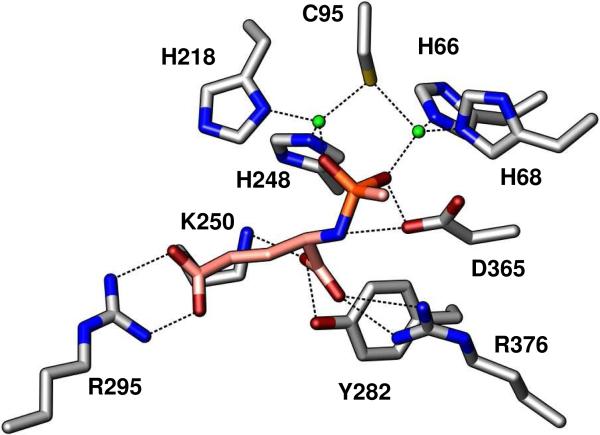
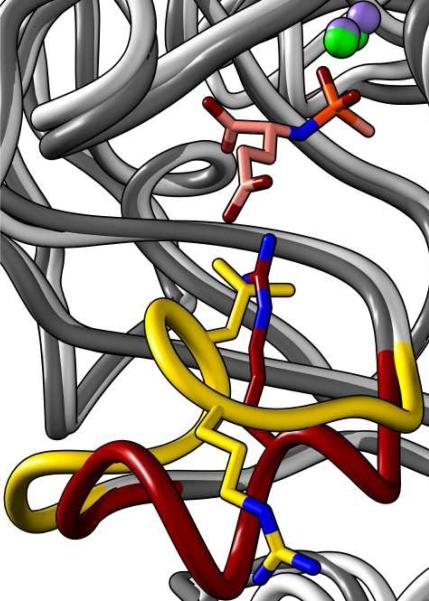
The second Bb3285 structure (Table 1) has well defined density for two Zn2+, and one inhibitor molecule bound in the active site of both molecules in the asymmetric unit. The 3-dimensional structure was solved to 1.8 Å resolution as a binuclear Zn enzyme with compound 3, a tight-binding inhibitor of Bb3285, bound in the active site (Figure 9). The histidine ligands, His-66 and His-68, at the end of β-strand 1 were coordinated at 2.1 and 2.0 Å to the Zn in the α-site. The Znβ-metal was ligated by His-218 and His-248 as in the native structure. The two metals were bridged by Cys-95 (2.4 and 2.3 Å) and each phosphoryl oxygen of compound 3 to either Znα (2.0 Å) or Znβ (2.0 Å). The C-terminal carboxylate of 3 was within hydrogen-bonding distance to Lys-250 (2.8 Å), Tyr-282 (2.7 Å) and Arg-376 (2.7 Å). The oxygens of the side-chain carboxylate of d-glutamate were 2.7−2.8 Å from the guanidino group of Arg-295. Though not shown, for the purpose of clarity, the hydroxyl oxygen of Tyr-190 is 2.7 Å from the phosphoryl oxygen which coordinates to Znβ. The catalytic aspartate at the end of the 8th β-strand is 3.5 and 2.6 Å from the amide nitrogen of 3 and phosphoryl oxygen that is coordinated to Znα.
Figure 9.
Binuclear Zn (green spheres) active site of Bb3285 with bound inhibitor (pink carbons, orange phosphorus). Enzyme-substrate contacts within 2.0−3.5 Å are indicated by dashed lines. For clarity, the 2.7 Å contact between the hydroxyl group of Y190 and the phosphoryl oxygen coordinated to Znβ is not shown.
Network Analysis of N-acyl-d-amino Acid Deacylases
Approximately 250 N-acyl-d-amino-acid deacylase like sequences were identified in the NCBI database. Four of these sequences are from eukaryotes, two are from archaea, and the rest are from bacteria. At an E-value cutoff of 1 × 10−45, the sequences partition into four main clusters, as shown in Figure 10. Each node in the network represents a single sequence and each edge represents a pairwise connection between two sequences. Edges (lines) are drawn only if the BLAST score connecting two proteins is at least as good as E = 1 × 10−45. Lengths of edges are not meaningful except that sequences in tightly clustered groups are more similar to each other than sequences with few connections.
