Abstract
Identification of people often makes use of unique features of the face, fingerprints and retina. Beyond this, a similar identifying process can be applied to internal parts of the body that are not visible to the unaided eye. Here we show that knee X-rays can be used for the identification of individual persons. The image analysis method is based on the wnd-charm algorithm, which has been found effective for the diagnosis of clinical conditions of knee joints. Experimental results show that the rank-10 identification accuracy using a dataset of 425 individuals is ~56%, and the rank-1 accuracy is ~34%. The dataset contained knee X-rays taken several years apart from each other, showing that the identifiable features correspond to specific persons, rather than the present clinical condition of the joint.
Keywords: biometrics, X-ray, radiography, bone, joint
1 Introduction
Distinguishing characteristics used for biometric identification usually include faces, fingerprints and irises. However, while visual biometric identification usually utilizes features that can be distinguished by the human eye, it is also plausible that features that cannot be easily sensed by an unaided eye can be used to identify individuals. These can include structures of the human body that are imaged using plain radiography (X-ray), computed tomography (CT), or MRI.
Studies of the knee joint have shown that the radiographic shape and texture of the bone area around the joint can be informative for the detection and classification of osteoarthritis of the knee (Boniatis et al., 2006, 2007; Lynch, Hawkes & Buckland-Wright, 1991; Podsiadlo, P., Wolski, M. & Stachowiak, 2008; Podsiadlo et al., 2008; Shamir et al., 2008b), and reflect biochemical and biomechanical processes (Bolbos et al., 2008; Messent et al., 2005). Here we show that radiographic images of the knee joint can be used for biometric identification. In Section 2 we briefly describe the image analysis tool, in Section 3 we describe the dataset used for the experiment, and in Section 4 the experimental results are discussed.
2 Classification method
The image analysis method used in this study is wnd-charm (Orlov et al., 2008; Shamir et al., 2008a), which has been found effective for the analysis and detection of osteoarthritis using knee X-rays (Shamir et al., 2008b). Wnd-charm is a multi-purpose image classification method that makes use of a large set of image features, extracted from several image transforms and compound transforms. The image features used by wnd-charm can be classified into high-contrast features, textures (e.g., Haralick, Tamura), statistical distribution of the pixel values (e.g., multi-scale histogram, first four moments), and polynomial decomposition of the image. The image transforms that are used are Fourier transform, Chebyshev transform, Wavelet (symlet 5, level 1) transform, and edge transform. A detailed description of the image features and image transforms can be found in (Orlov et al., 2008; Shamir et al., 2008a).
While the major downside of wnd-charm is its computational complexity, its large set of image content descriptors allows it to apply a systematic search for the most informative features. This is done by assigning a Fisher score to each image feature, so that the different features are weighted by their informativeness. When a feature vector of a test image needs to be classified, a simple Weighted Nearest Neighbor rule is applied such that the Fisher scores are the feature weights. This simple classification method is described more thoroughly in (Orlov et al., 2008; Shamir et al., 2008a). The source code of wnd-charm is publicly available, and can be downloaded at (Shamir et al., 2008a).
3 Dataset
The experiment used a dataset of 1700 X-ray images of 425 individuals, such that each person in the dataset had four knee joint images. The film X-rays were digitized using a UMAX PowerLook 1100 scanner and saved as 16-bit 2550×3000 TIFF files. All images were then normalized to a fixed mean and standard deviation, and a 700×500 area of the center of the joint was separated from the image using a simple joint detection algorithm (Shamir et al., 2008b). Figure 1 shows a sample joint image used for the automated recognition.
Figure 1.
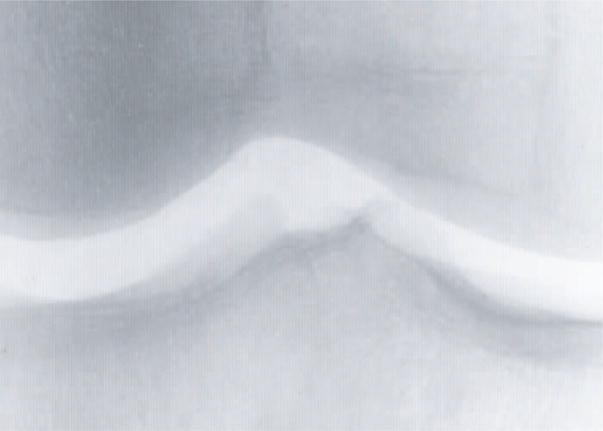
A knee joint X-ray image used for the automatic identification
All data were acquired as part of the BLSA (Baltimore Longitudinal Study of Aging) project (Shock et al., 1984), which is a longitudinal normative aging study. Due to the longitudinal nature of the project, each X-ray image was acquired in a different year, and when the person was of a different age. The average time difference between the acquisition of two X-rays is ~4.4 years, with standard deviation of ~1.9 years. The importance of this time difference is that it allows to perform the identification based on the actual biometric identifiers of the joint X-ray, rather than the status of the on-going biochemical and biomechanical processes that might be unique for that person only for that certain period of time.
