Abstract
The functional unit of the kidney is the nephron. During its organogenesis, the mammalian metanephric kidney generates thousands of nephrons over a protracted period of fetal life. All nephrons are derived from a population of self-renewing multi-potent progenitor cells, termed the cap mesenchyme. However, our understanding of the molecular and cellular mechanisms underlying nephron development is at an early stage. In order to identify factors involved in nephrogenesis, we performed a high-resolution, spatial profiling of a number of transcriptional regulators expressed within the cap mesenchyme and early developing nephron. Our results demonstrate novel, stereotypic, spatially defined cellular sub-domains within the cap mesenchyme, which may, in part, reflect induction of nephron precursors. These results suggest a hitherto unappreciated complexity of cell states that accompany the assembly of the metanephric kidney, likely reflecting diverse regulatory actions such as the maintenance and induction of nephron progenitors.
Keywords: cap mesenchyme, metanephros, kidney, progenitor, transcription factor, nephrogenesis
Introduction
The mammalian metanephric kidney, in its simplest sense, consists of an array of tubules, termed nephrons, connected to a branched collecting system, the ureteric epithelium and served by a highly organized vascular network. Nephrons are the basic functional unit of the kidney, the number of which varies from tens of thousands to hundreds of thousands, depending on the mammalian species (Cullen-McEwen et al., 2001; Nyengaard and Bendtsen, 1992). Nephrogenesis, the de novo formation of nephrons, occurs over an extensive period of both fetal and early post-natal life to generate the full complement of nephrons (Saxén, 1987; Hartman et al., 2007). The main body of the nephron, the renal tubule, comprises an epithelium segmented into distinct specialized domains along a proximal (glomerular) to distal (collecting duct) axis (El-Dahr et al., 2008; Kopan et al., 2007). These different regions are responsible for resorption and modification of the plasma filtrate that enters at the glomerulus and exits the kidney as urine via the ureter.
A program of repetitive reciprocal inductive interactions between the intermediate mesoderm (IM) mesenchyme and the IM-derived ureteric epithelium drives the assembly of the metanephric kidney (Grobstein, 1953; Grobstein, 1953; Gruenweld, 1952). At the onset of mouse metanephric development at embryonic day 10.5 (E10.5), the posterior IM mesenchyme expresses Gdnf. GDNF induces the adjacent ureteric bud to invade and branch within the mesenchyme (Costantini, 2006; Costantini and Shakya, 2006; Pepicelli et al., 1997; Vega et al., 1996). We have recently demonstrated that the mesenchymal cells of the E10.5 posterior IM largely consist of two sub-populations, an outer population of Foxd1+ cells, termed the cortical interstitial mesenchyme, and an inner core of Six2+ cells termed the cap mesenchyme (Mugford et al., 2008b). Throughout the remainder of metanephric development, the cap mesenchyme maintains Gdnf expression and remains closely associated with the tips of the branching ureteric epithelium in the cortical zone of the kidney (Pepicelli et al., 1997). This expression domain ensures the outward growth of the ureteric epithelium and ultimately the establishment of the arborized network of the collecting duct system.
Concurrent with each branching event, the ureteric epithelium induces a portion of the cap mesenchyme to undergo a mesenchymal to epithelial transition via the canonical Wnt signaling pathway (Carroll et al., 2005; Park et al., 2007). Recent studies demonstrate that the cap mesenchyme is a multipotent, self-renewing population of nephron tubule progenitors and is maintained in a progenitor state by the transcriptional regulator Six2 (Boyle et al., 2008; Kobayashi et al., 2008; Self et al., 2006). Upon induction, cells of the cap mesenchyme down-regulate Six2, activate Fgf8, Wnt4, Lhx1 and Pax8 and form the pretubular aggregate, the precursor to the renal vesicle, on the ventral (medullary) side of each branching tip (Bouchard et al., 2002; Grieshammer et al., 2005; Kobayashi et al., 2005b; Perantoni et al., 2005; Stark et al., 1994). In response to a number of recently defined regulatory pathways, the renal vesicle undergoes a series of poorly understood morphological changes in order to elaborate into the main body of the mature nephron and connect back to the ureteric epithelium (Cheng et al., 2007; Cheng et al., 2003; Grieshammer et al., 2005; Kobayashi et al., 2005a; Nakai et al., 2003; Perantoni et al., 2005; Wang et al., 2003).
In order to shed additional light on the genetic programs regulating nephrogenesis, we have addressed the relationship among domains of regulatory gene expression and cellular organization by examining the spatial expression of transcriptional regulators previously annotated within the cap mesenchyme and the induced renal vesicle (www.gudmap.org). Our results demonstrate that the cap mesenchyme, all Six2+ cells lying between the ureteric tip and the cortical-most nephrogenic interstitium, is not a homogeneous population. In this, gene expression stereotypically divides the Six2+ nephron progenitor compartment into three sub-domains, defined here as the inner capping mesenchyme, the outer capping mesenchyme and the induced mesenchyme. The likely domain of uninduced nephron progenitors is refined to a sub-domain that is both negative for factors associated with nephron induction and also likely refractory to the primary inductive action of canonical Wnt signaling. Interestingly, our analysis demonstrates evidence of a canonical Wnt response in the developing interstitial mesenchyme suggesting a broader role for Wnt action beyond the cap mesenchyme. Finally, the data demonstrate that, upon induction, the pretubular aggregate is already polarized with respect to specific molecular markers, prior to formation of the epithelial renal vesicle; polarization is maintained throughout the early development of the nephron. By refining our understanding of gene expression within the cap mesenchyme and developing nephron, these data suggest that metanephric development involves coordinated gene activity in order to maintain a balance of progenitor self-renewal, nephron induction and cell fate decisions during nephron patterning.
Results
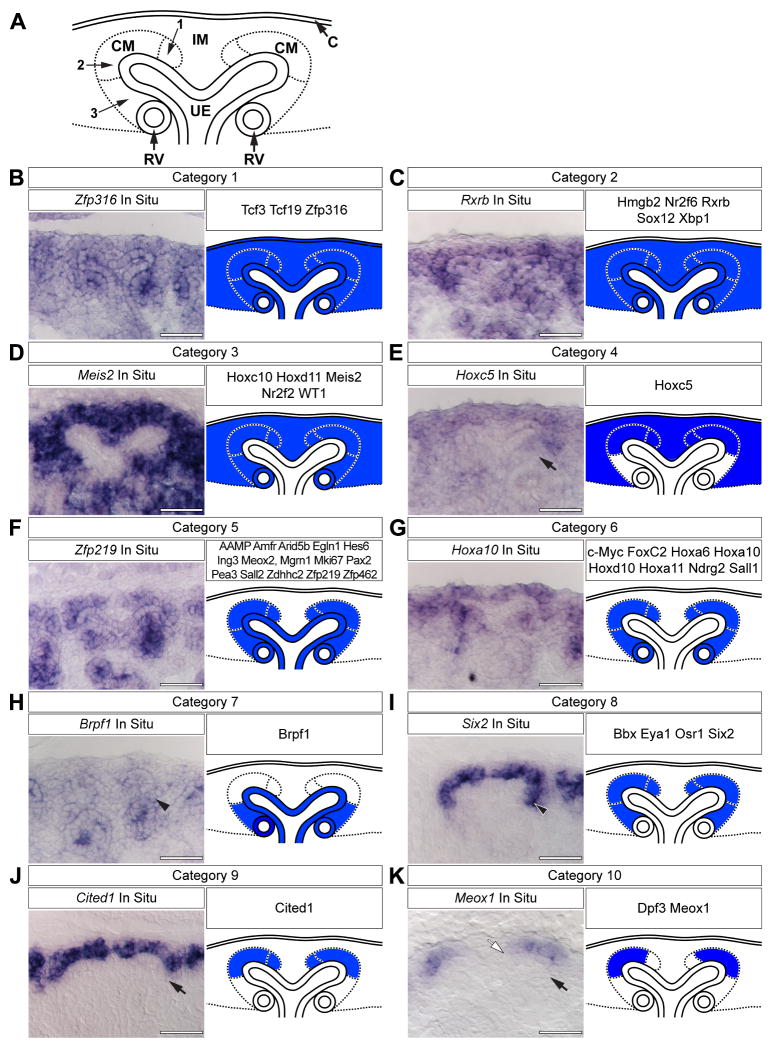
Gene expression patterns can be classified into ten different categories
In order to map the expression domains of transcriptional regulators at high resolution in the E15.5 cap mesenchyme, we examined the results of a genome scale low-resolution whole-mount in situ hybridization (WISH) screen of mammalian transcriptional regulators available through the GUDMAP resource (Little et al., 2007; McMahon et al., 2008; Yu et al., in preparation; www.gudmap.org). Forty-five genes (listed throughout Fig.1) were identified that were expressed within the cap mesenchyme, though we did not eliminate genes expressed in other populations. We then performed section in situ hybridization (SISH) for each of these and annotated expression in the following structures according to following established ontologies: the renal capsule, the cortical interstitial mesenchyme, the cap mesenchyme, the ureteric epithelia and the renal vesicle (Little et al., 2007). Based upon these annotations, transcripts were grouped into categories and a representative schematic of each category was created (Fig.1 A). In total, ten distinct expression categories were identified (Fig.1 B-K).
Figure 1. Categorized gene expression within the cap mesenchyme.
(A) Schematic representation of the nephrogenic zone. Numbers indicate sub-domains within the cap mesenchyme. (B-K) SISH for representative transcriptional regulators and the schematic interpretation and categorization of all expression patterns examined. Black arrows (E, K, J) indicate lack of expression in aggregating mesenchyme. Black arrowheads (H, I) indicate expression in aggregating mesenchyme. White arrow (K) indicates lack of expression in mesenchyme near the cleft of the ureteric epithelium. C = renal capsule, IM = interstitial mesenchyme, CM = cap mesenchyme, RV = renal vesicle and UE = ureteric epithelium. Scale bars = 50 μm.