With the exception of Gox1177, which was functionally characterized here, all of the sequences that have been shown experimentally to catalyze the deacylation of N-acyl-d-amino acids are found in cluster 3 (9, 17, 24, 39). Cluster 2 includes the enzyme Bb2785, for which specificity could not be determined on the basis of library screening using the N-acetyl-d-amino acid library. Cluster 1, which is clearly distinct from the other clusters in the network, contains no characterized enzymes.
Discussion
Five genes coding for putative d-aminoacylases were cloned and four of these enzymes were successfully expressed in E. coli: Bb3285, Bb2785, Sco4986 and Gox1177. Each of these enzymes was tested as a catalyst for the hydrolysis of N-acyl-d/l-Xaa substrates contained within a series of well-defined libraries of N-substituted amino acid derivatives. Using ninhydrin and/or HPLC-based assays, it was possible to measure the rate of hydrolysis of each library component by quantifying the specific amino acids liberated within these libraries as a function of time or enzyme concentration. Each enzyme was screened against more than 400 compounds.
Specificity of Bb3285
Bb3285 exclusively hydrolyzes derivatives of d-glutamate, where this amino acid is substituted with a simple acyl group or another amino acid. This enzyme will hydrolyze N-acetyl-, N-formyl- and N-succinyl-derivatives of d-glutamate but not the N-acyl-derivatives of any other d- or l-amino acid. Bb3285 hydrolyzes a variety of l-Xaa-d-Glu dipeptides but the best substrates are N-formyl- and N-acetyl-d-Glu. The N-formyl-d-Glu substrate has the highest value of kcat (2200 s−1) but the values for kcat/Km with this substrate and N-acetyl-d-Glu are essentially the same. The enzyme is less stringent regarding the identity of the l-amino acid at the amino-terminus of dipeptide substrates, but it does exhibit a preference for leucine or methionine derivatives of d-Glu.
Specificity of Gox1177
The second enzyme examined in this investigation, Gox1177, was found to deacetylate a broad range of N-acetyl-hydrophobic d-amino acids. Changing the acyl group from N-acetyl- to N-formyl-d-Leu results in a decrease in kcat and an increase in Km. With the l-Xaa-d-Xaa dipeptides, Gox1177 has a clear preference for the larger hydrophobic and aromatic residues (Tyr, Trp, Phe, Met and Leu) at the amino terminus. The relatively high Km values for Gox1177 with the substrates identified in this investigation calls into question whether N-acetyl-d-amino acids are necessarily the physiological substrates for this enzyme. Perhaps the native substrate of Gox1177 has a different functionality attached to the d-amino acid.
Specificity of Sco4986
The gene for Sco4986 was expressed in E. coli Rosetta 2 cells but the protein could not be purified to homogeneity because the protein was largely insoluble. However, the deacetylase activity of this protein could be detected in whole cell lysates. The relative rates of substrate hydrolysis clearly demonstrate that Sco4986 hydrolyzes many of the N-acetyl-d-amino acid derivatives with a preference for hydrophobic and aromatic amino acids. The Michaelis constants for the best substrates found with Sco4986 are ∼ 5-fold lower than the best substrates for Gox1177.
Activity of Bb2785
We were unable to identify any catalytic activity for Bb2785. Many of the residues whose side chains are expected to bind the substrate could not be identified in sequence alignments and thus if this protein has enzymatic activity the substrates were not included in our screening libraries.
