Abstract
Two proteins from the amidohydrolase superfamily of enzymes were cloned, expressed and purified to homogeneity. The first protein, Cc0300, was from Caulobacter crescentus CB-15 (Cc0300) while the second one (Sgx9355e) was derived from an environmental DNA sequence originally isolated from the Sargasso Sea (gi| 44371129). The catalytic functions and the substrate profiles for the two enzymes were determined with the aid of combinatorial dipeptide libraries. Both enzymes were shown to catalyze the hydrolysis of L-Xaa-L-Xaa dipeptides where the amino acid at the N-terminus was relatively unimportant. These enzymes were specific for hydrophobic amino acids at the C-terminus. With Cc0300, substrates terminating in isoleucine, leucine, phenylalanine, tyrosine, valine, methionine, and tryptophan were hydrolyzed. The same specificity was observed with Sgx9355e but this protein was also able to hydrolyze peptides terminating in threonine. Both enzymes were able to hydrolyze N-acetyl and N-formyl derivatives of the hydrophobic amino acids and tripeptides. The best substrates identified for Cc0300 were L-Ala-L-Leu with values of kcat and kcat/Km of 37 s−1 and 1.1 × 105 M−1 s−1, respectively, and N-formyl-L-Tyr with values of kcat and kcat/Km of 33 s−1 and 3.9 × 105 M−1 s−1, respectively. The best substrate identified for Sgx9355e was L-Ala-L-Phe will values of kcat and kcat/Km of 0.41 s−1 and 5.8 × 103 M−1 s−1. The three-dimensional structure of Sgx9355e was determined to a resolution of 2.33 Å with L-methionine bound in the active site. The α-carboxylate of the methionine is ion-paired to His-237 and also hydrogen bonded to the backbone amide groups of Val-201 and Leu-202. The α-amino group of the bound methionine interacts with Asp-328. The structural determinants for substrate recognition were identified and compared with other enzymes in this superfamily that hydrolyze dipeptides with different specificities.
Complete bacterial genomes are now being sequenced at an extraordinary rate (1). This influx of new information has provided an unprecedented view of the evolutionary relationships that exist among enzymes that recognize different substrates and catalyze diverse functional transformations. The pace for the deposition of new three dimensional structures in the Protein Data Bank (PDB1) has also risen sharply in recent years (2). However, it is apparent that a significant fraction of the newly sequenced genes have an unknown, uncertain or incorrect catalytic functional assignment (3). This reality suggests that a considerable number of new metabolic transformations remain to be elucidated and that the annotation of function from sequence and structural information alone is a challenging problem. Nevertheless, the vast amount of structure and sequence information provides a unique opportunity for the elucidation of the mechanisms for the evolution of new catalytic functions from pre-existing structural templates.
One approach to the assignment of function for enzymes with an unknown substrate profile is the utilization of enzyme superfamilies. Comprehensive amino acid sequence alignments and three dimensional structural comparisons can provide valuable insights toward the identification of the catalytic and structural motifs that dictate the substrate and reaction profiles (4). One such superfamily that has proven to be amenable to this approach is the amidohydrolase superfamily (5, 6). This superfamily was first identified by Holm and Sander who recognized the structural and functional relationships among adenosine deaminase, urease, and phosphotriesterase (7). All enzymes from this superfamily adopt a (β/α)8-barrel structural fold with their active sites located at the C-terminal face of the β-barrel (5). Within the active site is a mono- or binuclear metal center that functions to activate solvent water for nucleophilic attack and/or stabilization of the transition state. Most of the enzymes within this superfamily catalyze the hydrolysis of C-O, C-N, or P-O bonds (5). However, some members at the periphery of the superfamily catalyze decarboxylation, hydration, and isomerization reactions (8).
In this paper we have interrogated the catalytic functions for two enzymes within the amidohydrolase superfamily. The first of these enzymes, Cc0300 from Caulobacter crescentus CB15, is currently annotated by NCBI as an Xaa-Pro dipeptidase. The second enzyme, derived from an environmental DNA sequence isolated from the Sargasso Sea and labeled here as Sgx9355e, is 39% identical in amino acid sequence with Cc0300. We have recently determined the catalytic functions for two other putative Xaa-Pro dipeptidases from C. crescentus, Cc2672 and Cc3125, that are ~40% identical in sequence to Cc0300 (9). These two enzymes do not catalyze the hydrolysis of Xaa-Pro dipeptides but they do catalyze the hydrolysis of L-Xaa-LArg/Lys dipeptides and thus the current functional annotations for these enzymes are incorrect. The X-ray structure of a catalytic homologue to Cc2672 and Cc3125 has unveiled the structural motif responsible for the recognition of the cationic side chains of arginine and lysine containing substrates in these enzymes. In Cc0300, this recognition motif is missing, suggesting that the substrate specificity for Cc0300 differs from that determined previously for Cc2672 and Cc3125. The substrate profiles for Cc0300 and Sgx9355e were determined by screening multiple dipeptide libraries containing nearly all possible combinations of L-Xaa-L-Xaa dipeptides made from the 20 common amino acids. The X-ray structure of the methionine-bound form of Sgx9355e has been determined to a resolution of 2.33 Å.
Materials and Methods
Materials
Genomic DNA from C. crescentus CB15 was purchased from ATCC (American Type Culture Collection). The synthesis of oligonucleotides and DNA sequencing reactions were performed by the Gene Technology Laboratory of Texas A&M University. The pET-30a(+) expression vector was obtained from Novagen. T4 DNA ligase and various restriction enzymes were acquired from New England Biolabs. Platinum Pfx DNA polymerase was purchased from Invitrogen. The Wizard Plus SV Mini-Prep DNA purification kit was obtained from Promega. Chromatographic columns and resins were from G.E. Healthcare. Chelex-100 resin was purchased from BioRad. ICP standards were purchased from Inorganic Ventures Inc. The tripeptides, L-Gly-L-Phe-L-Arg and L-Gly-L-Ala-L-Tyr, were purchased from Aroz Technologies LLC. NAD and formate dehydrogenase from Candida boidinii were purchased from Sigma. The N-methyl phosphonate derivative of L-leucine was synthesized according to the method described by Xu et al. (10). The structure of this compound is presented in Scheme 1. All other buffers, purification reagents and other chemicals used in this work were purchased from Sigma-Aldrich, unless otherwise stated.
Scheme 1.
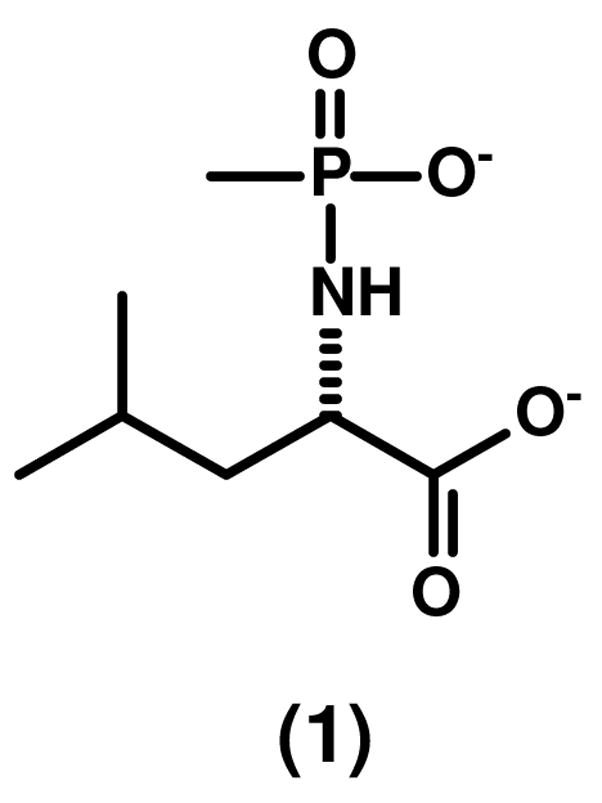
Synthesis of Dipeptides and Dipeptide Libraries
Syntheses of the L,L-dipeptide libraries were conducted as reported previously (9). Mass spectrometric analysis (ESI, positive and negative mode) was employed to verify the components of each library. The individual dipeptides L-Ala-L-Leu, L-Ala-L-Phe and L-Asp-L-Leu were prepared in a manner similar to the synthesis of the dipeptide libraries.