Genes within category 1, exemplified by Zfp316 (Fig.1 B), were expressed throughout the nephrogenic zone at varying levels, including the cap mesenchyme, cortical interstitial mesenchyme precursors, ureteric epithelium, renal capsule and renal vesicles. Rxrb exemplifies expression domain category 2, which is similar to that of category 1, but excludes expression in the renal capsule (Fig.1 C). Meis2 is representative of category 3 genes; no expression was detected in either the renal capsule or ureteric epithelium (Fig.1 D). Hoxc5 fell into its own category; expression was absent from cells of the renal vesicle, ureteric epithelium and a small portion of mesenchyme beneath the ureteric tip (Fig.1 E, arrow). This latter mesenchyme (CM region 3 in Fig.1 A) is likely the site of pretubular aggregate formation and will be referred to as the induced mesenchyme.
The majority of the genes examined fell into categories 5 and 6 (Fig.1 F, G). Category 5 genes, as exemplified by Zfp219, were expressed in the ureteric epithelium, renal vesicles and cap mesenchyme (Fig.1 F) whereas genes such as Hoxa10 were only expressed in the latter two cell populations (Fig.1 G). The expression pattern of Brpf1 (Fig.1 H) was unique. Expression was detected in the ureteric epithelium, renal vesicle and the induced mesenchyme (Fig.1 H, arrowhead); a reciprocal pattern to that of Hoxc5.
Finally, genes within categories 8 through 10 (Fig.1 I-K) were expressed only within the cap mesenchyme, though qualitative differences were observed among these expression domains. Six2 (Fig.1 I) is representative of a class of genes expressed throughout the entire cap mesenchyme (regions 1 through 3 in Fig.1 A). In contrast, Cited1 expression appears to be excluded from induced mesenchyme (compare arrowhead in Fig.1 I and arrow in Fig.1 J). Category 10 genes, as represented by Meox1, were expressed in a small subset of cap mesenchyme cortical to the ureteric tip (CM regions 2 in Fig.1 A), but excluded from the induced mesenchyme and the mesenchyme close to the cleft between adjacent tips of a single ureteric branch (CM regions 3 and 1 in Fig.1 A, respectively; Fig.1 K, black arrow and white arrow, respectively).
The nephrogenic zone consists of distinct cellular domains
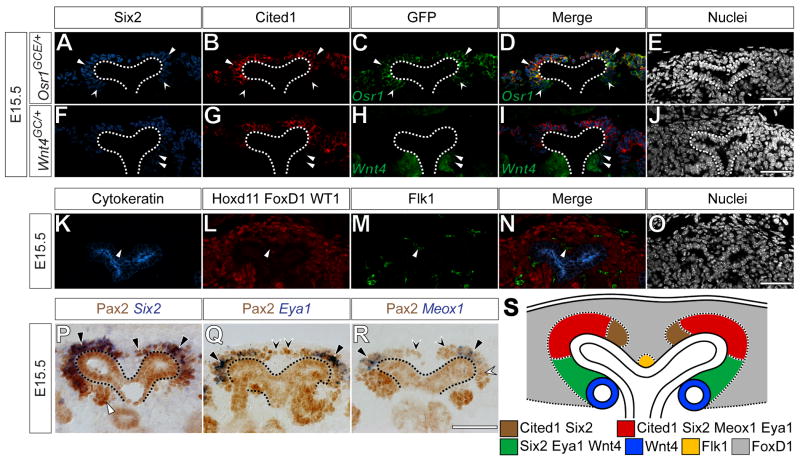
Since nephron progenitors lie within the cap mesenchyme (Boyle et al., 2008; Kobayashi et al., 2008; Self et al., 2006), we focused on defining the domains represented by categories 8 through 10 at, or close to, single cell resolution. Osr1 is expressed in a similar domain to Six2 (James et al., 2006; Mugford et al., 2008b) (Fig.1 I) and Wnt4 is expressed in induced cap mesenchyme (Stark et al., 1994). We have previously reported the generation of an eGFPCreERT2 knock-in to the Osr1 locus (Osr1m1(cre/ERT2)Amc, referred to here as Osr1GCE/+) (Mugford et al., 2008b) and have also generated an eGFPCre knock-in to the Wnt4 locus (Wnt4GC/+) (A. Kobayashi and A.P. McMahon, unpublished reagent). Consequently, the activity of the Osr1 and Wnt4 loci can be monitored by the presence of GFP in the E15.5 metanephric kidney of Osr1GCE/+ or Wnt4GC/+ embryos, respectively.
As suggested by the ISH results, Osr1-driven GFP and Six2 were completely overlapping in the cap mesenchyme of E15.5 Osr1GCE/+ embryos (Fig.2 A, C, D, arrowheads and concave arrowheads). Confirming our ISH results, Cited1 was absent specifically in Six2+, GFP+ cells of the induced mesenchyme (Fig.2 A-D, concave arrowheads). All other Six2+, GFP+ cells were Cited1+ (Fig.2 A-D, arrowheads). Furthermore, in E15.5 Wnt4GC/+ embryos, Six2+, GFP+ cells were observed within cells of the induced pretubular aggregate ventral to the ureteric bud; however, Six2 levels are notably reduced as compared to Six2+, GFP- cells. As expected, Six2+, GFP+ cells in Wnt4GC/+ kidneys were Cited1- (Fig.2 F-J, arrowheads). Thus, in nephron progenitors, Six2 is present within cells that have undergone induction, but is gradually down-regulated as the inductive process progresses. In contrast, there is a sharp border between Cited1+ and Cited1- cells that discriminates between non-induced and induced nephron progenitors. From this point forward, we refer to Six2+, Cited1+, Wnt4- mesenchyme as the “capping mesenchyme” and Six2+, Cited1-, Wnt4+cells as “induced mesenchyme”. The entire Six2+ mesenchyme is considered the “cap mesenchyme”.
Figure 2. Combinatorial analysis of expression patterns reveals new domains within the cap mesenchyme.
(A-O) Confocal immunofluorescence of E15.5 Osr1GCE/+, (A-E), Wnt4GC/+ (F-J) or wild type (K-O) metanephric kidneys immunostained for Six2 (A, F), Cited1 (B, G), GFP (C, H) Cytokeratin (K), Hoxd11, Foxd1, WT1 (L) and Flk1 (M). Nuclei stained for Hoechst 33258 (E, J, O). Merged images (D, I, N). (P-R) Immunohistochemistry for Pax2 combined with In situ hybridization for Six2 (P), Eya1 (Q) and Meox1 (R). (S) Schematic representation of novel domains within the cap mesenchyme. Dashed lines (A-J, P-R) indicate the ureteric epithelium. Arrowheads and concave arrowheads (A-D) indicate Six2+, Cited1+, GFP+ and Six2+, Cited1-, GFP+ cells, respectively. Arrowheads (F-I) indicate Six2+, Cited1-, GFP+ cells. Arrowheads (K-N) indicate Flk1+ endothelium. Black and white arrowheads (P) indicate Pax2+, Six2 expressing mesenchyme and Pax2+ pretubular aggregate, respectively. Arrowheads and concave arrowheads (Q) indicate Pax2+, Eya1 expressing mesenchyme and Pax2+ mesenchyme, respectively. Arrowheads, white concave arrowheads and black concave arrowheads (R) indicate Pax2+, Meox1 expressing mesenchyme and Pax2+ mesenchyme near the nephric duct cleft and cortical to the renal vesicle, respectively. Scale bars = 50 μm.
We have previously demonstrated that Pax2 is present in all Six2+ cells and that these Six2+, Pax2+ cells are not part of the Foxd1+ cortical interstitial mesenchyme population (Mugford et al., 2008b). Pax2 immunostaining was used in combination with SISH for Six2, Eya1, Bbx, Dpf3 and Meox1 to establish the spatial relationships among these genes and Six2 expressing cells. As expected, Six2 was expressed in all Pax2+ cap mesenchyme cells (Fig.2 P, black arrowheads) but not in the Pax2+ pre-tubular aggregate or ureteric epithelium (Fig.2 P, white arrowhead and dashed line, respectively). Eya1 (Fig.2 Q) and Bbx (data not shown) were expressed in the majority of Pax2+ cap mesenchyme (Fig.2 Q, arrowheads), but were excluded from a small portion of Pax2+ capping mesenchyme adjacent to the cleft of the branching ureteric epithelium and the Pax2+ ureteric epithelium (Fig.2 Q, concave arrowheads and dashed line, respectively). We also noted that not all Pax2+ mesenchymal cells were Eya1+ at E10.5, E11.5 or E12.5 (data not shown). Therefore, though we had initially assigned Six2 and Eya1 to the same expression category by single SISH (Fig.1 I), upon closer inspection with additional markers, Eya1 and Bbx in fact form a separate expression domain within the capping mesenchyme, distinct from Six2 and Osr1. Finally, Meox1 (Fig.2 R) and Dpf3 (data not shown) were expressed in a limited number of Pax2+ capping mesenchyme cells. Neither was expressed in either the Pax2+ capping mesenchyme adjacent to the cleft of the branching ureteric epithelium, the Pax2+ induced mesenchyme or the Pax2+ ureteric epithelium (Fig.2 R, black concave arrowheads, white concave arrowheads and dashed line, respectively). From this point forward, we define the Pax2+, Eya1-, Meox1- capping mesenchyme adjacent to the cleft of the branching ureteric epithelium as the “inner capping mesenchyme”, whereas the Pax2+, Eya1+, Meox1+ capping mesenchyme as the “outer capping mesenchyme”.
Hoxd11 and WT1 were expressed throughout the interstitial and cap mesenchyme (Fig.1 D). Furthermore, Foxd1 is expressed exclusively in the cortical interstitial mesenchyme (Hatini et al., 1996) and Cytokeratin is present within the nephric duct epithelium (Fig.2 K) (Fleming and Symes, 1987). Interestingly, immunostaining for the combination of Cytokeratin, Hoxd11, WT1 and Foxd1 labeled most, but not all cells within the nephrogenic zone; one to three cells residing among the interstitial mesenchyme between the branching ureteric epithelium were not labeled (Fig.2 K, L arrowhead). These remaining cells were positive for the vascular markers Flk1 (Fig.2 M, arrowhead) and PECAM (data not shown), indicating that these cells are likely vascular endothelial progenitors.