Structural Basis for Substrate Specificity
The three-dimensional X-ray structures of Bb3285 in the presence of potent inhibitor 3, and of the acetate/formate complex, have revealed the structural determinants for enzyme specificity of this enzyme. In the complex with acetate and two formate molecules, one of the formate molecules is ion-paired with Arg-376 and Lys-250 in addition to a hydrogen bonding interaction with the phenolic side chain of Tyr-282. The arginine and tyrosine residues are fully conserved in Bb3285, Gox1177, and Sco4986 and thus these two residues are likely required for recognition of the α-carboxylate group at the C-terminus of the substrate. The arginine residue is also fully conserved among all of the enzymes that are ≥40% identical in amino acid sequence to Bb3285, Gox1177 or Sco4986. The tyrosine residue is semi-conserved but the substitution is limited to a histidine in these proteins. The lysine is conserved in those enzymes that are >40% identical in sequence to Bb3285 but is not conserved in those sequences that are more similar to Gox1177 or Sco4986.
The second formate in the active site of Bb3285 is ion-paired with the side chain guanidino group of Arg-295 and thus this interaction is likely required for recognition of the side chain carboxylate of the C-terminal d-glutamate. This assignment is confirmed by the structure of Bb3285 in the presence of compound 3, a mimic of the tetrahedral reaction intermediate (Figure 9). In Bb3285, Arg-295 is found in a loop that starts after the end of β-strand 7. This is the same loop that was proposed to serve as the specificity loop for the DAA from A. faecalis DA1, based upon a computational model of N-acetyl-d-methionine bound in the active site (21). The hydrophobic side chain of d-methionine was postulated to interact with Leu-298 in this enzyme. However, the corresponding residue, from a sequence alignment with Bb3285, is Glu-297 (see Figure 1) and thus it was not so clear how the side chain carboxylate of d-glutamate substrates would be able to interact with Glu-297 in Bb3285. The answer to this dilemma is found in a structural overlay of Bb3285 with the DAA from A. faecalis as shown in Figure 11 (38). The specificity loops in these two proteins adopt distinct conformations. In the A. faecalis structure, Leu-298 is pointed towards the active site and the side chain of Arg-296 is pointed away from the active site. However, in the Bb3285 structure, Arg-295 is pointed toward the active site. The conformational differences in these two loops are likely the result of a twist in the loop that is initiated by Pro-293 in Bb3285. Presumably, the side chain of N-acetyl-d-aspartate is not long enough to interact with Arg-295 and thus Bb3285 is unable hydrolyze substrates with a terminal d-aspartate.
Figure 11.
Stuctural overlay of Bb3285 (dark grey ribbon, green Zn) with the d-aminoacylase (PDB ID: 1M7J) from A. faecalis (light grey ribbon, purple Zn) showing the conformational differences between the loops in the two proteins that determine the differences in substrate specificity. The loop, colored yellow, from the d-aminoacylase from A. faecalis is for the residues 292 to 302. In this structure Leu-298 is pointing toward the active site and Arg-296 is pointing away from the active site. The loop colored red from Bb3285 is for residues 291 to 302. The phosphonamidate inhibitor is colored with pink carbons and an orange phosphorus. In this structure Arg-295 is pointing toward the active site and ion-paired with the guanidino group of the bound inhibitor.
Prediction of Functional Specificity for Uncharacterized Members of the N-acyl-d-Xaa Deacetylase Group
Conservation of functionally important residues in multiple sequence alignments is often used to infer similarity of functional characteristics. However, prediction of reaction specificity for uncharacterized proteins in the large multiple alignment representing the proteins in Figure 10 is equivocal. For example, two residues in the “specificity loops” of Bb3285, Pro293 and Arg295, likely play a role in the exclusive preference for derivatives of d-glutamate in this protein. However, only 7 sequences in Cluster 3 of the network show conservation of these residues, suggesting that only a few of these unknowns may share this preference.
All but one of the experimentally characterized deacylases represented in Figure 10 are in Cluster 3; their functional specificities vary and are in most cases broad, suggesting that substrate preferences for the many uncharacterized sequences in this cluster (small circles) may be difficult to infer from multiple alignments or protein similarity networks. A network generated using a more stringent E-value cut-off of 10−135 results in many more discrete clusters (network not shown). Clusters from this network that contain experimentally characterized proteins were examined for correlation with other properties, including organismal lineage, genome context, the identity of the bridging metal ligand, and variations in sequence length and the positions of associated inserts. No correlations could be made that cleanly support an assignment of reaction specificity to any of those clusters. One explanation for this result is that excluding sequence pairs that are >90% identical, there are 45 species (76 unique strains) with multiple N-acyl-d-amino acylase-like sequences among the ∼250 we identified. As is the case for the uronate isomerases, another group in the AHS for which the substrate specificities were recently reported (3), the multiple representatives of this sequence group within a single organism may have different substrate specificities, making it difficult to predict their functions without experimental and structural characterization of additional proteins in this sequence group.