Gene cloning
Standard molecular cloning techniques were employed throughout (11). A 1287-bp fragment containing the entire open reading frame of Cc0300 was amplified from the genomic DNA of C. crescentus CB15 using the primer pair 5′ AGAACTTCCATA TGCGTAAATTGATGGCGGGCGCCTGC 3′ (forward) and 5′ ACGGAATTCTTACTTGTAGACGA CGCCGCCCTTGATGAC 3′ (reverse). The forward and reverse primers were designed to introduce an NdeI restriction site and an EcoRI restriction site, respectively. The PCR product, purified using a PCR clean-up system (Promega) and digested with NdeI and EcoRI, was ligated into the same sites of the expression vector pET-30a(+). The cloned fragment was completely sequenced to verify the fidelity of the PCR amplification. Based upon the reported sequence of gi|44371129 deduced from a DNA sample originally isolated from the Sargasso Sea, the New York SGX Research Center for Structural Genomics (NYSGXRC, www.nysgxrc.org) cloned and expressed Sgx9355e as a His-tagged protein following codon-optimization and gene synthesis by Codon Devices, Inc (Cambridge, MA). NYSGXRC will make the clone (9355e1BCt10p) available through the Protein Structure Initiative Material Repository (PSI-MR), accessible from the PSI Knowledgebase (http://kb.psi-structuralgenomics.org/KB/psi_resources.html).
Protein Purification
The plasmid for Cc0300 was constructed for expression under control of the IPTG-inducible T7 RNA polymerase. BL21(DE3) Star cells (Novagen) were transformed with the plasmid containing the gene for Cc0300. A single colony was used to inoculate a 5-mL overnight culture of LB medium containing 50 μg/mL kanamycin. The 5-mL overnight culture was then used to inoculate 1.0 L of LB medium containing 50 μg/mL kanamycin. The cells were grown at 30 °C and induced by adding IPTG at a final concentration of 0.5 mM when an OD600 of the culture reached 0.6. Zn(OAc)2, 1.0 mM, was added to the culture and the cells harvested by centrifugation (6,000 rpm for 15 minutes) 14 hours after IPTG induction and stored at −80 °C. To purify Cc0300, 10 g of frozen cells were thawed and resuspended in 50 mL of 50 mM Hepes, pH 8.0. The cells were disrupted by sonication in an ice bath and the soluble fraction was isolated by centrifugation. The supernatant solution was treated with 2% (w/v) protamine sulfate to precipitate nucleic acids, which were removed by centrifugation. The supernatant solution was treated with ground ammonium sulfate until 60% saturation. The pellet from the ammonium sulfate fractionation step was dissolved in a minimal volume of 50 mM Hepes, pH 8.0, and then passed through a 0.2 μm pore filter prior to loading onto a pre-equilibrated Hi Load 26/60 Superdex 200 prep grade gel filtration column (Amersham Biosciences). Fractions containing the target protein were pooled based on SDS-PAGE analysis. The protein was further purified by loading onto a Resource Q anion exchange column (Amersham Biosciences) and eluted with a linear gradient of NaCl in 20 mM Hepes, pH 8.0. The appropriate fractions were pooled after analysis by SDS-PAGE. Protein concentrations were determined using the calculated extinction coefficient of 31,860 M−1cm−1 at 280 nm (12).
For purification of Sgx9355e, E. coli BL21 (DE3) Star cells were transformed with the plasmid encoding the gene for Sgx9355e. One liter of LB medium was inoculated with a 5 mL overnight culture. The inoculated culture was grown with agitation at 30 °C to an OD600 of 0.6 and then supplemented with 1.0 mM Zn(OAc)2 and induced with IPTG at a final concentration of 0.5 mM. The cells were grown at 30 °C for another 14 h and then harvested by centrifugation (6000 rpm for 15 minutes). The cell pellet was resuspended in binding buffer (20 mM Hepes, pH 7.9, 0.5 M NaCl and 5 mM imidazole). The cells were disrupted by sonication and the insoluble debris was removed by centrifugation (10,000 rpm for 15 minutes). The clarified cell extract was applied to a 24 mL column of chelating Sepharose Fast Flow resin (Amersham Biosciences) charged with Ni2+ and pre-equilibrated with binding buffer. The column was washed thoroughly with 1000 mL of binding buffer until the absorbance of the flow-through at 280 nm did not change. The His-tagged protein, Sgx9355e, was eluted with a linear gradient of elution buffer (10 mM Hepes, pH 7.9, 0.25 M NaCl and 0.5 M imidazole). The protein obtained was further purified by application to a Resource Q anion exchange column after the NaCl was removed by dialysis. Sgx9355e was eluted with a linear gradient of NaCl in 20 mM Hepes, pH 8.0. The protein was greater than 95% pure as judged by SDS-PAGE analysis.
Amino Acid Sequence Verification
N-terminal amino acid sequence analysis of the purified Cc0300 was conducted by the Protein Chemistry Laboratory at Texas A&M University. The amino acid sequence obtained for the first 8 amino acid residues of Cc0300 was QATFVQAG, which indicates that the first 23 amino acid residues of the protein were either post-translationally removed via proteolysis or the protein was initially expressed from the 24th amino acid residue of the reported gene sequence.
Metal Analysis
Apo-Sgx9355e was made by incubating the enzyme with 2.0 mM 1,10-phenanthroline for 24 h at 4 °C. The chelator was removed by loading the protein/chelator mixture onto a PD-10 column (GE Health care) and eluting with 50 mM metal-free Hepes buffer, pH 8.0. The metal-free Hepes buffer was made by passing through a Chelex-100 column to remove possible metal salts present in the buffer. All glassware used for making metal-free buffer was soaked with 10 mM EDTA overnight and then rinsed thoroughly with deionized water. The apo-Sgx9355e was reconstituted with 2 equivalents of ZnCl2 in 50 mM metal-free Hepes, pH 8.0, in the presence of 10 mM potassium bicarbonate. The metal content of as-purified and reconstituted proteins was determined by inductively coupled plasma emission-mass spectrometry (ICP-MS) (13).
Assay Methods for Peptidase Activity
A modified colorimetric ninhydrin assay method was used to screen the dipeptidase activity of Cc0300 and Sgx9355e (14). The same method was used to measure the hydrolysis of tripeptides and the hydrolytic activity with N-acetyl- and N-formyl derivatives of L-amino acids. The absorbance at 507 nm was recorded with a SPECTRAmax plate reader from Molecular Devices. Quantitative analysis of the liberated amino acids was determined by the Protein Chemistry Lab at Texas A&M University.