Taken together, our results provide evidence for an unappreciated spatial and molecular complexity to the organization of mesenchyme within the nephrogenic zone of the developing kidney (Fig.2 S). The mesenchyme immediately in contact with the ureteric epithelium (the cap mesenchyme) can be resolved into three distinct compartments, the inner capping mesenchyme, the outer capping mesenchyme and the induced mesenchyme. These compartments may, in part, reflect induction of nephron precursors. Interstitial progenitors fill up much of the remaining cellular space between the capsule and cap mesenchyme, while a small population of vascular progenitors lie within the cleft of each branching ureteric bud.
Pea3, Lef1 and Wnt4 co-expression identifies induced cap mesenchyme cells
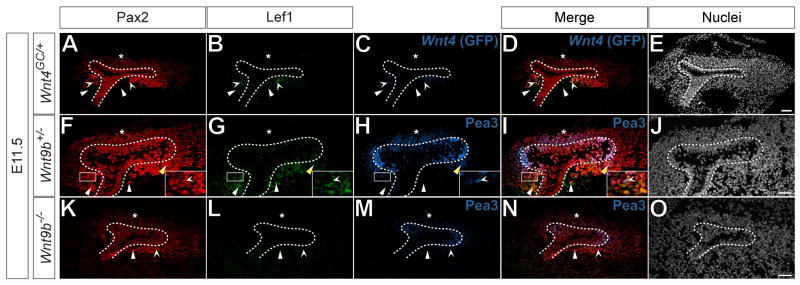
We next sought to identify a combination of molecular markers that definitively mark the first inductive responses within the cap mesenchyme. Wnt9b acts through the canonical Wnt signaling pathway to induce the cap mesenchyme and is upstream of all known markers of nephrogenesis, including Wnt4, Fgf8, Pax8 and Lhx1 (Carroll et al., 2005; Park et al., 2007). The transcriptional regulator Lef1 is a known downstream target of canonical Wnt signaling and acts as a feedforward component of the canonical Wnt pathway (Driskell et al., 2004; Filali et al., 2002). Furthermore, Lef1 is up-regulated in isolated rat metanephric mesenchyme in response to Wnt agonists (Kuure et al., 2007). Fgf8 is genetically upstream of Wnt4 and required for the maturation of the induced nephron (Grieshammer et al., 2005; Perantoni et al., 2005). This indicates a required role for continued FGF signaling within the pretubular aggregate for the progression of inductive process and the establishment of the renal vesicle. Additionally, FGF signaling is potentially required for the survival of the cap mesenchyme (Poladia et al., 2006). The transcriptional regulator Pea3 is a known target of Gdnf signaling (Haase et al., 2002) and a subset of FGF signals, predominantly Fgf8 (Roehl and Nusslein-Volhard, 2001). Up regulation within the branching tip of the ureteric epithelium likely reflects Gdnf induced branching morphogenesis. In contrast, Gdnf signaling in the mesenchyme has not been documented and the GDNF receptor c-Ret is not expressed within the cap mesenchyme (Majumdar et al., 2003). Therefore, Pea3 presence within the mesenchyme likely reflects an FGF response within the mesenchyme, enabling FGF signaling to be visualized within the cap mesenchyme and induced nephron. Consequently, Lef1, Pea3 and Wnt4 activity can potentially identify ongoing responses to critical signaling inputs within the nephron progenitor compartment.
In E11.5 Wnt4GC/+ metanephric kidneys, Lef1 was present in GFP+, Pax2+ cells on the future medullary aspect beneath the initial branch of the ureteric epithelium, but not the future cortical side (Fig.3 A-E, concave arrowheads and asterisk, respectively). In phenotypically wild-type E11.5 Wnt9b+/- metanephric kidneys, Pea3+, Lef1-, Pax2+ cap mesenchyme was present on the cortical side of the ureteric bud (Fig.3 F-J, asterisk), while on the medullary side, Lef1, Pea3 and Pax2 were present in the same cells (Fig.3 F-J, yellow arrowhead). Lef1-, Pax2+, Pea3+ (Fig.3 F-J inset, concave arrowheads) and Lef1+, Pax2+, Pea3- cells were also present in this position (Fig.3 F-J white arrowheads). Conversely, in E11.5 Wnt9b-/- metanephric kidneys, where a Wnt inductive response is absent (Carroll et al, 2005), Lef1 was not present in Pax2+ mesenchyme cells (Fig.3 K-O, arrowhead), though Pea3 and Pax2 co-label a subset of mesenchyme cells in both cortical and medullary positions (Fig.3 K-O, asterisk and concave arrowhead, respectively). These data suggest that Lef1 provides an appropriate readout of canonical Wnt induction of the cap mesenchyme and that a combination of Pea3, Lef1 and Wnt4 expression can be used to spatially characterize inductive responses within the cap mesenchyme.
Figure 3. Lef1, Pea3 and Wnt4 denote induced cap mesenchyme cells.
(A-O) Confocal immunofluorescence of E11.5 Wnt4GC/+ (A-E), Wnt9b+/- (F-J) and Wnt9b-/- (K-O) metanephric kidneys immunostained for Pax2 (A, F, K), Lef1 (B, G, L), GFP (C) and Pea3 (H, M). Nuclei stained for Hoechst 33258 (E, J, O). Merged images (D, I, N). Dashed lines (A-O) indicate the ureteric epithelium. Asterisks (A-D, F-I, K-N) indicate cortical Pax2+ cap mesenchyme. Arrowheads and concave arrowheads (A-D) indicate Lef1+, GFP-, Pax2- and Lef1+, GFP+, Pax2+ cells, respectively. Arrowheads, yellow arrowheads and concave arrowheads (F-J) indicate Lef1+, Pea3-, Pax2+ cells, Lef1+, Pax2+, Pea3+ cells and Lef1-, Pax2+, Pea3+ cells, respectively. Solid boxes (F-J) indicate inset area. Arrowheads and concave arrowheads (K-N) indicate Lef1-, Pea3-, Pax2+ and Lef1-, Pea3+, Pax2+ cells, respectively. Scale bars = 50 μm.
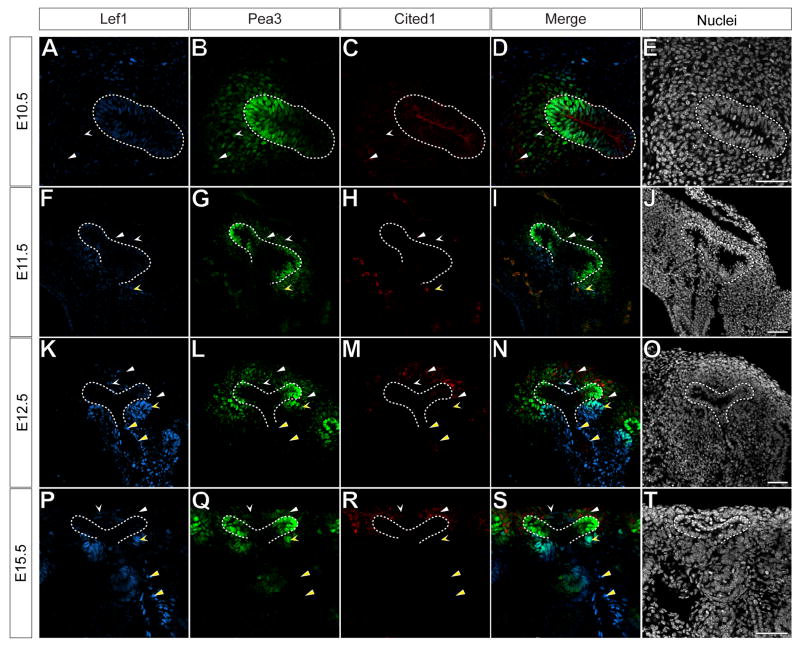
Inductive markers are not present in Cited1+cells
As noted earlier, no activation of Wnt4 was observed in E15.5 Cited1+ capping mesenchyme cells, suggesting that Cited1 distinguishes between induced and non-induced nephron progenitors. We next assessed the relationship of Pea3 and Lef1 to Cited1. As previously reported (Boyle et al., 2007), Cited1 was present in the nephric duct epithelia and in only a few cells of the metanephric mesenchyme at E10.5 (Fig.4 C, D, dashed line and arrowhead, respectively). Cited1+ cells accumulated within the capping mesenchyme between E10.5 and E12.5 (Fig.4 H, I, M, N, arrowheads) such that by E12.5, Cited1 was present throughout the capping mesenchyme, a pattern maintained throughout nephrogenesis (Fig.2 R, S, arrowheads). Pea3 was present in all Cited1+ cells at all time points examined (Fig.4 A-T, arrowheads), whereas no overlap was detected between Cited1 and Lef1. As nephrogenesis commenced, Lef1+, Pea3+ cells were detected in induced nephron structures, though Pea3 was confined to distal nephron structures (Fig.4 F-T, yellow concave arrowheads and data not shown).
Figure 4. Cited1+ cells do not demonstrate a response to inductive signals.
(A-T) Immunofluorescent confocal microscopy of transverse sections of the E10.5 metanephric blastema (A-E), saggital sections of E11.5 (F-J) and E12.5 metanephric kidneys (K-O) and frontal sections of E15.5 wild type (P-T) metanephric kidneys. Samples immunostained for Lef1 (A, F, K, P), Pea3 (B, G, L, Q) and Cited1 (C, H, M, R). Nuclei stained for Hoechst 33258 (E, J, O, T). Merged images (D, I, N, S). Dashed lines (A-S) indicate ureteric epithelium. White arrowheads and concave arrowheads (A-D, F-I, K-N, P-S) indicate Cited1+, Pea3+, Lef1- cap mesenchyme and Cited1-, Pea3+, Lef1+ stromal mesenchyme, respectively. Yellow concave arrowheads (F-I, K-N, P-S) indicate Cited1-, Pea3+, Lef1+ induced cap mesenchyme. Yellow arrowheads (K-N, P-S) indicate Lef1+ cells adjacent to the ureteric epithelium. Scale bars = 50μm.