4 Experimental results
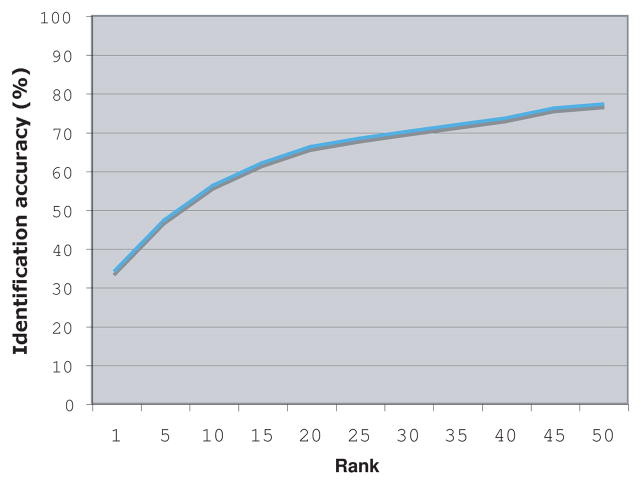
The identification accuracy was tested using 1275 images for training (three images per person) and 425 for testing (1 per person). Each experiment was repeated 20 times, such that in each run different images of each person were randomly assigned for training and test sets. Figure 2 shows the resulting identification accuracy.
Figure 2.
Identification accuracy (N=425)
As the figure shows, the rank-1 recognition accuracy of the X-rays is 34.1%, and the rank-10 accuracy is 56%. Although these numbers are not comparable to recent reports on iris and fingerprint identification, they are substantially higher than random, and provide an indication that different persons can be identified by their knee X-rays.
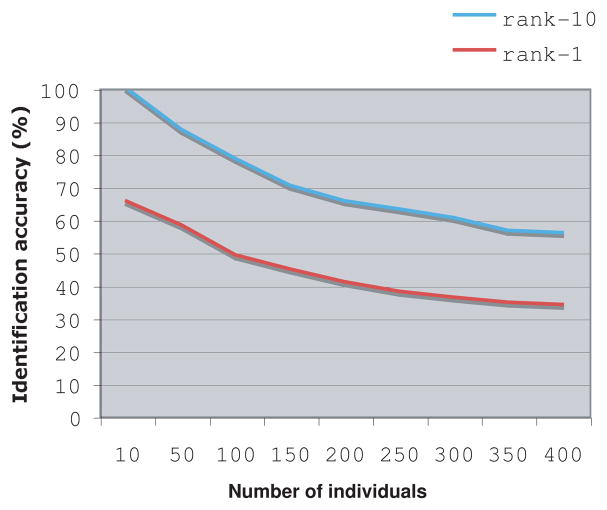
Another experiment aimed to test how the identification accuracy changes as the number of individuals in the dataset gets larger. Figure 3 shows the rank-1 and rank-10 identification accuracy as a function of the number of individuals in the dataset. As expected, the recognition accuracy dropped as the number of individuals in the system increased.
Figure 3.
Identification accuracy as a function of the number of individuals
5 Conclusion
Here we showed that X-ray images of knee joints can be used for the identification of different persons. The accuracy of the method used in this study is not comparable to recognition rates reported by recent studies of iris and fingerprint identification. However, the recognition accuracy using X-rays is significantly higher than random, and demonstrates that the knee joint is recognizably different for different people. This shows that visual features that cannot be sensed by the unaided eye can be informative for the identification of different persons.
Moreover, biometrics systems that utilize internal body structures imaged by instruments such as CT and MRI are less vulnerable to deceptive manipulation than standard biometrics such as facial features, and therefore potentially appealing for the purpose of reliable biometric identification.
Acknowledgments
This research was supported entirely by the Intramural Research Program of the NIH, National Institute on Aging.
Biographies
Lior Shamir is a research fellow in the Image Informatics Unit of Laboratory of Genetics, National Institute on Aging, National Institute of Health.