Screening of Dipeptide Libraries
Cc0300 and Sgx9355e were tested for dipeptidase activity with the entire dipeptide library (361 compounds in all). Each dipeptide library consisted of a mixture of dipeptides with an identical L-amino acid at the N-terminus but 19 different amino acids at the C-terminus. L-Cysteine was not included in any of the libraries. Screening of the dipeptide libraries was initiated by mixing a fixed concentration of the dipeptide library (containing ~0.1 mM of each dipeptide) and various amount of enzyme over a concentration range of 10 nM to 1000 nM. All reactions were conducted in 50 mM Hepes, pH 8.0 in 96-well microplates. The reactions were incubated at 30 °C for a fixed time period and then quenched by adding the Cd-ninhydrin reagent. The quenched reactions were incubated for 5 minutes at 80 °C for color development. Control reactions without enzyme or without the dipeptide library were conducted simultaneously. The relative rates were determined by a fit of the data to equation 1, where y is the change in absorbance at 507 nm, x is concentration of enzyme and b is the relative rate constant. In these measurements the enzyme concentration was varied but the reaction time was fixed.
| (1) |
Amino Acid Analysis
The specific amino acids that were released from the dipeptide libraries were quantitatively identified by HPLC. The substrate specificity for either Cc0300 or Sgx9355e was determined by assay with selected dipeptide libraries. The hydrolysis reactions were conducted in 25 mM ammonium bicarbonate buffer, pH 8.0. For each dipeptide library assay, a mixture of 19 dipeptides at ~0.1 mM was subjected to hydrolysis with variable concentrations of enzyme ranging from 1 nM to 1000 nM. The identity of the amino acids released from the hydrolysis reactions was determined by amino acid analysis. Two dipeptide libraries (L-Ala-L-Xaa and L-Asp-L-Xaa) were assayed with Cc0300 and two dipeptide libraries (L-Ala-L-Xaa and L-Gln-L-Xaa) were used in the assays with Sgx9355e. The enzyme and dipeptide library were incubated at 30 °C for 14 h in a 96-well microplate and then the enzyme was removed by passage through a filter. Quantitative amino acid analysis was conducted after the samples were dried under reduced pressure and reconstituted with water. The relative reaction rates were determined by fitting the change in amino acid concentration as a function of enzyme concentration to equation 1.
Kinetic Measurements for Individual Substrates
Selected dipeptides (L-Ala-L-Leu, L-Ala-L-Phe, L-Asp-L-Leu), N-formyl-L-amino acids (N-formyl-L-Leu, N-formyl-L-Ile, N-formyl-L-Phe, N-formyl-L-Tyr, N-formyl-L-Met, N-formyl-L-Val), N-acetyl-L-amino acids (N-acetyl-L-Leu, N-acetyl-L-Ile, N-acetyl-L-Phe, N-acetyl-L-Tyr, N-acetyl-L-Met, N-acetyl-L-Val) and the tripeptide, L-Gly-L-Ala-L-Tyr, were used as substrates for measurement of the kinetic parameters for Cc0300 and Sgx9355e. The kinetic assays were conducted in 50 mM Hepes, pH 8.0, and the products were quantified using the ninhydrin-based assay. The kinetic constants, kcat, Km and kcat/Km, were determined by fitting the initial velocity data to equation 2, where ν is the initial velocity, kcat is the turnover number, Et is the enzyme concentration, A is the substrate concentration, and Km is the Michaelis constant.
| (2) |
Inhibition by Tetrahedral Analogue
The inhibitory properties of the N-methyl phosphonate derivative of L-leucine (1) with Cc0300 were determined using N-formyl-L-leucine as the substrate. The assays were conducted by monitoring the rate of formate production using a formate dehydrogenase coupling system in the presence of NAD. The reaction was followed spectrophotometrically at 340 nm, using a SPECTRAmax (Molecular Devices) plate reader. All assays were conducted in a 96-well UV plate in a volume of 250 μL. The buffer used in all assays was 50 mM Hepes, pH 8.0. The concentrations of N-formyl-L-leucine and the N-methyl phosphonate derivative of L-leucine were varied in the range of 50–200 μM and 1–100 μM, respectively. For data analysis, the initial rates for substrate hydrolysis in the presence of the inhibitor at different concentrations of the substrate were fitted to equation 3 for competitive inhibition. In this equation, νis the initial velocity. A is the substrate concentration, I is the inhibitor concentration, kcat is the turnover number, Et is the enzyme concentration, A is the substrate concentration. Km is the Michaelis constant for the substrate, and the Ki is the competitive inhibition constant.
| (3) |
Crystallization and Data Collection for Sgx9355e
Crystals of the selenomethionine form of Sgx9355e protein were grown using the sitting drop vapor diffusion method at 20 °C. Equal volumes of protein (10–15 mg/mL) and a solution containing 30% MPD, 0.1 M Hepes, 0.2 M NaCl, pH 7.0, were mixed and equilibrated against a 800 μL reservoir containing the same precipitant solution. A single crystal was flash frozen by adding 20% glycerol to the mother liquor before the crystal was loop mounted. SAD data extending to 2.33 Å were collected at the selenium absorption edge (λ = 0.979 Å) from a single crystal at liquid nitrogen temperature using the ADSC QUANTUM 315 detector at the National Synchrotron Light Source (NSLS) using beamline X29. X-ray diffraction images were processed with HKL2000 and the data collection statistics are given in Table 1 (15). The crystals belong to the monoclinic space group I4 with cell dimensions, a = b = 144.9, c = 101.0 Å (Table 1). Assuming two molecules of 45,781 Da per asymmetric unit, the Matthews coefficient is 2.0 Å3 Da−1 corresponding to an estimated solvent content of 47% by volume of the unit cell.
Table 1.
Crystal data, phasing and refinement statistics for Sgx9355e
Cell dimensions: a = b =144.9, c = 101.0 Å and α = β = γ = 90°; Space group I4
| Data set | Peak |
|---|---|
| Wavelength (Å) | 0.9791 |
| Resolution range (Å) | 50.0-2.33(2.41-2.33) |
| Unique reflections | 44,488 |
| Completeness (%) | 99.8(98.4) |
| Redundancy | 19(12.5) |
| Rmerge1 | 0.10(0.29) |
| Phasing power2 (ano) | 2.95 |
| FOM3: SAD (centric/acentric) | 0.18/0.55 |
| After solvent flattening | 0.95 |
| Refinement Statistics | |
| R-factor4/R-free | 0.21/0.23 |
| Resolution range (Å) | 50.0–2.33 |
| Number of atoms | |
| Proteins | 6165 |
| Waters | 285 |
| Heterogen atoms | 20 |
| RMS deviation from ideality | |
| Bonds (Å) | 0.007 |
| Angles (°) | 1.3 |
Rmerge = Σj(|Ih−<I>h|)/ΣIh, where <Ih> is the average intensity over symmetry equivalents
Phasing power and
FOM (Figure of merit) are as defined in SHARP
R-factor =Σ|Fobs−Fcalc|/Σ|Fobs|
Structure Determination and Refinement of Sgx9355e
The structure was determined by the single-wavelength anomalous dispersion method. Twenty two out of the possible twenty four selenium positions in the asymmetric unit were located using SHELXD (16). These selenium positions were refined and phases calculated with SHARP followed by solvent flattening (17, 18). The resulting experimental electron density map was of excellent quality and revealed practically all of the secondary structural elements comprising the dimer. The automated model building program ARP/wARP was then used to build the model into the electron density map (19). This process built about 87% of each protomeric polypeptide chain and the remainder of the model was completed using the graphics program ‘O’ (20, 21). The structure was refined with the program Crystallography and NMR system (CNS) (22). Cycles of manual rebuilding and structure refinement were continued until the convergence of R and Rfree. Phasing and refinement statistics are provided in Table 1. The final model was validated with the program PROCHECK (23). Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB id: 2qs8).
1. Results
Purification of Cc0300 and Sgx9355e
Cc0300 from C. crecentus was expressed in reasonable quantities after the plasmid containing the gene for this enzyme was transformed in BL21(DE3) Star cells and allowed to grow in an LB medium supplemented with 1.0 mM Zn(OAc)2. The enzyme was purified and SDS-PAGE analysis showed a single protein band which was slightly smaller than the theoretical molecular weight of 44 kDa as calculated from the reported gene sequence. N-terminal protein sequence analysis of the purified Cc0300 indicated that the first 23 amino acids were missing. This result indicates that either these residues were lost through proteolysis or that this gene has an alternate initiation site. The as-purified protein was found to contain an average of 1.6 equivalents of Zn2+ per subunit. Sgx9355e was purified as a His-tagged protein and was found to contain 0.2 equivalents of Zn per subunit. However, the Zn content could be increased to an average of 1.2 equivalents per subunit by incubating apo-Sgx9355e with 2 equivalents of ZnCl2 in the presence of 10 mM bicarbonate overnight.