These results indicate that while the Cited1 capping mesenchyme domain is established between E10.5 and E12.5, Cited1+ cells are likely actively responding to FGF, but not canonical Wnt signaling. Indeed, the reciprocal relationship between Cited1 and Lef1/Wnt4 expression provides a distinct separation between induced and uninduced nephron progenitors.
Inhibition of GSK-3b does not cause ectopic induction in the Cited1 domain
We have demonstrated that in its normal context, the capping mesenchyme does not demonstrate a detectable inductive response. We next asked if inductive markers could be activated in these cells by ectopically activating the canonical Wnt pathway. In the absence of a canonical Wnt signal, β-catenin, an essential transcriptional co-activator of the canonical Wnt pathway, is sequestered in the cytoplasm and marked for degradation by GSK-3β. In the presence of a canonical Wnt input, β-catenin degradation is relieved, allowing for its translocation into and accumulation within the nucleus. β-catenin then forms an activating complex with Lef/TCF DNA binding components to regulate Wnt targets (Logan and Nusse, 2004; Wang and Wynshaw-Boris, 2004).
Permanent expression of a non-degradable form of β-catenin within Six2+ population causes ectopic pre-tubular aggregate formation, though these aggregates do not epithelialize properly (Park et al., 2007). This experimental strategy activates β-catenin around E11.0, prior to the full establishment of the novel sub-domains we have identified (Fig.2). It is therefore possible that the establishment of these sub-domains plays a role in the attenuation of canonical Wnt signals, in effect controlling which cells are induced, and which cells remain in a progenitor state.
To test this hypothesis, we cultured E11.5 and E15.5 wild-type metanephric kidneys for 8 hours in the presence of the GSK-3β inhibitor 6-bromoindirubin-3′-oxime (BIO) or carrier (DMSO). BIO effectively mimics a canonical Wnt signaling response inhibiting GSK-3β-mediated β-catenin turnover (Kuure et al., 2007; Meijer et al., 2003). We also monitored the activity of the Axin2 promoter using an Axin2tm1Jbeh (Axin2-LacZ) allele, where a β-galactosidase (β-gal) coding sequence was inserted into the endogenous Axin2 locus (Lustig et al., 2002). Like Lef1, Axin2 is a direct target of the canonical Wnt pathway in several circumstances; its presence is thought to function as a negative regulator of canonical Wnt signaling (Fagotto et al., 1999; Jho et al., 2002).
X-gal staining of E11.5 (Fig.5 A) or E15.5 (Fig.5 I) Axin2-LacZ metanephric kidneys treated with DMSO (control) demonstrated Axin2 promoter activity in a few cells of the ureteric epithelium, cortical stroma, cap mesenchyme and induced pre-tubular aggregates (Fig.5 A, I, asterisk, dashed line, arrowhead and concave arrowhead, respectively). In addition, immunostaining for Pax2, Cytokeratin and Lef1 in wild-type control E11.5 metanephric kidneys showed a normal distribution of all proteins, where Lef1 was only present in Pax2+ pre-tubular aggregates (Fig.5 C-D, concave arrowheads) and Cytokeratin+ branching ureteric epithelium (Fig.5 C-D). Lef1 was not present in Pax2+ capping mesenchyme (Fig.5 C-D, arrowhead). Similarly, in E15.5 controls immunostained for Cited1, Cytokeratin and Lef1, Cited1+ cells were Lef1- (Fig.5 J-L, arrowheads), whereas the tips of the Cytokeratin+ ureteric epithelium and developing nephrons were Lef1+ (Fig.5 J-L, concave arrowheads).
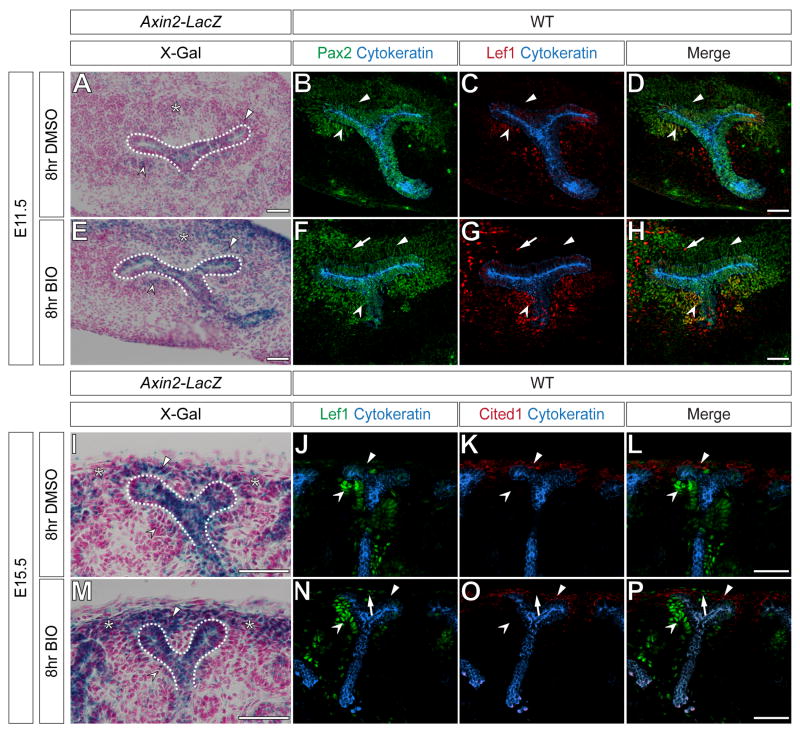
Figure 5. Global activation of canonical Wnt signaling does not ectopically induce markers of induction within the cap mesenchyme.
(A-H) DMSO treated (A-D) or BIO treated (E-H) E11.5 metanephric kidneys of Axin2-LacZ (A, E) or wild-type mice (B-D, F-H). Sections stained for X-gal (A, E), immunostained for Pax2 (green) and Cytokeratin (blue) (B, F) or immunostained for Lef1 (red) and Cytokeratin (blue) (C, G). Merged images (D, H). (I-P) DMSO treated (I-L) or BIO treated (M-P) E15.5 metanephric kidneys of Axin2-LacZ (I, M) or wild-type mice (J-L, M-O). Sections stained for X-gal (I, M), immunostained for Lef1 (green) and Cytokeratin (blue) (J, N) or immunostained for Cited1 (red) and Cytokeratin (blue) (K, O). Merged images (L, P). Dashed lines, asterisks, arrowheads and concave arrowheads (A, E, I, M) indicate X-gal+ ureteric epithelium, cortical interstitium, cap mesenchyme and induced cap mesenchyme, respectively. Arrowheads and concave arrowheads (B-D, F-H, J-L, N-P) indicate Lef1- cap mesenchyme and Lef1+ induced cap mesenchyme, respectively. Arrows (F-H, N-P) indicated elevated Lef1 levels in cortical interstitium after BIO treatment. Scale bars = 50 μm.
When treated with BIO, Axin2 promoter activity appeared to increase in the E11.5 and E15.5 cortical interstitium (Fig.5 E, M, asterisks). Whereas, X-gal activity increased in the E11.5, ureteric epithelium and cap mesenchyme (Fig. E, dashed line and arrowheads, respectively), a similar increase over controls was not detected in these areas at E15.5 (Fig. E, dashed line and arrowheads, respectively). No appreciable increase was seen in induced nephrons at either stage (Fig.5 E, M, concave arrowhead). Immunostaining of BIO treated metanephric kidneys at E11.5 for Pax2, Cytokeratin and Lef1 or at E15.5 for Cited1, Cytokeratin and Lef1 revealed that Lef1 was not ectopically activated in the E11.5 Pax2+ or E15.5 Cited1+ capping mesenchyme (Fig.5 F-G, N-P, arrowheads). Both the E11.5 and E15.5 Cytokeratin+ branching ureteric epithelium (Fig.5 F-G, N-P) and developing nephrons were Lef1+ (Fig.5 F-G, N-P, concave arrowheads). Interestingly, BIO treatment appeared to increase Lef1 levels in the E11.5 Pax2- or E15.5 Cited1- cortical interstitium (Fig.5 F-G, N-P, arrows). Ectopic nephrons were not detected at either stage after BIO treatment (Fig.5 E-H, M-P). Taken together these results suggest that the capping mesenchyme, but not the cortical interstitium, is normally refractory to artificially increased canonical Wnt signaling.
Lef1 is present in the interstitium and tips of the branching nephric duct
In addition to the developing nephrons, Lef1+ cells were detected in the interstitial cleft mesenchyme and in the tips of the invading or branching ureteric epithelium at E10.5, E11.5, E12.5 and E15.5 (Fig.4 A-T concave arrowheads and dashed lines, respectively). Furthermore, at E11.5 (Fig.3 A-E, arrowheads), E12.5 and E15.5 (Fig.4 K-T, yellow arrowheads), Lef1+ cells were observed surrounding the stalk of the ureteric epithelium. Canonical Wnt signaling is required within the branching ureteric epithelium (Bridgewater et al., 2008; Marose et al., 2008) and recent studies in our laboratory have demonstrated a direct requirement for canonical Wnt signaling in the development of the central nervous system vasculature (Stenman et al., 2008), raising the possibility that Wnts may also regulate kidney vascular progenitor populations.
To determine if Lef1+, Pea3+, Cited1- cells were interstitial or endothelial cells, we used a Cre driver line, Foxd1eGFPCre/+ (Foxd1GC/+) (A Kobayashi and AP McMahon, unpublished data), and the Gt(ROSA)26tm1Sor (R26R) (Soriano, 1999) reporter allele to activate β-gal in all renal interstitial cells, enabling these lineages to be distinguished. In E15.5 Foxd1GC/+;R26R kidneys, Lef1+, β-gal+ cells were detected within the cleft of the branching nephric duct and surrounding the stalk of the ureteric epithelium (Fig.S1, concave arrowheads and yellow arrowheads, respectively). In contrast, no Lef1+, Flk1+ cells were detected (Fig.S1, white arrowheads), indicating that interstitial mesenchyme and not vascular progenitors are a likely Wnt target. Thus, canonical Wnt signaling may play a role in several distinct cellular compartments; descendants of the cap mesenchyme, the branching ureteric epithelium and the developing renal interstitium.