For Cluster 2, Bb2785, the only protein that has been experimentally screened, showed no catalytic activity with N-acetyl-d-amino acid compounds. From the multiple alignment in Figure 1, this protein appears to be an outlier. Further, it is missing the last two histidine metal binding ligands (Figure 1), which could suggest that it catalyzes another, as yet unrecognized, reaction. The reaction specificity of none of the enzymes in Clusters 1 and 2 has been experimentally determined, raising questions about whether Bb2785 functions as an N-acyl-d-Xaa deacylase, based on the information currently available.
With respect to the physiological functions of the Cluster 3 sequence group, genome context does provide some possible clues. The gene for Sco4986 is adjacent to another open reading frame (Sco4987) that is currently annotated as a d-amino acid deaminase. The top five BLAST hits to the Sco4986 gene from different organisms show approximately 60% identity to this gene. All five are also adjacent or nearby to an ORF annotated as a d-amino acid deaminase, amino acid racemase-like protein or d-amino acid aldolase and these proteins are 38−50% identical in sequence to Sco4987. The similarities in genome context for homologues of these two genes among multiple organisms could suggest that the deacylase is liberating a free d-amino acid and then a second enzyme, the gene product of Sco4987, is racemizing or deaminating the d-amino acid. Acetyltransferases for d-amino acids are known and it is possible that this small cluster of genes is a simple two-enzyme system to metabolize N-acetyl-d-amino acids.
Footnotes
This work was supported by the NIH (GM 71790), the Robert A. Welch Foundation (A-840) and the Hackerman Advanced Research Program (010366−0034−2007). The X-ray coordinates and structure factors for Bb3285 have been deposited in the Protein Data Bank (PDB accession codes: 3gip and 3giq)
Abbreviations: AHS, amidohydrolase superfamily; DAA, d-aminoacylase; IPTG, isopropyl thiogalactopyranoside; PMSF, phenylmethanesulphonylfluoride.
References
- 1.Friedberg I, Jambon M, Godzik A. New avenues in protein function prediction. Protein Sci. 2006;15:1527–1529. doi: 10.1110/ps.062158406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marti-Arbona R, Xu C, Steele S, Weeks A, Kuty GF, Seibert CM, Raushel FM. Annotating Enzymes of Unknown Function: N-Formimino-l-glutamate Deiminase Is a Member of the Amidohydrolase Superfamily. Biochemistry. 2006;45:1997–2005. doi: 10.1021/bi0525425. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen TT, Brown S, Fedorov AA, Fedorov EV, Babbitt PC, Almo SC, Raushel FM. At the Periphery of the Amidohydrolase Superfamily: Bh0493 from Bacillus halodurans Catalyzes the Isomerization of d-Galacturonate to d-Tagaturonate. Biochemistry. 2008;47:1194–1206. doi: 10.1021/bi7017738. [DOI] [PubMed] [Google Scholar]
- 4.Hermann JC, Marti-Arbona R, Fedorov AA, Fedorov E, Almo SC, Shoichet BK, Raushel FM. Structure-based activity prediction for an enzyme of unknown function. Nature. 2007;448:775–779. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seibert CM, Raushel FM. Structural and Catalytic Diversity within the Amidohydrolase Superfamily. Biochemistry. 2005;44:6383–6391. doi: 10.1021/bi047326v. [DOI] [PubMed] [Google Scholar]