Crystal Structure of Sgx9355e
The structure of Sgx9355e is shown in Figures 1 and 2. Within the crystal, the protein occurs as a homo octamer with approximately 3759 Å2 buried surface area (~19% of the total surface area) per protein-protein interaction (Figure 1). Each subunit consists of two domains, a small predominantly β-stranded domain (residues 7–64 and 379–412) and a large TIM barrel domain (residues 65–370) as presented in Figure 2. The active site is located at the bottom of the cone-shaped structure of each subunit and is open to solvent.
Figure 1.
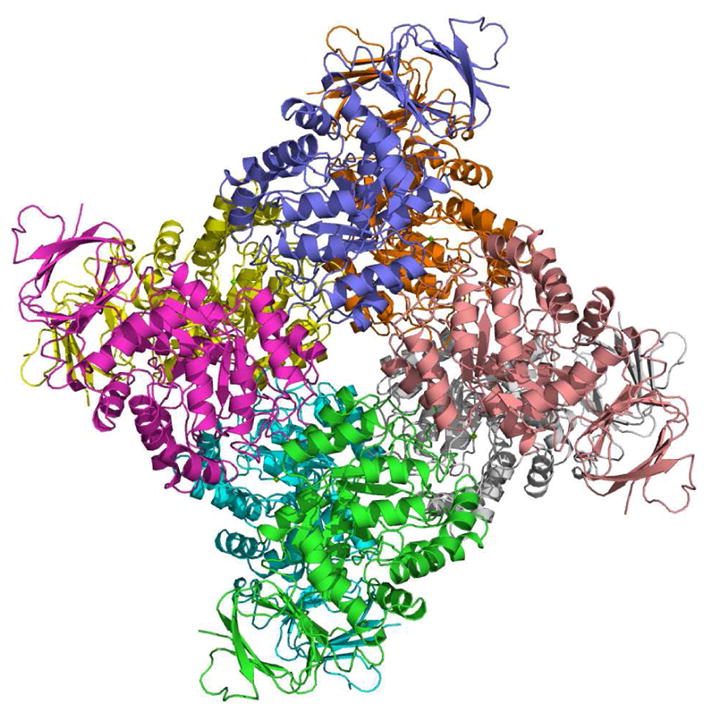
Structure of Sgx9355e. Ribbon representation of the Sgx9355e homo-octamer with different subunits shown in green, yellow, blue, magenta, cyan, gold, brown and grey.
Figure 2.
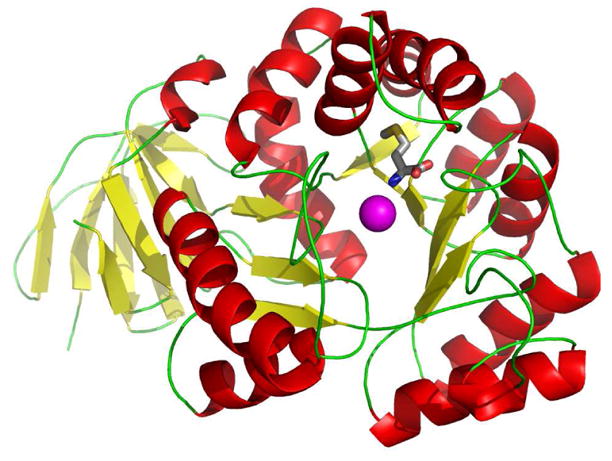
Ribbon representation of a single subunit of Sgx9355e with the bound methionine shown as a stick figure. The magnesium ion is shown as a magenta sphere.
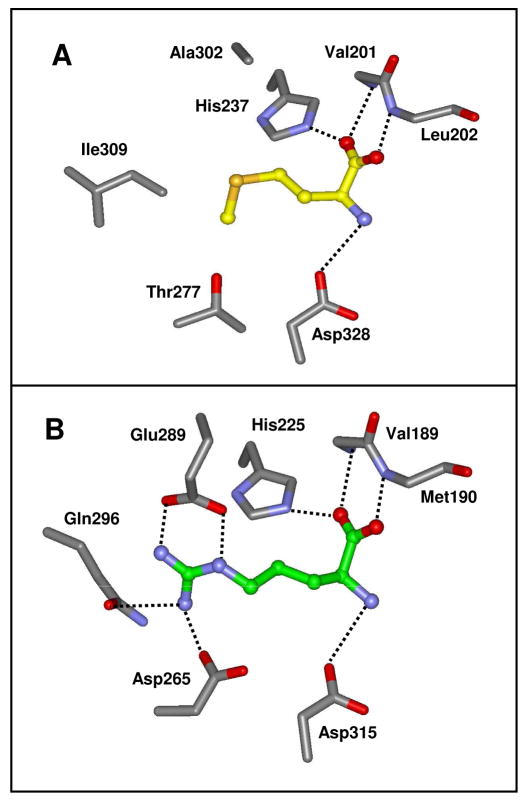
The residues that are expected to bind the two divalent cations are His-69, His-71, His-235, His-255, Lys-194 and Asp-328 based upon the known homology with other members of the amidohydrolase superfamily (5). The relative orientations of these presumptive metal binding residues are presented in Figure 3. However, the enzyme crystallized without zinc and a single magnesium ion was found ligated to the α-metal site. The magnesium is coordinated to His-69, His-71 and Asp-328. Interestingly, methionine, from the crystallization medium, is bound in the active site close to the position normally occupied by the more solvent exposed β-metal. The α-amino group of the methionine interacts with the side chain carboxylate of Asp-328. The α-carboxylate of the bound methionine is ion-paired with His-237 and hydrogen bonded with the backbone amide groups of Val-201 and Ile-202 as illustrated in Figure 4. The side chain of methionine is found within a pocket formed by the side chains of Thr-277 and Ile-309.
Figure 3.
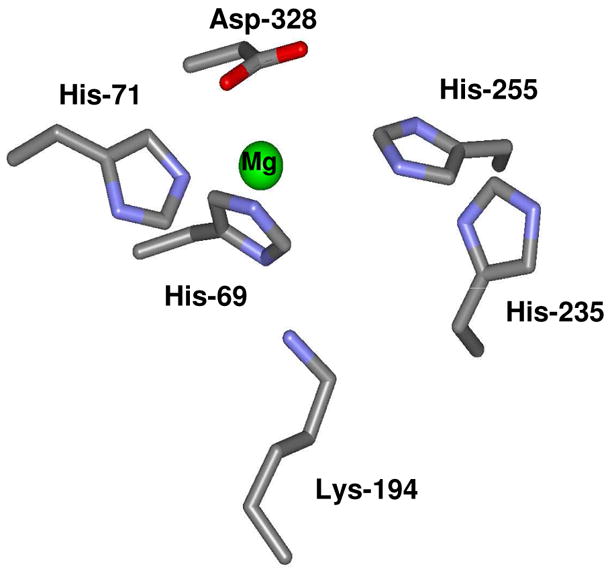
Active site geometry of residues expected to form the binuclear metal center of Sgx9355e.
Figure 4.
(A) Structure of Sgx9355e with L-methionine bound within the active site. (B) Structure of Sgx9359b with L-arginine bound within the active site.