Pretubular aggregates are polarized structures
Induced cap mesenchyme cells form the pretubular aggregate, which is the precursor to the renal vesicle. We noted in our previous analysis that Wnt4, Pea3 and Lef1 were present in developing nephrons, but that their distribution was not homogenous (Fig.4). Previous reports have demonstrated that the renal vesicle is a polarized structure (Grieshammer et al., 2005; Nakai et al., 2003; Perantoni et al., 2005; Piscione et al., 2004), but the onset of polarization with respect to induction has not been clearly established. E-cadherin is expressed weakly within cells of the renal vesicle, and in cells of the nephric duct epithelium and maturing nephron structures, but not in the pretubular aggregate (Fig.6 A, D, F, I, K, N) (Park et al., 2007). Furthermore, unlike mature renal vesicles, pre-tubular aggregates lack a laminin+ basement membrane (Kobayashi et al., 2008; Self et al., 2006). In either E-cadherin- pretubular aggregates (Fig.6 A-D, asterisk) or E-cadherin+ renal vesicles (Fig.6 A-D, dashed line) of E15.5 Wnt4GC/+ kidneys, levels of Lef1 and GFP were distributed unequally. Cells containing higher levels of Lef1 tended to have lower levels of GFP and vice versa (compare concave arrowheads with arrowheads in Fig.6 A-D). This unequal distribution was also seen in laminin- pre-tubular aggregates (Fig.S2) and became more apparent as the nephron matured into the E-cadherin+ S-Shaped body (Fig.6 F-J). Cells in distal structures of the S-Shaped body were E-cadherin+, Lef1+, GFP- (Fig.6 F-I, concave arrowhead), whereas, cells of the mid to proximal structures had lower levels of Lef1 and were GFP+ (Fig.6 F-I, arrowhead).
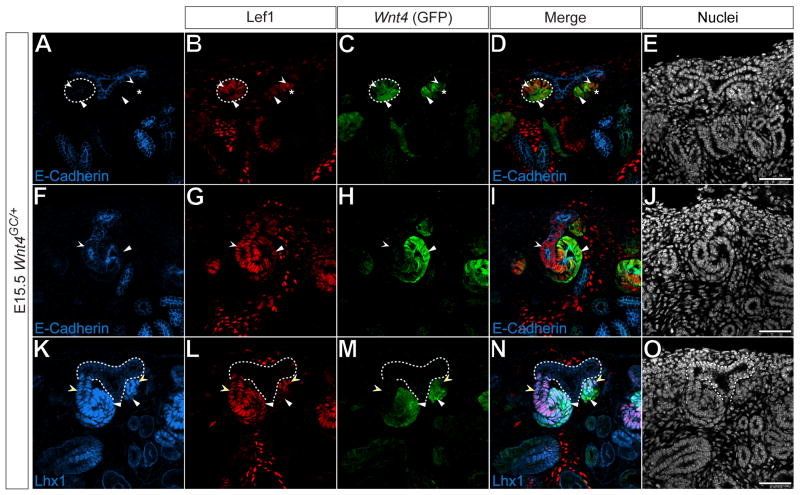
Figure 6. Differential gene expression in pretubular aggregates, renal vesicles and developing S-shaped bodies.
(A-O) Confocal immunofluorescence of E15.5 Wnt4GC/+ metanephric kidneys immunostained for E-cadherin (A, F), Lef1 (B, G, L), GFP (C, H, M) and Lhx1 (K). Nuclei stained for Hoechst 33258 (E, J, O). Merged images (D, I, N). Dashed lines and asterisks (A-D) indicate E-cadherin+ renal vesicle and E-cadherin- pretubular aggregate, respectively. Dashed lines (K-O) indicate the ureteric epithelium. White arrowheads and concave arrowheads (A-D) indicate Lef1high, GFPlow and Lef1low, GFPhigh cells, respectively. White arrowheads and concave arrowheads (F-I) indicate Lef1+, GFP- and Lef1low, GFP+ cells, respectively. White arrowheads and concave arrowheads (A-D) indicate Lef1low, Lhx1+ and Lef1high, Lhx1+ cells, respectively. Scale bars = 50 μm.
Lhx1 is present at low levels throughout the ureteric epithelium (Fig.6 K-O, dashed line), and up-regulated within both the pre-tubular aggregate and throughout the developing nephron (Fig.6 K-N). Furthermore, during nephrogenesis, it is required downstream of Wnt4 and Fgf8 in distal nephron structure formation (Grieshammer et al., 2005; Kobayashi et al., 2005a; Perantoni et al., 2005; Tsang et al., 2000). Within the developing nephron of Wnt4GC/+ E15.5 kidneys, Lhx1 was present in both Lef1+, GFP- distal structures (Fig.6 K-N, concave arrowheads) and Lef1+, GFP+ mid to proximal structures (Fig.6 K-N, arrowheads). Thus, it is evident that these markers are unequally distributed in the pretubular aggregate prior to its morphogenesis into an epithelial renal vesicle providing the earliest evidence of nephron polarity. Furthermore, the distribution of Wnt4 and Lef1 suggests that Wnt signaling could itself play a role in regulating nephron polarity during the morphogenesis of epithelial nephron precursors.
Discussion
Molecularly and spatially distinct populations within the cap mesenchyme define the nephron progenitor pool
Previous studies have demonstrated that the Six2+ cap mesenchyme population incorporates the nephron progenitor pool (Kobayashi et al., 2008). Six2 itself is required for self-renewal of nephron progenitors through an as yet undefined inhibition of canonical Wnt signaling (Kobayashi et al., 2008; Self et al., 2006). The Wnt mediated inductive process activates a nephrogenic program within cap mesenchyme (Carroll et al., 2005; Park et al., 2007), leading to the activation of a Wnt4+, non-renewing population that is committed to a nephron forming fate (Kobayashi et al., 2008).
In the simplest model, Six2 activity would be sufficient to both represses induction and maintain nephron progenitors. However, we identified a population of GFP+ (Wnt4+), Six2+ cells in a sub-domain of cap mesenchyme, the induced mesenchyme, just ventral to the branching ureteric bud, a population that likely represents an early stage of the inductive process whereby cells retaining Six2 are transitioning to the pretubular aggregate. Thus the presence of Six2 is not sufficient to attenuate renal vesicle induction suggesting the involvement of other regulatory factors.
Consistent with this hypothesis, a Cited1+ sub-domain occupies the majority of the Six2+ cap mesenchyme, the capping mesenchyme. This population is negative for both markers of induction (Wnt4 and Lhx1) and indicators of canonical Wnt signaling (Lef1). Additionally, we demonstrate that upon BIO-mediated activation of canonical Wnt signaling, Cited1+ cells activate negative regulators of the canonical Wnt pathway (Axin2), but not feed-forward components of the pathway (Lef1). A recent report demonstrates that descendants of Cited1+ cells give rise to all cells within the main body of the nephron and, as with Six2+ cells, are capable of self-renewal (Boyle et al., 2008). Cited1 has been shown to physically interact in vitro with both β-catenin and Smad proteins, acting as a bi-functional co-factor that attenuates Wnt signaling while promoting BMP signaling (Plisov et al., 2005). Both Cited1 and Cited2 are expressed within the cap mesenchyme and Cited1;Cited2 double mutant kidneys have reduced nephron numbers, though the mechanism underlying this phenotype has not been explored (Boyle et al., 2007). Interestingly, reduced nephron number is reminiscent of Bmp7 mutants where cap mesenchyme survival is diminished (Dudley et al., 1995; Dudley and Robertson, 1997).
Interestingly, Cited1 is gradually acquired within the Six2+ capping mesenchyme, several days after the Six2+ cap mesenchyme and Foxd1+ interstitial mesenchyme progenitors segregate from an Osr1+ intermediate mesoderm pool (Boyle et al., 2007; Mugford et al., 2008b; this study). Thus, neither expression nor function of Cited1 correlates with specification of nephron progenitors. Rather, these data suggest that Cited proteins may contribute to the maintenance of the self-renewing capping mesenchyme. As the ureteric epithelium branches, the number of capping mesenchyme cells per branching tip is reduced (compare Pax2+ cap mesenchyme cells in Fig.3 A with those in Fig.2 P). Furthermore, we previously demonstrated that the total number of Six2+ cells increases from ∼10000 to ∼180000 from E11.5 to post-natal day 1 (P1), while the number of UB tips increases from 2 to ∼3000 over the same period (Kobayashi et al., 2008). This equates to an approximate reduction of nephron progenitors per UB tip by ∼83-fold (∼5000 cells/tip at E11.5 compared to ∼60 cells/tip at P1). It is therefore likely that a tighter control of nephron induction is required to ensure that nephron progenitors are maintained throughout kidney organogenesis. Taken together, these data suggest that the Cited family of transcriptional co-factors may fine-tune the response of the capping mesenchyme to both survival and inductive signals and suggests that Cited1 marks the definitive nephron precursor domain.
Interestingly, the E11.5 and E15.5 capping mesenchyme both appear to be refractory to BIO-mediated activation of canonical Wnt signaling in explant culture. In contrast, activation of a non-degradable form of β-catenin within the Six2+ domain causes ectopic induction within cap mesenchyme cells, though these ectopic induced structures do not epithelialize (Park et al., 2007). These results may reflect simple technical differences such as the availability of BIO within the Six2+ capping mesenchyme in cultured kidneys. Alternatively, they may suggest other mechanisms that are GSK-3β independent that may regulate β-catenin activity in this compartment.
A dual role for Eya1 during metanephric development
The presence of Eya1 in only a sub-domain Six2+ cells, the outer capping mesenchyme, is of particular interest. The metanephric kidney does not develop in Eya1 mutants, as both Six2 and Pax2 expression are absent and the metanephric mesenchyme undergoes apoptosis (Sajithlal et al., 2005; Xu et al., 1999; Xu et al., 2002). Therefore, it is surprising that an apparent Pax2+, Six2+, Eya1- sub-domain exists. However, Eya1 is expressed in the IM mesenchyme prior to the development of the metanephros (Mugford et al., 2008a; Sajithlal et al., 2005), so the phenotype observed in Eya1 deficient embryos may be due to global defects in IM development and specification that occur prior to metanephric specification.