Shari Ling is currently a Staff Clinician in the Clinical Research Branch of the National Institute on Aging Intramural Research Program. She also holds an appointment in the Division of Geriatric Medicine, Gerontology and Rheumatology at Johns Hopkins University School of Medicine, and is on the clinical faculty at the University of Maryland. Her area of interest is aging, osteoarthritis, the identification and development of novel diagnostic and prognostic indicators of arthritis development in the elderly.
Salim Rahimi is studying towards his Bachelors degree in Computer Engineering at the State University of New York, Farmingdale.
Luigi Ferrucci received his medical degree at the University of Florence, where he also earned his doctor of philosophy degree in Biology and Pathophysiology of Aging. Dr. Ferrucci is the Director of the Baltimore Longitudinal Study of Aging and a senior investigator at the Clinical Research Branch of the National Institute on Aging, National Institutes of Health. His research interests involve frailty and mobility disability in the elderly.
Ilya G. Goldberg received his BS in Biochemistry from the University of Wisconsin-Madison in 1990, and his PhD. in Biochemistry and Cell Biology from the Johns Hopkins University School of Medicine in 1997. After a post-doc in crystallography at Harvard, he started the Open Microscopy Environment (OME) project as a post-doc at MIT. In 2002, he returned to Baltimore and joined the Laboratory of Genetics at the National Institute on Aging, where he started a group dedicated to quantitative morphology and systematic functional genomics.
References
- Bolbos RI, Zuo J, Banerjee S, Link TM, Ma CB, Li X, Majumdar S. Relationship between trabecular bone structure and articular cartilage morphology and relaxation times in early OA of the knee joint using parallel MRI at 3T. Osteoarthritis Cartilage. doi: 10.1016/j.joca.2008.02.018. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniatis I, Costaridou L, Cavouras D, Kalatzis I, Panagiotopoulos E, Panayiotakis G. Osteoarthritis severity of the hip by computer-aided grading of radiographic images. Med & Bio Eng & Comp. 2006;44:793–803. doi: 10.1007/s11517-006-0096-3. [DOI] [PubMed] [Google Scholar]
- Boniatis I, Cavouras D, Costaridou L, Kalatzis I, Panagiotopoulos E, Panayiotakis G. Computer-aided grading and quantification of hip osteoarthritis severity employing shape descriptors of radiographic hip joint space. Comp Bio & Med. 2007;37:1786–1795. doi: 10.1016/j.compbiomed.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Lynch JA, Hawkes DJ, Buckland-Wright JC. Analysis of texture in macroradiographs of osteoarthritic knee using the fractal signature. Phys Med Biol. 1991;36:709–722. doi: 10.1088/0031-9155/36/6/001. [DOI] [PubMed] [Google Scholar]
- Messent EM, Ward RJ, Tonkin CJ, Buckland-Wright JC. Cancellous bone difference between knees with early, definite and advanced joint space loss: A comparative quantitative macroradiographic study. Osteoarthritis Cartilage. 2005;13:39–47. doi: 10.1016/j.joca.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Orlov N, Shamir L, Johnston J, Macura T, Eckley DM, Goldberg IG. WND-CHARM: Multi-purpose image classification using compound image transforms. Pattern Recognition Letters. 2008;29:1684–1693. doi: 10.1016/j.patrec.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podsiadlo P, Wolski MM, Stachowiak GW. Automated selection of trabecular bone regions in knee radiographs. Med Phys. 2008;35:1870–1883. doi: 10.1118/1.2905025. [DOI] [PubMed] [Google Scholar]
- Podsiadlo P, Dahl L, Englund M, Lohmander LS, Stachowiak GW. Differences in trabecular bone texture between knees with and without radiographic osteoarthritis detected by fractal methods. Osteoarthritis Cartilage. 2008;16:323–329. doi: 10.1016/j.joca.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Shamir L, Orlov N, Macura T, Eckley DM, Johnston J, Goldberg IG. Wndchrm - An Open Source Utility for Biological Image Analysis. BMC - Source Code for Biology and Medicine. 2008;3:13. doi: 10.1186/1751-0473-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamir L, Ling S, Scott W, Bos A, Orlov N, Macura T, Eckley DM, Goldberg IG. Knee X-ray image analysis method for automated detection of Osteoarthritis. IEEE Transactions on Biomedical Engineering. doi: 10.1109/TBME.2008.2006025. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shock NW, et al. Normal human aging: the Baltimore Longitudinal Study of Aging. Washington, DC: Government Printing Office; 1984. NIH Publication No. 84–2450. [Google Scholar]