Screening of Dipeptide Libraries
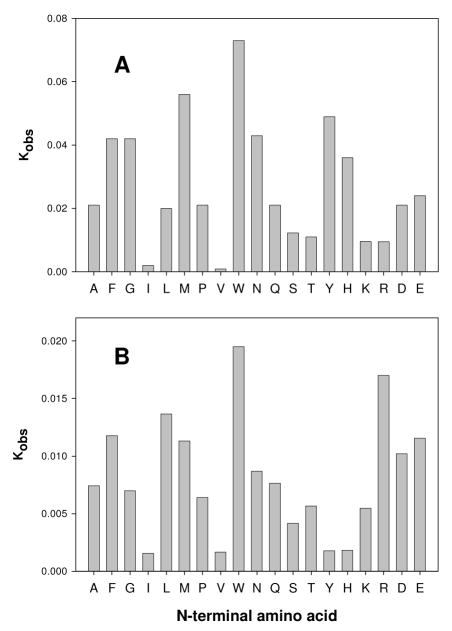
To determine the substrate profile of Cc0300 and Sgx9355e, a total of 19 dipeptide libraries were tested for catalytic activity using the Cd-ninhydrin based assays. The relative preferences for the N-terminal amino acid residue within these dipeptide libraries are depicted in Figure 5. Cc0300 and Sgx9355e exhibit broad catalytic activity for the hydrolysis of these peptide libraries with different N-termini but the relative rates of hydrolysis vary from library to library. These two enzymes show relatively high reaction rates with L-Trp-L-Xaa and low reaction rates with L-Ile-L-Xaa and L-Val-L-Xaa. Cc0300 shows a higher reaction rate with L-Tyr-L-Xaa and L-His-L-Xaa compared with Sgx9355e. Conversely, Sgx9355e exhibits a higher turnover rate with L-Lys-L-Xaa and L-Arg-L-Xaa compared with Cc0300.
Figure 5.
Relative substrate activities for hydrolysis of 19 dipeptide libraries of the type L-Xaa-L-Xaa where a single amino acid is at the N-terminus (defined on the x-axis) and 19 different amino acids were at C-terminus (not including L-cysteine). The results for Cc0300 are presented in panel A while those for Sgx9355e are shown in panel B. Additional details are found in the text.
Specificity of Cc0300 and Sgx9355e
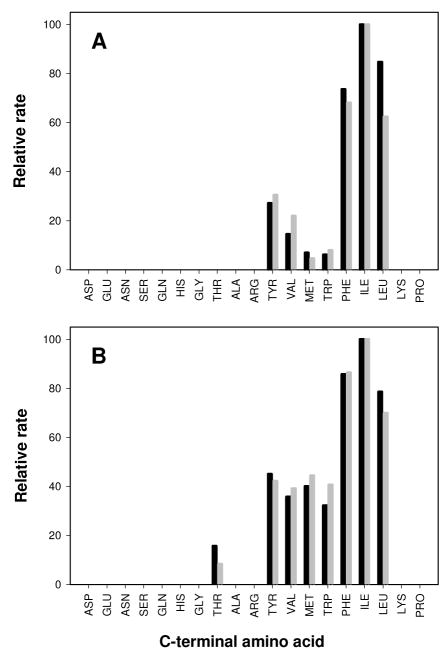
Quantitative amino acid analysis (AAA) was employed to determine the relative rates of hydrolysis for specific dipeptides within selected dipeptide libraries. Cc0300 was assayed with the L-Ala-L-Xaa and L-Asp-L-Xaa dipeptide libraries. The amino acid products, detected from the reaction mixture of Cc0300 with the L-Ala-L-Xaa library, were limited to alanine, leucine, isoleucine, phenylalanine, tryptophan, methionine, tyrosine and valine. The amino acid products detected from the hydrolysis reaction using the L-Asp-L-Xaa library as a substrate were aspartate, leucine, isoleucine, phenylalanine, tryptophan, methionine, tyrosine and valine. Since alanine and aspartate were only detected when libraries containing these amino acids were at the N-terminus, it can be concluded that the C-terminal specificity of Cc0300 with dipeptides is limited to the hydrophobic amino acid residues leucine, isoleucine, phenylalanine, tryptophan, methionine, tyrosine and valine. Two libraries, L-Ala-L-Xaa and L-Gln-L-Xaa, were selected for quantitative analysis with Sgx9355e for specificity at the C-terminus. Amino acid analysis showed that the C-terminal specificity of Sgx9355e with these dipeptide substrates is very similar to that found for Cc0300 except that Sgx9355e can hydrolyze dipeptides with threonine at the C-terminus. The relative reaction rates for Cc0300 and Sgx9355e are presented in Figure 6. The relative rates of hydrolysis for the other dipeptides in these libraries are less than 5% for both Cc0300 and Sgx9355e.
Figure 6.
Relative rates of hydrolysis for the hydrolysis of selected dipeptide libraries by Cc0300 and Sgx9355e. (A) Cc0300 with L-Ala-L-Xaa (black) and L-Asp-L-Xaa (grey); (B) Sgx9355e with L-Ala-L-Xaa (black) and L-Gln-L-Xaa (grey). The relative rates were determined by a fit of the data to equation 1. Additional details are found in the text.
Hydrolysis of N-substituted Amino Acids and Tripeptides
Cc0300 and Sgx9355e were screened for catalytic activity with N-acetyl-L-Xaa and N-formyl-L-Xaa derivatives. These two enzymes showed very similar catalytic specificities with these two types of substituted amino acid derivatives. The hydrolyzed substrates were limited to the N-acetyl- and N-formyl derivatives of L-hydrophobic amino acids leucine, isoleucine, phenylalanine, tryptophan, methionine, tyrosine and valine. Cc0300 and Sgx9355e were also tested with two tripeptides: L-Gly-L-Phe-L-Arg and L-Gly-L-Ala-L-Tyr. These two enzymes were unable to hydrolyze L-Gly-L-Phe-L-Arg but were able to hydrolyze the C-terminal L-tyrosine from L-Gly-L-Ala-L-Tyr.
Kinetic Constants for Cc0300 and Sgx9355e
The kinetic constants for Cc0300 were determined using the following substrates: L-Ala-L-Leu, L-Ala-L-Phe, L-Asp-L-Leu, N-formyl-L-Leu, N-formyl-L-Ile, N-formyl-L-Phe, N-formyl-L-Met, N-formyl-L-Tyr, N-formyl-L-Val, N-acetyl-L-Leu, N-acetyl-L-Ile, N-acetyl-L-Phe, N-acetyl-L-Met, N-acetyl-L-Tyr and N-acetyl-L-Val. Also tested was the tripeptide, L-Gly-L-Ala-L-Tyr. The kinetic data were fit to equation 2 and the values of kcat, Km, and kcat/Km for the hydrolysis of the selected substrates are presented in Table 2. The turnover numbers ranged from 1.7 −1 for the hydrolysis of N-Acetyl-L-Met to 37 s−1 for the hydrolysis of L-Ala-L-Leu. The kinetic constants for the hydrolysis of selected compounds by Sgx9355e are presented in Table 2. In general, the kinetic constants, kcat and kcat/Km, for Cc0300 are greater than those measured for Sgx9355e.
Table 2.