The Eya proteins are co-factors that, together with Six, Pax, Dach, and potentially Hox family members, form evolutionary conserved transcriptional complexes that function in many developmental contexts, including the eye, ear and kidney (Brodbeck and Englert, 2004; Gong et al., 2007; Hanson, 2001). However, as Eya proteins have no apparent DNA binding domain, Eya transcriptional target specificity hinges upon the specific combination of DNA binding transcriptional regulators present within each specific complex (Ohto et al., 1999). Furthermore, a previous study demonstrates that Eya proteins contain phosphatase activity and act directly on Six proteins to alter their transcriptional activity from that of a repressor to an activator (Li et al., 2003).
Both Six1 and Six2 are required for the development of the metanephric kidney, though their mutant phenotypes suggest they have different functions (Kobayashi et al., 2008; Self et al., 2006; Xu et al., 2003). Six1 is expressed in the IM prior to metanephric development and Six1 mutants have a similar phenotype to that of Eya1 mutants (Xu et al., 2003). Additionally, Six1 is down-regulated in the cap mesenchyme after E11.5 (Mugford et al., 2008a; Xu et al., 2003), indicating that its function is not required after the initial specification of the cap mesenchyme. Conversely, Six2 is not required for the specification of the cap mesenchyme; rather it acts downstream of both Eya1 and Six1 and functions in the self-renewal of nephron progenitors (Sajithlal et al., 2005; Self et al., 2006; Xu et al., 1999; Xu et al., 2003; Xu et al., 2002). As their transcriptional targets have not been identified, their precise molecular actions are not clear.
Further, Eya1 may play distinct roles during metanephric organogenesis dependent upon the Six protein it associates with; Six1 in the initial specification of a nephric mesenchyme anlage and Six2 in the maintenance of this population in the developing kidney thereafter. In this latter function, it is tempting to speculate that the presence or absence of Eya phosphatase activity may discriminate distinct cell populations within the nephron progenitor pool such as those that might continue to repopulate the capping mesenchyme from those that establish a permissive state for progression to renal vesicle induction. Based on our expression data, these populations may be represented by the inner capping mesenchyme and the outer capping mesenchyme, respectively. It is also possible that Meox1, Bbx and Dpf3 play specific roles in this process; however, the function of these genes within kidney development is unknown. New genetic tools that enable these populations to be isolated, manipulated and their fates traced will likely be informative.
A potential model for nephron vascularization
Our analysis of the mesenchyme population has also detected a population of vascular cells stereotypically located within the cleft of the ureteric bud. The developing podocytes within the proximal S-shaped body activate Vegf, a potent mitogen and chemo-attractant for endothelial cells (Coultas et al., 2005; Kitamoto et al., 1997; Loughna et al., 1997; Yancopoulos et al., 2000). Conflicting reports suggest an extra-renal anlagen (Sariola et al., 1983) and a renal anlagen origin (Loughna et al., 1997) for the kidney vasculature that may be reconciled if there are multiple modes by which regional vascular structures originate within different regions of developing kidney. The presence of vascular cells that are not organized into mature endothelial networks within the cortical domain where nephron precursors are emerging suggests a model whereby a resident cluster of endothelial cells is maintained within the cleft of each branching ureteric epithelium. In this model, vascular invasion of each Bowman's capsule is ensured by an appropriately positioned cluster of endothelial cells located in close proximity to newly forming nephrons.
Evidence for canonical Wnt signaling in the cortical renal interstitium
Though canonical Wnt signaling has been shown to play a critical roles in the ureteric epithelial, medullary interstitium and nephron progenitor populations (Carroll et al., 2005; Marose et al., 2008; Yu et al., 2009), the demonstration of likely Wnt activity in the cortical interstitium is a novel observation. Here, non-vascular Lef1+ cells reside within cortical interstitial progenitors located within the cleft of the branching tips of the ureteric epithelium. Further, Lef1 levels are increased upon BIO addition, a treatment expected to enhance β-catenin mediated canonical Wnt signaling. A recent report demonstrates that the genetic removal of β-catenin from the Foxd1+ interstitium and its descendants phenocopies mutations in Wnt7b activity and abolishes renal medulla formation (Yu et al., 2009). It is therefore, possible that the cortical cells identified in this study are responding to Wnt7b; however, Wnt7b is not expressed in the branching UB tips, rather it is restricted to the stalk of the collecting duct epithelium. Furthermore, other Wnt ligands are expressed in the branching ureteric epithelium, such as Wnt6 (Itaranta et al., 2002). It is therefore possible that these cortical cells are out of physical range of Wnt7b and responding to a different Wnt ligand. It would be of interest to evaluate Lef1 presence in cortical interstitium and the subsequent consequences to interstitial development in the absence of these other Wnt ligands.
Gene expression within pretubular aggregates and developing nephrons reveal potential roles for coordinated Fgf and canonical Wnt signaling in nephron pattern specification
The polarized distribution of Wnt4, Pea3 and Lef1 prior to E-cadherin activation suggests a polarization in nephron precursors that precedes formation on an epithelial, renal vesicle. Given a later observed polarization that clearly correlates with proximal-distal polarity in the nephron, it is tempting to speculate that pre-epithelial polarity also reflects an emerging proximo-distal axis. Given, the repetitive nature of nephrogenesis, definitive proof of this conjecture will be difficult. We demonstrate that Pea3 is present within presumptive distal nephron precursor cells. Previous reports have shown that Fgf8 is also expressed in these cells and is required for the activation of transcriptional regulator Lhx1 (Grieshammer et al., 2005; Perantoni et al., 2005). Lhx1, in turn, is upstream of Brn1 (Kobayashi et al., 2005a), a transcriptional regulator that is both expressed in presumptive distal nephron precursor cells and required for the development of distal nephron structures (Nakai et al., 2003). However, in addition to Pea3, Lef1, but not Wnt4, is present at high levels throughout distal nephron segments, suggesting a possible role for canonical Wnt signaling in this population. The localized expression of Pea3 and Lef1 may reflect responses to Fgf8 (autocrine) and Wnt4 (paracrine) signaling, respectively in the precursors of distal nephron segments. The convergence of these two signaling pathways within the most cortical cells of the pretubular aggregate, closest to the ureteric epithelium, correlates with Lhx1 expression and may be critical in the Lhx1-directed development of distal nephron structures (Kobayashi et al., 2005a).
Wnt4 is also expressed in mid to proximal structures of the developing S-shaped body. Here Lef1 is present, but at lower levels. The Notch signaling pathway is both necessary and sufficient for the development of proximal nephron structures (Cheng et al., 2007; Cheng et al., 2003; Piscione et al., 2004; Wang et al., 2003), however the genetic interactions between the Wnt and Notch pathways has not been addressed and this data may reflect a potential convergence of both pathways in proximal nephron development.
In conclusion, transcriptional analysis has defined new cellular domains within the kidney progenitor compartments. Furthermore, virtually all cells within the nephrogenic zone can be accounted for by the capping mesenchyme, induced mesenchyme, interstitial mesenchyme precursors, ureteric epithelium, vascular cells and their respective descendants. We suggest new models as to how the capping mesenchyme may be repopulated; the signals and transcriptional regulators that maintain the cap mesenchyme; mechanical models involving the vascularization of glomeruli; differentiation of interstitial stroma; and patterning of nephron segments. These descriptions provide the basis for the building of precise, testable hypotheses to refine our understanding of the mechanisms governing kidney organogenesis.
Materials and Methods
Animals and genotyping
Animal care and research protocols were performed in accordance with Harvard University's institutional guidelines, following approval by Harvard University's institutional committee on animal use. For staging of embryos, the morning of vaginal plug was designated as embryonic day 0.5 (E0.5). Gt(ROSA)26tm1Sor mice (Jackson Laboratories) (R26R) were genotyped as previously described (Soriano, 1999). Wnt9b+/- and Wnt9b-/- embryos were generated by intercrossing Sox2-cre;Wnt9b+/- and Wnt9bc/- lines and were genotyped as previously described (Carroll et al., 2005). Osr1m1(cre/ERT2)Amc (Osr1GCE/+) (Mugford et al., 2008a) and Axin2tm1Jbeh (Axin2-LacZ) mice were genotyped as previously described (Lustig et al., 2002). The generation and genotyping of the Foxd1GC/+ (A Kobayashi and AP McMahon, unpublished reagent), Wnt4GC/+ (A Kobayashi and AP McMahon, unpublished reagent) will be described in future publications.
Metanephric culture
E11.5 or E15.5 WT or Axin2-LacZ metanephric kidneys were isolated and cultured (8 hours, 37°C, 5.9% CO2) in DMEM + 10%FBS, 0.29mg/ml L-Glutamine, 100units/ml Penicillin, 100mg/ml Streptomycin with either 4μM BIO in DMSO or DMSO only. Samples were then washed 3× in PBS, fixed for 1hr on ice in cold 4%PFA in PBS and washed thoroughly in PBS.
Histological analysis
A more detailed description of all histological methodologies and modifications can be found in the Supplemental Material. All section in situ hybridizations were carried out as previously described (Little et al., 2007; McMahon et al., 2008) with slight modifications. In situ hybridization and X-gal stained samples were imaged on a Nikon Eclipse 90i compound microscope with a Nikon DXM1200C camera (Nikon Instruments). Information regarding in situ probe design may be obtained at www.gudmap.org. Immunofluorescence was carried out as previously described (Mugford et al., 2008a). The following antibodies and dilutions were used: anti-Cited1 (LabVison, 1:500), anti-Foxd1 (1:2000) (Mugford et al., 2008b), anti-Six2 (1:1000) (Kobayashi et al., 2008), anti-Pax2 (1:250, Covance), anti-GFP (1:500, AvesLabs), anti-pan-cytokeratin (1:500, Sigma), anti-b-galactosidase (1:500, AbCam), anti-Lhx1 (1:50, DSHB), anti-E-cadherin (1:200, Zymed) anti-Flk1 (1:1000, BDPharmigen), anti-Pea3 (1:1000), anti-Laminin (1:300, Sigma) and anti-Lef1 (1:100, Cell Signaling). Appropriate Cy2, Cy5 (1:500, Jackson Immuno), Alexa488, Alexa568, Alexa647 or HRP (1:500, Invitrogen) conjugated secondary antibodies were used to detect primary antibodies. Nuclei stained with Hoechst 33342 (Invitrogen). Experiments requiring the simultaneous use of two rabbit antibodies were conducted using Zenon kits (Invitrogen). Confocal images were acquired at 1μm optical slices on a Zeiss LSM510 META confocal microscope (Zeiss) or a Leica SP2 ABOS confocal microscope (Leica).