- 6.Pegg SC-H, Brown SD, Ojha S, Seffernick J, Meng EC, Morris JH, Chang PJ, Huang CC, Ferrin TE, Babbitt PC. Leveraging Enzyme Structure-Function Relationships for Functional Inference and Experimental Design : The Structure-Function Linkage Database. Biochemistry. 2006;45:2545–2555. doi: 10.1021/bi052101l. [DOI] [PubMed] [Google Scholar]
- 7.Li T, Iwaki H, Fu R, Hasegawa Y, Zhang H, Liu A. α-Amino-β-carboxymuconic-ε-semialdehyde Decarboxylase (ACMSD) Is a New Member of the Amidohydrolase Superfamily. Biochemistry. 2006;45:6628–6634. doi: 10.1021/bi060108c. [DOI] [PubMed] [Google Scholar]
- 8.Roodveldt C, Tawfik DS. Shared Promiscuous Activities and Evolutionary Features in Various Members of the Amidohydrolase Superfamily. Biochemistry. 2005;44:12728–12736. doi: 10.1021/bi051021e. [DOI] [PubMed] [Google Scholar]
- 9.Moriguchi M, Sakai K, Katsuno Y, Maki T, Wakayama M. Purification and Characterization of Novel N-Acyl-d-aspartate Amidohydrolase from Alcaligenes xylosoxydans subsp. xylosoxydans A-6. Biosci. Biotech. Biochem. 1993;57:1145–1148. doi: 10.1271/bbb.57.1145. [DOI] [PubMed] [Google Scholar]
- 10.Sugie M, Suzuki H. Purification and Properties of d-Aminoacylase of Streptomyces olivaceus. Agric. Biol. Chem. 1978;42:107–113. [Google Scholar]
- 11.Yang H, Zheng G, Peng X, Qiang B, Yuan J. d-Amino acids and d-Tyr-tRNATyr deacylase: stereospecificity of the translation machine revisited. FEBS Lett. 2003;552:95–98. doi: 10.1016/s0014-5793(03)00858-5. [DOI] [PubMed] [Google Scholar]
- 12.Soutourina O, Soutourina J, Blanquet S, Plateau P. Formation of d-Tyrosyl-tRNATyr Accounts for the Toxicity of d-Tyrosine toward Escherichia coli. J. Biol. Chem. 2004;279:42560–42565. doi: 10.1074/jbc.M402931200. [DOI] [PubMed] [Google Scholar]
- 13.Baltz RH, Miao V, Wrigley SK. Natural products to drugs: Daptomycin and related lipopeptide antibiotics. Nat. Prod. Rep. 2005;22:717–741. doi: 10.1039/b416648p. [DOI] [PubMed] [Google Scholar]
- 14.Sakai K, Oshima K, Moriguchi M. Production and Characterization of N-Acyl-d-Glutamate Amidohydrolase from Pseudomonas sp. Strain 5f-1. Appl. Environ. Microbiol. 1991;57:2540–2543. doi: 10.1128/aem.57.9.2540-2543.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakai K, Imamura K, Sonoda Y, Kido H, Moriguchi M. Purification and characterization of N-acyl-d-glutamate deacylase from Alcaligenes xylosoxydans subsp. xylosoxydans A-6. FEBS Lett. 1991;289:44–46. doi: 10.1016/0014-5793(91)80904-h. [DOI] [PubMed] [Google Scholar]
- 16.Chen H, Wu S, Wang K. d-Aminoacylase from Alcaligenes faecalis Possesses Novel Activities on d-Methionine. Bioorg. Med. Chem. 1994;2:1–5. doi: 10.1016/s0968-0896(00)82195-1. [DOI] [PubMed] [Google Scholar]
- 17.Lin P, Su S, Tsai Y, Lee C. Identification and characterization of a new gene from Variovorax paradoxus Iso1 encoding N-acyl-d-amino acid amidohydrolase responsible for d-amino acid production. Eur. J. Biochem. 2002;269:4868–4878. doi: 10.1046/j.1432-1033.2002.03190.x. [DOI] [PubMed] [Google Scholar]
- 18.Wakayama M, Kitahata S, Manoch L, Tachiki T, Yoshimune K, Moriguchi M. Production, purification and properties of d-aminoacylase from a newly isolated Trichoderma sp. SKW-36. Process Bioch. 2004;39:1119–1124. [Google Scholar]