Kinetic parameters for Cc0300 and Sgx9355e
| enzyme | substrate | Kcat (s−1) | Km (mM) | Kcat/Km (M−1s−1) |
|---|---|---|---|---|
| Cc0300 | L-Ala-L-Leu | 37 ± 2 | 0.35 ± 0.04 | (1.1 ± 0.1) × 105 |
| L-Ala-L-Phe | 19 ± 1 | 0.66 ± 0.05 | (2.9 ± 0.3) × 104 | |
| L-Asp-L-Leu | 29 ± 1 | 0.22 ± 0.04 | (1.3 ± 0.2) × 105 | |
| N-acetyl-L-Leu | 2.5 ± 0.1 | 0.15 ± 0.03 | (1.6 ± 0.3) × 104 | |
| N-acetyl-L-Ile | 2.6 ± 0.1 | 0.14 ± 0.02 | (1.9 ± 0.2) × 104 | |
| N -acetyl-L-Phe | 5.3 ± 0.2 | 0.31 ± 0.03 | (1.7 ± 0.2) × 104 | |
| N -acetyl-L-Tyr | 4.6 ± 0.2 | 0.17 ± 0.02 | (2.7 ± 0.3) × 104 | |
| N -acetyl-L-Val | 2.0 ± 0.2 | 1.1 ± 0.1 | (1.8 ± 0.2) × 104 | |
| N -acetyl-L-Met | 1.7 ± 0.1 | 0.66 ± 0.05 | (2.6 ± 0.2) × 103 | |
| N -formyl-L-Leu | 39 ± 2 | 0.13 ± 0.01 | (3.0 ± 0.3) × 105 | |
| N -formyl-L-Ile | 20 ± 1 | 0.07 ± 0.01 | (2.9 ± 0.4) × 105 | |
| N-formyl-L-Phe | 52 ± 2 | 0.26 ± 0.03 | (2.0 ± 0.2) × 105 | |
| N-formyl-L-Tyr | 33 ± 1 | 0.09 ± 0.01 | (3.9 ± 0.4) × 105 | |
| N-formyl-L-Val | 18 ± 1 | 0.08 ± 0.01 | (2.2 ± 0.3) × 105 | |
| N-formyl-L-Met | 12 ± 1 | 0.41 ± 0.03 | (2.9 ± 0.2) × 104 | |
| L-Gly-L-Ala-L-Tyr | 0.87 ± 0.05 | 0.39 ± 0.02 | (2.2 ± 0.2) × 103 | |
| Sgx9355e | L-Ala-L-Leu | 0.34 ± 0.01 | 0.25 ± 0.03 | (1.3 ± 0.2) × 103 |
| L-Ala-L-Phe | 0.41 ± 0.01 | 0.07 ± 0.01 | (5.8 ± 0.6) × 103 | |
| N-acetyl-L-Leu | 0.032 ± 0.01 | 0.11 ± 0.01 | (2. 9 ± 0.4) × 102 | |
| N-formyl-L-Leu | 0.072 ± 0.003 | 0.26 ± 0.04 | (2.8 ± 0.4) × 102 | |
| L-Gly-L-Ala-L-Tyr | 0.10 ± 0.01 | 0.30 ± 0.04 | (3.3 ± 0.4) × 103 |
Inhibition by N-methyl Phosphonate Derivative (1)
Compound 1 was tested as an inhibitor of Cc0300. This compound was found to be a competitive inhibitor of this enzyme when N-formyl-L-leucine was used as the substrate. An inhibition constant of 2.3 ± 0.2 μM was obtained from a fit of the data (not shown) to equation 3.
Discussion
Elucidation of the substrate specificity for any enzyme given the amino acid sequence and/or three-dimensional structure alone is a difficult and demanding problem for most proteins. The rapid sequencing of whole bacterial genomes has provided an explosion of uncharacterized enzymes whose functions cannot be reliably annotated based upon a traceable homology to proteins of known function. Our approach to this problem is to address an entire enzyme superfamily simultaneously in an attempt to reveal the evolutionary lineages for the emergence of new catalytic functions and to provide a structural framework for the deciphering of substrate profiles from the amino acid sequences alone. Over 6,000 proteins with unique amino acid sequences have been classified as members of the amidohydrolase superfamily (24). To date more than 30 distinct reactions have been shown to be catalyzed by members of this superfamily using amino acid, carbohydrate, and nucleic acid derived substrates (5, 7, 24).
The substrate specificity for two related enzymes was examined in this investigation. Cc0300 from C. crecentus and Sgx9355e from an unknown bacterium found in the Sargasso Sea are currently annotated as Xaa-Pro dipeptidase in the NCBI. Sequence comparisons have established that both proteins belong to the amidohydrolase superfamily. In this study the two enzymes were purified to homogeneity and the enzyme from the Sargasso Sea was crystallized and its three dimensional structure determined. Cc0300 is a zinc metalloprotein and the active site is likely to be populated by a binuclear metal center, similar to that found previously for urease and phosphotriesterase (25–27). In this binuclear metal center, the two metal ions are coordinated by an aspartic acid and four histidine residues. The metal ions are also bridged by a carboxylated lysine residue and a hydroxide from solvent. The structure of Sgx9355e is consistent with this conclusion but, unfortunately, the metal ions were lost during purification and/or crystallization.
The substrate specificity for Cc0300 and Sgx9355e were determined using small libraries of potential dipeptide substrates. Contrary to initial expectations neither of these enzymes was able to hydrolyze any peptide that contained proline at the C-terminus. However, both enzymes were able to catalyze the hydrolysis of dipeptides that terminated in a hydrophobic amino acid but were not specific for the amino acid at the N-terminus. N-acetyl and N-formyl derivatives of L-hydrophobic amino acids were hydrolyzed by these two enzymes. Limited experiments with tripeptides demonstrated that hydrophobic amino acids from the C-terminus can be hydrolyzed and thus these two enzymes are more accurately classified as carboxypeptidases with a requirement for a hydrophobic amino acid at the C-terminus.
The X-ray structure of Sgx9355e was determined with L-methionine bound in the active site. The positioning of methionine in the active site has identified those amino acid residues that are responsible for the structural determinants of the substrate specificity for this enzyme. In this complex L-methionine is positioned as the product derived from the hydrolysis of the C-terminal end of an oligopeptide. The α-carboxylate of methionine is ion-paired with His-237, a residue that originates from the loop that follows β-strand 5. The α-carboxylate of L-methionine is also hydrogen bonded to the backbone amide groups of Val-201 and Leu-202. The α-amino group of the bound L-methionine interacts with Asp-328. This residue is found at the end of β-strand 8 and is also expected to ligate the divalent metal ion in the Mα position. The positioning of the α-amino group of the C-terminal derived amino acid product with Asp-328 is consistent with this residue functioning as a general acid/base catalyst in the transfer of the proton from the bridging hydroxide to the leaving group amine (28). The side chain of the L-methionine is found in a small pocket that is formed from Thr-277, Ala-302, Val-305, and Ile-309. Thr-277 is found in a very short loop that immediately follows β-strand 7, whereas Ala-302, Val-305, and Ile-309 are found in a pair of helices that follow this loop. All of these residues, with the exception of Ile-309, are conserved in the amino acid sequence of Cc0300. In Cc0300 the isoleucine is substituted with a methionine.
We have previously determined the substrate profile for two other peptidases from C. crecentus, Cc2672 and Cc3125 (9). Cc2672 is specific for the hydrolysis of oligopeptides that terminate in either L-lysine or L-arginine. The specificity of Cc3125 is more restrictive in the sense that only dipeptides (but not longer peptides) that terminate in L-lysine or L-arginine represent productive substrates. In addition to these two proteins we have determined the structure and substrate specificity of Sgx9359b from the Sargasso Sea (gi| 44368820). This enzyme is specific for the hydrolysis of oligopeptides that terminate in L-arginine. The mode of binding of L-arginine to the active site of Sgx9359b is presented in Figure 4B. In this structure the α-carboxylate is ion paired to His-225 and hydrogen bonded to the backbone amide groups from Val-189 and Met-190. The α-amino group interacts with the α-carboxylate of Asp-315. These residues are homologous to those residues found to interact with L-methionine in the structure of Sgx9355e (Figure 4A). As expected, the structural determinants for the recognition of the guanidino group of the bound L-arginine in Sgx9359b are different. The guanidino group is hydrogen bonded with the side chain amide group of Gln-296 and ion paired with the side chain carboxylates of Glu-289 and Asp-265. These resides occur in positions similar to those of three amino acids that define the substrate recognition in Sgx9355e (Ile-309, Ala-302, and Thr-277).