Accepted histological properties of cells within the nephrogenic zone (as described in Little et al. (2007)) were used to assign gene expression to a specific cell type. To eliminate bias due to location of any branching tip, care was taken to examine multiple kidneys (>3) in numerous cortical locations when scoring any individual expression pattern.
Supplementary Material
Acknowledgments
We thank Dr. Walter Birchmeier for the kind gift of Axin2-LacZ embryos, Dr. Silvia Arber for the kind gift of the anti-Pea3 antibody, and Benjamin Ruback for technical assistance. A.K. was supported by fellowships from the National Kidney Foundation and an HSCI Seed Grant. The primary screen leading to the identification of the sub-set of transcriptional regulators followed up in detail herein was performed as part of the GUDMAP consortium with funding to A.P.M. (U01 DK070181). All data is visible at www.gudmap.org. Work in A.P.M.'s laboratory is funded by NIH grant R37 DK054364.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bouchard M, Souabni A, Mandler M, Neubuser A, Busslinger M. Nephric lineage specification by Pax2 and Pax8. Genes Dev. 2002;16:2958–70. doi: 10.1101/gad.240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S, Misfeldt A, Chandler KJ, Deal KK, Southard-Smith EM, Mortlock DP, Baldwin HS, de Caestecker M. Fate mapping using Cited1-CreERT2 mice demonstrates that the cap mesenchyme contains self-renewing progenitor cells and gives rise exclusively to nephronic epithelia. Dev Biol. 2008;313:234–45. doi: 10.1016/j.ydbio.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S, Shioda T, Perantoni AO, de Caestecker M. Cited1 and Cited2 are differentially expressed in the developing kidney but are not required for nephrogenesis. Dev Dyn. 2007;236:2321–30. doi: 10.1002/dvdy.21242. [DOI] [PubMed] [Google Scholar]
- Bridgewater D, Cox B, Cain J, Lau A, Athaide V, Gill PS, Kuure S, Sainio K, Rosenblum ND. Canonical WNT/beta-catenin signaling is required for ureteric branching. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Brodbeck S, Englert C. Genetic determination of nephrogenesis: the Pax/Eya/Six gene network. Pediatr Nephrol. 2004;19:249–55. doi: 10.1007/s00467-003-1374-z. [DOI] [PubMed] [Google Scholar]
- Carroll TJ, Park JS, Hayashi S, Majumdar A, McMahon AP. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell. 2005;9:283–92. doi: 10.1016/j.devcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Cheng HT, Kim M, Valerius MT, Surendran K, Schuster-Gossler K, Gossler A, McMahon AP, Kopan R. Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron. Development. 2007;134:801–11. doi: 10.1242/dev.02773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HT, Miner JH, Lin M, Tansey MG, Roth K, Kopan R. Gamma-secretase activity is dispensable for mesenchyme-to-epithelium transition but required for podocyte and proximal tubule formation in developing mouse kidney. Development. 2003;130:5031–42. doi: 10.1242/dev.00697. [DOI] [PubMed] [Google Scholar]
- Costantini F. Renal branching morphogenesis: concepts, questions, and recent advances. Differentiation. 2006;74:402–21. doi: 10.1111/j.1432-0436.2006.00106.x. [DOI] [PubMed] [Google Scholar]
- Costantini F, Shakya R. GDNF/Ret signaling and the development of the kidney. Bioessays. 2006;28:117–27. doi: 10.1002/bies.20357. [DOI] [PubMed] [Google Scholar]
- Coultas L, Chawengsaksophak K, Rossant J. Endothelial cells and VEGF in vascular development. Nature. 2005;438:937–45. doi: 10.1038/nature04479. [DOI] [PubMed] [Google Scholar]
- Cullen-McEwen LA, Drago J, Bertram JF. Nephron endowment in glial cell line-derived neurotrophic factor (GDNF) heterozygous mice. Kidney Int. 2001;60:31–6. doi: 10.1046/j.1523-1755.2001.00767.x. [DOI] [PubMed] [Google Scholar]
- Driskell RR, Liu X, Luo M, Filali M, Zhou W, Abbott D, Cheng N, Moothart C, Sigmund CD, Engelhardt JF. Wnt-responsive element controls Lef-1 promoter expression during submucosal gland morphogenesis. Am J Physiol Lung Cell Mol Physiol. 2004;287:L752–63. doi: 10.1152/ajplung.00026.2004. [DOI] [PubMed] [Google Scholar]
- Dudley AT, Lyons KM, Robertson EJ. A requirement for bone morphogenetic protein-7 during development of the mammalian kidney and eye. Genes Dev. 1995;9:2795–807. doi: 10.1101/gad.9.22.2795. [DOI] [PubMed] [Google Scholar]
- Dudley AT, Robertson EJ. Overlapping expression domains of bone morphogenetic protein family members potentially account for limited tissue defects in BMP7 deficient embryos. Dev Dyn. 1997;208:349–62. doi: 10.1002/(SICI)1097-0177(199703)208:3<349::AID-AJA6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- El-Dahr SS, Aboudehen K, Saifudeen Z. Transcriptional control of terminal nephron differentiation. Am J Physiol Renal Physiol. 2008;294:F1273–8. doi: 10.1152/ajprenal.00562.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F, Jho E, Zeng L, Kurth T, Joos T, Kaufmann C, Costantini F. Domains of axin involved in protein-protein interactions, Wnt pathway inhibition, and intracellular localization. J Cell Biol. 1999;145:741–56. doi: 10.1083/jcb.145.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filali M, Cheng N, Abbott D, Leontiev V, Engelhardt JF. Wnt-3A/beta-catenin signaling induces transcription from the LEF-1 promoter. J Biol Chem. 2002;277:33398–410. doi: 10.1074/jbc.M107977200. [DOI] [PubMed] [Google Scholar]
- Fleming S, Symes CE. The distribution of cytokeratin antigens in the kidney and in renal tumours. Histopathology. 1987;11:157–70. doi: 10.1111/j.1365-2559.1987.tb02619.x. [DOI] [PubMed] [Google Scholar]
- Gong KQ, Yallowitz AR, Sun H, Dressler GR, Wellik DM. A hox-eya-pax complex regulates early kidney developmental gene expression. Mol Cell Biol. 2007;27:7661–8. doi: 10.1128/MCB.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieshammer U, Cebrian C, Ilagan R, Meyers E, Herzlinger D, Martin GR. FGF8 is required for cell survival at distinct stages of nephrogenesis and for regulation of gene expression in nascent nephrons. Development. 2005;132:3847–57. doi: 10.1242/dev.01944. [DOI] [PubMed] [Google Scholar]
- Grobstein C. Inductive Epithelio-mesenchymal Interaction in Cultured Organ Rudiments of the Mouse. Science. 1953;118:52–5. doi: 10.1126/science.118.3054.52. [DOI] [PubMed] [Google Scholar]
- Grobstein C. Inductive interactions in the development of the mouse metanephros. J Exp Zool. 1955;13:319–40. doi: 10.1002/jez.1401690105. [DOI] [PubMed] [Google Scholar]
- Gruenweld P. Development of the Excretory System. Vol. 55. Annals of the New York Academy of Sciences; 1952. pp. 142–6. [DOI] [PubMed] [Google Scholar]
- Haase G, Dessaud E, Garces A, de Bovis B, Birling M, Filippi P, Schmalbruch H, Arber S, deLapeyriere O. GDNF acts through PEA3 to regulate cell body positioning and muscle innervation of specific motor neuron pools. Neuron. 2002;35:893–905. doi: 10.1016/s0896-6273(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Hanson IM. Mammalian homologues of the Drosophila eye specification genes. Semin Cell Dev Biol. 2001;12:475–84. doi: 10.1006/scdb.2001.0271. [DOI] [PubMed] [Google Scholar]
- Hartman HA, Lai HL, Patterson LT. Cessation of renal morphogenesis in mice. Dev Biol. 2007;310:379–87. doi: 10.1016/j.ydbio.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatini V, Huh SO, Herzlinger D, Soares VC, Lai E. Essential role of stromal mesenchyme in kidney morphogenesis revealed by targeted disruption of Winged Helix transcription factor BF-2. Genes Dev. 1996;10:1467–78. doi: 10.1101/gad.10.12.1467. [DOI] [PubMed] [Google Scholar]
- Itaranta P, Lin Y, Perasaari J, Roel G, Destree O, Vainio S. Wnt-6 is expressed in the ureter bud and induces kidney tubule development in vitro. Genesis. 2002;32:259–68. doi: 10.1002/gene.10079. [DOI] [PubMed] [Google Scholar]
- James RG, Kamei CN, Wang Q, Jiang R, Schultheiss TM. Odd-skipped related 1 is required for development of the metanephric kidney and regulates formation and differentiation of kidney precursor cells. Development. 2006;133:2995–3004. doi: 10.1242/dev.02442. [DOI] [PubMed] [Google Scholar]
- Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–83. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto Y, Tokunaga H, Tomita K. Vascular endothelial growth factor is an essential molecule for mouse kidney development: glomerulogenesis and nephrogenesis. J Clin Invest. 1997;99:2351–7. doi: 10.1172/JCI119416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Kwan KM, Carroll TJ, McMahon AP, Mendelsohn CL, Behringer RR. Distinct and sequential tissue-specific activities of the LIM-class homeobox gene Lim1 for tubular morphogenesis during kidney development. Development. 2005a;132:2809–23. doi: 10.1242/dev.01858. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, McMahon AP. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3:169–81. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Tanaka H, Kuwana H, Inoshita S, Teraoka H, Sasaki S, Terada Y. Wnt4-transformed mouse embryonic stem cells differentiate into renal tubular cells. Biochem Biophys Res Commun. 2005b;336:585–95. doi: 10.1016/j.bbrc.2005.08.136. [DOI] [PubMed] [Google Scholar]