- 19.Kumagai S, Kobayashi M, Yamaguchi S, Kanaya T, Motohashi R, Isobe K. A new d-aminoacylase from Defluvibacter sp. A 131−3. J. Mol. Catal. B: Enzym. 2004;30:159–165. [Google Scholar]
- 20.Lai W, Chou L, Ting C, Kirby R, Tsai Y, Wang AH, Liaw S. The Functional Role of the Binuclear Metal Center in d-aminoacylase: One-metal activation and second-metal attenuation. J. Biol. Chem. 2004;279:13962–13967. doi: 10.1074/jbc.M308849200. [DOI] [PubMed] [Google Scholar]
- 21.Liaw S, Chen S, Ko T, Hsu C, Chen C, Wang AH, Tsai Y. Crystal Structure of d-Aminoacylase from Alcaligenes faecalis DA1. J. Biol. Chem. 2003;278:4957–4962. doi: 10.1074/jbc.M210795200. [DOI] [PubMed] [Google Scholar]
- 22.Zhang P, Hao Z, Li Y. Synthesis and Steric Structure of α-Amino-β-lactam Derivative of 1,5-Benzothiazepines. Chem. J. Chin. Univ. 2002;23:2101–2105. [Google Scholar]
- 23.Sakai A, Xiang D, Xu C, Song L, Yew WS, Raushel FM, Gerlt JA. Evolution of Enzymatic Activities in the Enolase Superfamily: N-Succinylamino Acid Racemase and a New Pathway for the Irreversible Conversion of d- to l-Amino Acids. Biochemistry. 2006;45:4455–4462. doi: 10.1021/bi060230b. [DOI] [PubMed] [Google Scholar]
- 24.Xu C, Hall R, Cummings J, Raushel FM. Tight Binding Inhibitors of N-Acyl Amino Sugar and N-Acyl Amino Acid Deacetylases. JACS. 2006;128:4244–4245. doi: 10.1021/ja0600680. [DOI] [PubMed] [Google Scholar]
- 25.Castanie M-P, Berges H, Oreglia J, Prere M-F, Fayet O. A set of pBR322-compatible plasmids allowing the testing of chaperone-assisted folding of proteins overexpressed in Escherichia coli. Anal. Biochem. 1997;254:150–152. doi: 10.1006/abio.1997.2423. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Long F, Vagin AA, Young P, Murshudov GN. BALBES: a molecular-replacement Pipeline. Acta Crystallogr. Sect. D: Biol. Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones TA. Interactive computer graphics: FRODO. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- 29.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D: Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Lamzin VS, Wilson KS. Automated refinement of protein models. Acta Crystallogr. Sect. D: Biol. Crystallogr. 1993;49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 31.Bailey S. The CCP4 suite: Programs for Protein Crystallography. Acta Crystallogr. Sect. D: Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 32.Doi E, Shibata D, Matoba T. Modified Colorimetric Ninhydrin Methods for Peptidase Assay. Anal. Biochem. 1981;118:173–184. doi: 10.1016/0003-2697(81)90175-5. [DOI] [PubMed] [Google Scholar]
- 33.Lazennec C, Meinnel T. Formate dehydrogenase-coupled spectrophotometric assay of peptide deformylase. Anal. Biochem. 1997;244:180–2. doi: 10.1006/abio.1996.9910. [DOI] [PubMed] [Google Scholar]
- 34.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atkinson HJ, Morris JH, Ferrin TE, Babbit PC. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS ONE. 2009;4:e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;13:2498–2504. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Wakayama M, Watanabe E, Takenaka Y, Miyamoto Y, Tau Y, Sakai K, Moriguchi M. Cloning, Expression, and Nucleotide Sequence of the N-Acyl-d-Aspartate Amidohydrolase Gene from Alcaligenes xylosoxydans subsp. xylosoxydans A-6. J. Ferment. Bioeng. 1995;80:311–317. [Google Scholar]