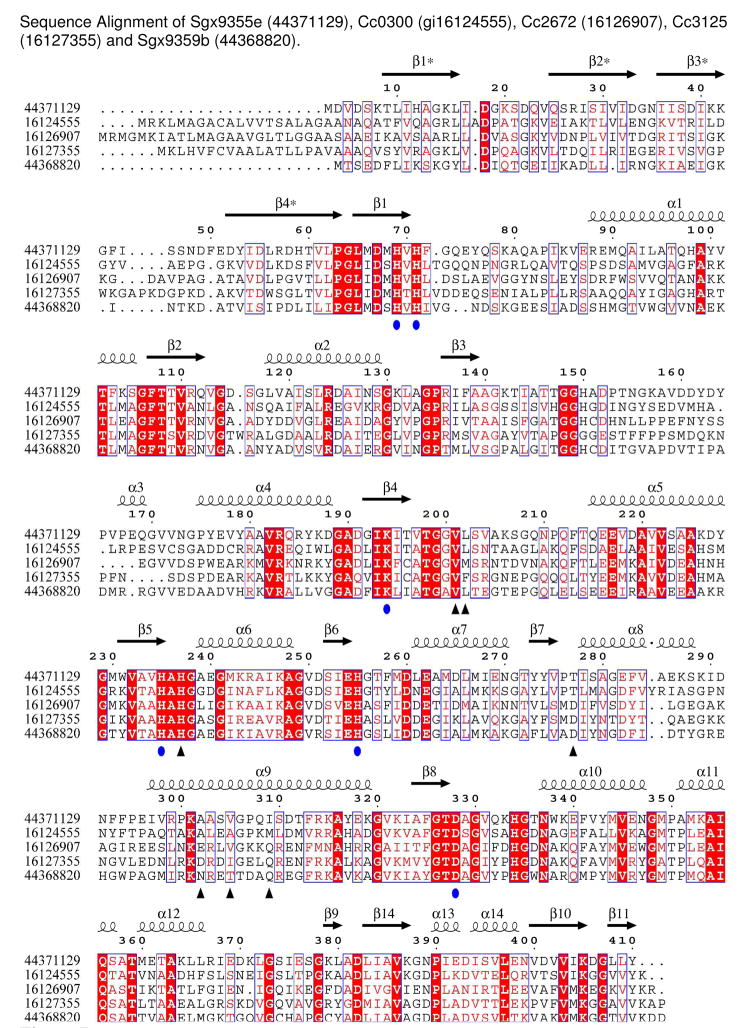
A sequence alignment of Sgx9355e, Cc0300, Cc2672, Cc3125, and Sgx9359b is presented in Figure 7. In this alignment there are a total of 71 residues that are fully conserved among the 5 proteins. All six of the amino acids expected to bind to the two divalent cations in the active site are conserved, as is the histidine that ion pairs with the C-terminal carboxylate of peptide substrates. The three enzymes that recognize positively charged amino acids at the C-terminus of potential substrates (Cc2672, Cc3125, Sgx9359b) have either anionic or highly polar residues at the binding site, indicated by black triangles in Figure 7. Conversely, the two enzymes that recognize hydrophobic side chains at the C-terminus of potential substrates (Sgx9355e and Cc0300) have less polar amino acids at the equivalent positions.
Figure 7.
Amino acid sequence comparison for Sgx9355e (gi| 44371129), Cc0300 (gi| 16124555), Cc2672 (gi| 16126907), Cc3125 (gi| 16127355) and Sgx9359b (gi|44368820). The predicted metal ligands for the two divalent metal ions are indicated with ovals. Residues in Sgx9355e, and the equivalent residues in Cc0300, that interact with the bound L-methionine are indicated with triangles. Residues in Sgx9359b, and the equivalent residues in Cc2672 and Cc3125, that interact with the bound L-arginine are at the equivalent positions. Other fully conserved residues in all five proteins are highlighted in red. The N-terminal residues that are removed via proteolysis during the expression of Cc0300, Cc2672, and Cc3125 are ANA/Q, ASA/A, and AAA/Q, respectively. The figure was produced with ESPript (29).
The discovery of function for Cc0300 and Sgx9355e, coupled with the structure elucidation of Sgx9355e with a bound product, permits annotation of other proteins in the amidohydrolase superfamily with greater confidence. A search of the NCBI database of completely sequenced bacterial genomes finds 29 other protein sequences that are now predicted by analogy to hydrolyze short oligopeptides that terminate in hydrophobic amino acids. All of these proteins have a conserved histidine equivalent to His-237, threonine and alanine residues equivalent to Thr-277 and Ala-302, and an isoleucine, methionine, leucine, or valine residue equivalent to Ile-309 in the structure of Sgx9355e. A list of these sequences is presented in Table 3.
Table 3.
Proteins predicted to share substrate specificity with Cc0300 and Sgx9355e.
| Gi number | Organism | Locus tag | Identity of residues equivalent to those in Sgx9355e (Thr277, Ala302, Val305 and Ile309) |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 44371129 | Environmental sequence | Sgx9355e | P | T | I | S | A | G | E……R | P | K | A | A | S | V | G | P | Q | I | S |
| 16124555 | C. crescentus CB15 | CC_0300 | P | T | L | M | A | G | D……T | A | K | A | L | E | A | G | P | K | M | L |
| 219674575 | O. alexandrii HTCC2633 | OA2633_13760 | P | T | V | M | A | G | E……A | A | K | A | R | V | V | G | P | L | M | L |
| 91794386 | S. denitrificans OS217 | Sden_3036 | P | T | L | L | A | G | A……R | A | K | A | I | R | V | G | A | D | M | Q |
| 116626946 | S. usitatus Ellin6076 | Acid_7923 | P | T | L | M | A | A | E……A | G | K | A | R | Q | A | M | A | A | L | N |
| 114561981 | S. frigidimarina NCIMB 400 | Sfri_0801 | P | T | L | L | A | G | D……K | A | K | A | I | R | V | G | G | D | M | M |
| 71280007 | C. psychrerythraea 34H | CPS_1348 | P | T | L | M | A | G | D……S | A | K | A | I | R | V | G | G | D | M | I |
| 221233241 | C. crescentus NA1000 | CCNA_00302 | P | T | L | M | A | G | D……T | A | K | A | L | E | A | G | P | K | M | L |
| 197106524 | P. zucineum HLK1 | PHZ_c3063 | P | T | L | M | A | G | D……T | A | K | A | L | Q | A | G | P | L | M | L |
| 127513920 | S. loihica PV-4 | Shew_2992 | P | T | L | L | A | G | D……K | Q | K | A | V | R | V | G | A | D | M | T |
| 212634704 | S. piezotolerans WP3 | swp_1881 | P | T | L | L | A | G | A……K | A | K | A | I | R | V | G | A | D | M | L |
| 209964193 | R. centenum SW | RC1_0865 | P | T | L | L | A | G | K……K | R | K | A | L | T | V | G | P | M | M | Q |
| 114798392 | H. neptunium ATCC 15444 | HNE_0934 | P | T | V | L | A | G | A……R | A | K | A | A | Q | V | G | P | L | M | L |
| 196186567 | B. sp. BAL3 | BBAL3_2700 | P | T | L | L | A | G | D……T | A | K | A | L | E | A | G | P | K | M | L |
| 148555139 | S. wittichii RW1 | Swit_2224 | P | T | L | L | V | G | Q……A | Q | K | A | I | A | I | A | P | M | L | Q |
| 170728069 | S. woodyi ATCC 51908 | Swoo_3741 | P | T | L | L | A | G | D……K | A | K | A | E | R | V | G | A | D | M | T |
| 83857796 | C. atlanticus HTCC2559 | CA2559_13368 | P | T | I | S | A | G | K……V | P | K | A | L | E | I | G | P | K | I | Q |
| 56461490 | I. loihiensis L2TR | IL2390 | P | T | I | M | A | G | K……R | P | K | A | R | A | I | G | P | Q | I | Q |
| 163753087 | K. algicida OT-1 | KAOT1_13042 | P | T | I | T | A | G | K……V | P | K | A | L | A | V | G | P | K | I | Q |
| 163786441 | F. bacterium ALC-1 | FBALC1_14687 | P | T | I | S | A | G | K……V | P | K | A | L | D | I | G | P | K | I | Q |
| 120434642 | G. forsetii KT0803 | GFO_0275 | P | T | I | T | A | G | K……V | P | K | A | R | E | I | G | P | K | I | Q |
| 88711102 | F. bacterium HTCC2170 | FB2170_03290 | P | T | I | T | A | G | K……A | K | K | A | A | E | I | G | P | L | I | Q |
| 124006459 | M. marina ATCC 23134 | M23134_02799 | P | T | I | S | A | G | E……V | P | K | A | L | K | I | G | S | Q | V | K |
| 88803709 | P. irgensii 23-P | PI23P_00415 | P | T | I | T | A | G | K……V | P | K | A | L | A | V | G | P | Q | I | Q |
| 89889513 | F. bacterium BBFL7 | BBFL7_01328 | P | T | I | T | A | G | K……V | P | K | A | R | T | V | G | P | Q | I | Q |
| 86141823 | L. blandensis MED217 | MED217_01790 | P | T | L | S | A | G | K……V | P | K | A | L | E | I | G | P | Q | L | Q |
| 86135565 | P. sp. MED152 | MED152_12694 | P | T | I | T | A | G | K……V | P | K | A | L | A | V | G | P | Q | I | Q |
| 91215139 | P. torquis ATCC 700755 | P700755_12617 | P | T | I | T | A | G | K……V | P | K | A | K | A | I | G | P | K | I | Q |
| 109900575 | P. atlantica T6c | Patl_4277 | P | T | V | M | A | G | N……R | P | K | A | A | T | I | G | P | L | I | L |
| 119773229 | S. amazonensis SB2B | Sama_0087 | P | T | L | L | A | G | E……R | P | K | A | A | A | I | G | P | K | I | Q |
| 196158836 | A. macleodii ‘Deep ecotype’ | MADE_04041 | P | T | I | L | A | G | K……R | P | K | A | A | A | I | G | P | L | I | Q |
Acknowledgments
We thank A. Mahmood and J. Levia for technical assistance.