- Kopan R, Cheng HT, Surendran K. Molecular insights into segmentation along the proximal-distal axis of the nephron. J Am Soc Nephrol. 2007;18:2014–20. doi: 10.1681/ASN.2007040453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuure S, Popsueva A, Jakobson M, Sainio K, Sariola H. Glycogen synthase kinase-3 inactivation and stabilization of beta-catenin induce nephron differentiation in isolated mouse and rat kidney mesenchymes. J Am Soc Nephrol. 2007;18:1130–9. doi: 10.1681/ASN.2006111206. [DOI] [PubMed] [Google Scholar]
- Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, Rosenfeld MG. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–54. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- Little MH, Brennan J, Georgas K, Davies JA, Davidson DR, Baldock RA, Beverdam A, Bertram JF, Capel B, Chiu HS, Clements D, Cullen-McEwen L, Fleming J, Gilbert T, Herzlinger D, Houghton D, Kaufman MH, Kleymenova E, Koopman PA, Lewis AG, McMahon AP, Mendelsohn CL, Mitchell EK, Rumballe BA, Sweeney DE, Valerius MT, Yamada G, Yang Y, Yu J. A high-resolution anatomical ontology of the developing murine genitourinary tract. Gene Expr Patterns. 2007;7:680–99. doi: 10.1016/j.modgep.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Loughna S, Hardman P, Landels E, Jussila L, Alitalo K, Woolf AS. A molecular and genetic analysis of renalglomerular capillary development. Angiogenesis. 1997;1:84–101. doi: 10.1023/A:1018357116559. [DOI] [PubMed] [Google Scholar]
- Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U, van de Wetering M, Clevers H, Schlag PM, Birchmeier W, Behrens J. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol Cell Biol. 2002;22:1184–93. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A, Vainio S, Kispert A, McMahon J, McMahon AP. Wnt11 and Ret/Gdnf pathways cooperate in regulating ureteric branching during metanephric kidney development. Development. 2003;130:3175–85. doi: 10.1242/dev.00520. [DOI] [PubMed] [Google Scholar]
- Marose TD, Merkel CE, McMahon AP, Carroll TJ. Beta-catenin is necessary to keep cells of ureteric bud/Wolffian duct epithelium in a precursor state. Dev Biol. 2008;314:112–26. doi: 10.1016/j.ydbio.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AP, Aronow BJ, Davidson DR, Davies JA, Gaido KW, Grimmond S, Lessard JL, Little MH, Potter SS, Wilder EL, Zhang P. GUDMAP: the genitourinary developmental molecular anatomy project. J Am Soc Nephrol. 2008;19:667–71. doi: 10.1681/ASN.2007101078. [DOI] [PubMed] [Google Scholar]
- Meijer L, Skaltsounis AL, Magiatis P, Polychronopoulos P, Knockaert M, Leost M, Ryan XP, Vonica CA, Brivanlou A, Dajani R, Crovace C, Tarricone C, Musacchio A, Roe SM, Pearl L, Greengard P. GSK-3-selective inhibitors derived from Tyrian purple indirubins. Chem Biol. 2003;10:1255–66. doi: 10.1016/j.chembiol.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Mugford JW, Sipila P, Kobayashi A, Behringer RR, McMahon AP. Hoxd11 specifies a program of metanephric kidney development within the intermediate mesoderm of the mouse embryo. Dev Biol. 2008a;319:396–405. doi: 10.1016/j.ydbio.2008.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford JW, Sipila P, McMahon JA, McMahon AP. Osr1 expression demarcates a multi-potent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev Biol. 2008b;324:88–98. doi: 10.1016/j.ydbio.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai S, Sugitani Y, Sato H, Ito S, Miura Y, Ogawa M, Nishi M, Jishage K, Minowa O, Noda T. Crucial roles of Brn1 in distal tubule formation and function in mouse kidney. Development. 2003;130:4751–9. doi: 10.1242/dev.00666. [DOI] [PubMed] [Google Scholar]
- Nyengaard JR, Bendtsen TF. Glomerular number and size in relation to age, kidney weight, and body surface in normal man. Anat Rec. 1992;232:194–201. doi: 10.1002/ar.1092320205. [DOI] [PubMed] [Google Scholar]
- Ohto H, Kamada S, Tago K, Tominaga SI, Ozaki H, Sato S, Kawakami K. Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol Cell Biol. 1999;19:6815–24. doi: 10.1128/mcb.19.10.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Valerius MT, McMahon AP. Wnt/beta-catenin signaling regulates nephron induction during mouse kidney development. Development. 2007;134:2533–9. doi: 10.1242/dev.006155. [DOI] [PubMed] [Google Scholar]
- Pepicelli CV, Kispert A, Rowitch DH, McMahon AP. GDNF induces branching and increased cell proliferation in the ureter of the mouse. Dev Biol. 1997;192:193–8. doi: 10.1006/dbio.1997.8745. [DOI] [PubMed] [Google Scholar]
- Perantoni AO, Timofeeva O, Naillat F, Richman C, Pajni-Underwood S, Wilson C, Vainio S, Dove LF, Lewandoski M. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development. 2005;132:3859–71. doi: 10.1242/dev.01945. [DOI] [PubMed] [Google Scholar]
- Piscione TD, Wu MY, Quaggin SE. Expression of Hairy/Enhancer of Split genes, Hes1 and Hes5, during murine nephron morphogenesis. Gene Expr Patterns. 2004;4:707–11. doi: 10.1016/j.modgep.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Plisov S, Tsang M, Shi G, Boyle S, Yoshino K, Dunwoodie SL, Dawid IB, Shioda T, Perantoni AO, de Caestecker MP. Cited1 is a bifunctional transcriptional cofactor that regulates early nephronic patterning. J Am Soc Nephrol. 2005;16:1632–44. doi: 10.1681/ASN.2004060476. [DOI] [PubMed] [Google Scholar]
- Poladia DP, Kish K, Kutay B, Hains D, Kegg H, Zhao H, Bates CM. Role of fibroblast growth factor receptors 1 and 2 in the metanephric mesenchyme. Dev Biol. 2006;291:325–39. doi: 10.1016/j.ydbio.2005.12.034. [DOI] [PubMed] [Google Scholar]
- Roehl H, Nusslein-Volhard C. Zebrafish pea3 and erm are general targets of FGF8 signaling. Curr Biol. 2001;11:503–7. doi: 10.1016/s0960-9822(01)00143-9. [DOI] [PubMed] [Google Scholar]
- Sajithlal G, Zou D, Silvius D, Xu PX. Eya 1 acts as a critical regulator for specifying the metanephric mesenchyme. Dev Biol. 2005;284:323–36. doi: 10.1016/j.ydbio.2005.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariola H, Ekblom P, Lehtonen E, Saxen L. Differentiation and vascularization of the metanephric kidney grafted on the chorioallantoic membrane. Dev Biol. 1983;96:427–35. doi: 10.1016/0012-1606(83)90180-x. [DOI] [PubMed] [Google Scholar]
- Saxén L. Organogenesis of the kidney. Cambridge University Press; Cambridge [Cambridgeshire]; New York: 1987. [Google Scholar]
- Self M, Lagutin OV, Bowling B, Hendrix J, Cai Y, Dressler GR, Oliver G. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. Embo J. 2006;25:5214–28. doi: 10.1038/sj.emboj.7601381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stark K, Vainio S, Vassileva G, McMahon AP. Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature. 1994;372:679–83. doi: 10.1038/372679a0. [DOI] [PubMed] [Google Scholar]
- Stenman JM, Rajagopal J, Carroll TJ, Ishibashi M, McMahon J, McMahon AP. Canonical Wnt signaling regulates organ-specific assembly and differentiation of CNS vasculature. Science. 2008;322:1247–50. doi: 10.1126/science.1164594. [DOI] [PubMed] [Google Scholar]
- Vega QC, Worby CA, Lechner MS, Dixon JE, Dressler GR. Glial cell line-derived neurotrophic factor activates the receptor tyrosine kinase RET and promotes kidney morphogenesis. Proc Natl Acad Sci U S A. 1996;93:10657–61. doi: 10.1073/pnas.93.20.10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wynshaw-Boris A. The canonical Wnt pathway in early mammalian embryogenesis and stem cell maintenance/differentiation. Curr Opin Genet Dev. 2004;14:533–9. doi: 10.1016/j.gde.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Wang P, Pereira FA, Beasley D, Zheng H. Presenilins are required for the formation of comma- and S-shaped bodies during nephrogenesis. Development. 2003;130:5019–29. doi: 10.1242/dev.00682. [DOI] [PubMed] [Google Scholar]
- Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23:113–7. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- Xu PX, Zheng W, Huang L, Maire P, Laclef C, Silvius D. Six1 is required for the early organogenesis of mammalian kidney. Development. 2003;130:3085–94. doi: 10.1242/dev.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PX, Zheng W, Laclef C, Maire P, Maas RL, Peters H, Xu X. Eya1 is required for the morphogenesis of mammalian thymus, parathyroid and thyroid. Development. 2002;129:3033–44. doi: 10.1242/dev.129.13.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos GD, Davis S, Gale NW, Rudge JS, Wiegand SJ, Holash J. Vascular-specific growth factors and blood vessel formation. Nature. 2000;407:242–8. doi: 10.1038/35025215. [DOI] [PubMed] [Google Scholar]
- Yu J, Carroll TJ, Rajagopal J, Kobayashi A, Ren Q, McMahon AP. A Wnt7b-dependent pathway regulates the orientation of epithelial cell division and establishes the cortico-medullary axis of the mammalian kidney. Development. 2009;136:161–71. doi: 10.1242/dev.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.