Footnotes
This work was supported by the NIH (GM71790 and GM74945) and the Hackerman Advanced Research Program (010366-0034-2007). The x-ray coordinates and structure factors for Sgx9355e have been deposited in the Protein Data Bank (PDB accession code: 2qs8)
Abbreviations: Protein Data Bank, PDB; American Type Culture Collection, ATCC; inductively coupled plasma emission-mass spectrometry, ICP-MS;
References
- 1.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Mazumader R, O’Donova C, Redaschi N, Suzek B. The universal protein resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman HM. The protein data bank: a historical perspective. Acta Crystallogr. 2008;A64:88–95. doi: 10.1107/S0108767307035623. [DOI] [PubMed] [Google Scholar]
- 3.Gerlt JA, Babbitt PC. Can sequence determine function? Genome Biol. 2000;5:1–10. doi: 10.1186/gb-2000-1-5-reviews0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hermann JC, Marti-Arbona R, Fedorov AA, Fedorov EV, Almo SC, Shoichet BK, Raushel FM. Structure-based activity prediction for an enzyme of unknown function. Nature. 2007;448:775–781. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seibert CM, Raushel FM. Structural and catalytic diversity within the amidohydolase superfamily. Biochemistry. 2005;44:6383–6391. doi: 10.1021/bi047326v. [DOI] [PubMed] [Google Scholar]
- 6.Pieper U, Chiang R, Seffernick JJ, Brown SD, Glasner ME, Kelly L, Eswar N, Sauder JM, Bonanno JB, Swaminathan S, Burley SK, Zheng X, Chance MR, Almo SC, Gerlt JA, Raushel FM, Jacobson MP, Babbitt PC, Sali A. Target selection and annotation for the structural genomics of the amidohydrolase and enolase superfamilies. J Struct Funct Genomics. 2009;10:0000–0000. doi: 10.1007/s10969-008-9056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holm L, Sander C. An evolutionary treasure: unification of a broad set of amidohydrolase related to urease. Proteins. 1997;28:72–82. [PubMed] [Google Scholar]
- 8.Nguyen TH, Brown S, Fedorov AA, Fedorov EV, Babbitt PC, Almo SC, Raushel FM. At the periphery of the Amidohydrolase superfamily: Bh0493 from bacillus halodurans catalyzes the isomerization of D-galacturonate to D-tagaturonate. Biochemistry. 2008;47:1194–1206. doi: 10.1021/bi7017738. [DOI] [PubMed] [Google Scholar]
- 9.Xiang D, Patskovsky Y, Chengfu X, Meyer AJ, Sauder JM, Burley SK, Almo SC, Raushel FM. Functional identification of incorrectly annotated prolidases from the amidohydrolase superfamily of enzymes. Biochemistry. 2009;48:0000–0000. doi: 10.1021/bi900111q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu C, Hall RS, Cummings J, Raushel FM. Tight binding inhibitors of N-acyl amino sugar and N-acyl amino acid deacetylases. J Am Chem Soc. 2006;128:4244–4245. doi: 10.1021/ja0600680. [DOI] [PubMed] [Google Scholar]
- 11.Russell PJ. iGenetics: A molecular approach. San Francisco, California United States of America: Pearson education; 2005. [Google Scholar]
- 12.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein science. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newman A. Elements of ICPMS. Analytical chemistry. 1996;68:46A–51A. [Google Scholar]
- 14.Doi E, Shibata D, Matoba T. Modified colorimetric ninhydrin methods for peptidase assay. Analytical Biochemistry. 1981;118:173–184. doi: 10.1016/0003-2697(81)90175-5. [DOI] [PubMed] [Google Scholar]
- 15.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 16.Sheldrick GM, Hauptman HA, Weeks CM, Miller M, Usón I. In: International tables for macromolecular crystallography. Arnold E, Rossmann M, editors. F. Dordrecht: Kluwer Academic Publishers; 2001. pp. 333–345. [Google Scholar]
- 17.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr. 2003;59:2023–30. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 18.De-La-Fortelle E, Bricogne G. Maximum-likelihood heavy atom parameter refinement in the MIR and MAD methods. Methods Enzymol. 1997;276:472–493. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 19.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nature Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 20.Jones TA. A graphics model building and refinement system for macromolecules. J Appl Crystallogr. 1978;11:268–272. [Google Scholar]
- 21.Jones TA, Zou J, Cowtan S, Kjeldgaard M. Improved methods in building protein models in electron density map and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 22.Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszwewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Somonsom T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 24.Pegg SC, Brown SD, Ojha S, Seffernick J, Menn EC, Morris JH, Chang PJ, Huang CC, Ferrin TE, Babbitt PC. Leveraging enzyme structure-function relationships for functional inference and experimental design: the structure-function linkage database. Biochemistry. 2006;45:2545–2555. doi: 10.1021/bi052101l. [DOI] [PubMed] [Google Scholar]
- 25.Aubert SA, Li Y, Raushel FM. Mechanism for the hydrolysis of organophosphates by the bacterial phosphotriesterases. Biochemistry. 2004;43:5707–5715. doi: 10.1021/bi0497805. [DOI] [PubMed] [Google Scholar]
- 26.Jabri E, Carr MB, Hausinger RP, Karplus PA. The crystal structure of urease from Klebsiella aerogenes. Science. 1995;268:998–1004. [PubMed] [Google Scholar]
- 27.Kim J, Tsai PC, Chen SL, Himo F, Almo SC, Raushel FM. Structure of diethyl phosphate bound to the binuclear metal center of phosphotriesterases. Biochemistry. 2008;47:9497–9504. doi: 10.1021/bi800971v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marti-Arbona R, Fresquet V, Thoden JB, Davis ML, Holden HM, Raushel FM. Mechanism of the reaction catalyzed by isoaspartyl dipeptidase from Escherichia coli. Biochemistry. 2005;44:7115–7124. doi: 10.1021/bi050008r. [DOI] [PubMed] [Google Scholar]
- 29.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]