DNA charge transport (CT) chemistry has been characterized in detail[1] owing to its utility in the construction of DNA-based sensors and nanoscale devices.[2–4] Interestingly, recent evidence also points to the application of this chemistry within the cell in the context of DNA damage and repair.[5] Both DNA hole transport (HT) and electron transport (ET) have been explored using spectroscopy,[6] biochemical assays,[7] and DNA electrochemistry.[8] In reporting on DNA-mediated HT, N2-cyclopropylguanine and N6-cyclopropyladenine (CPA) have been useful hole traps owing to their low oxidation potentials and their fast rates of ring opening (10−11 s) upon one-electron oxidation (Scheme 1).[9] Similarly, owing to their redox potentials,[10] the pyrimidines N4-cyclopropylcytosine (CPC)[11] and 5-bromouridine (BrU),[12] have been sensitive probes for DNA-mediated ET.
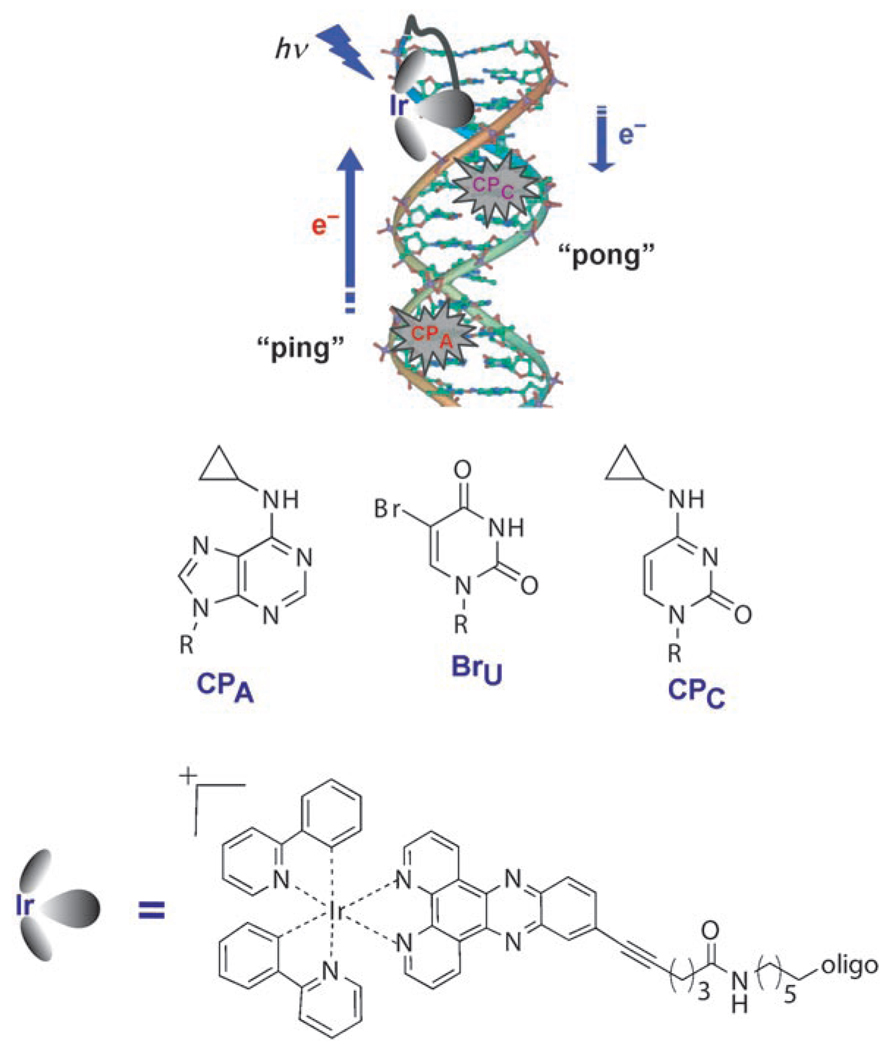
Scheme 1.
Representation of the ping-pong reaction generated through photoactivation of an Ir–DNA assembly along with modified base traps for hole (CPA) and electron (BrU and CPC) transport. Upon excitation, the excited IrIII complex irreversibly oxidizes a CPA base from a distance. The subsequent reduced metallic species is, in turn, able to reduce distal BrU or CPC bases.
Recently, direct comparisons of HT with ET were carried out using a charge injector strongly coupled to the base stack and able to oxidize or reduce bases from a distance within the same DNA assembly.[13, 14] Taking advantage of the powerful photochemistry of IrIII biscyclometalated complexes, we have shown that the [Ir(ppy)2(dppz′)]1+, functionalized through a modified dipyrido[3,2-a:2′,3′-c]phenazine (dppz′; ppy=2-phenylpyridine; Scheme 1), can serve both as a photooxidant and reductant of distal DNA bases.[13, 15] Strikingly, we found that DNA HT and ET have similar characteristics; both show a remarkably shallow distance dependence in their reactions as well as an equal sensitivity to perturbations in stacking of the intervening DNA bridge.
Significantly, although the excited state of the IrIII complex is sufficiently potent to oxidize guanine or adenine,[15] efficient electron injection into DNA required the use of the flash-quench technique.[16] Indeed, the excited state of the IrIII complex is not sufficiently long-lived (τ<10 ns) in water to photoreduce BrU from a distance in substantial yield.[13] When sodium ascorbate is used as an external reducing agent, however, photolysis of the tethered IrIII complex generates a long-lived ground-state reductant, which is in turn able to induce significant BrU decomposition from a distance.
Herein we describe a novel Ir system that is able to promote the reduction of pyrimidine bases from a distance without the presence of an external quencher. Instead, DNA-mediated ET is triggered by DNA-mediated HT. Thus, photoactivation of these Ir assemblies results in both a forward and a reverse pattern for charge migration, which we term “ping-pong” electron transfer through DNA (Scheme 1).
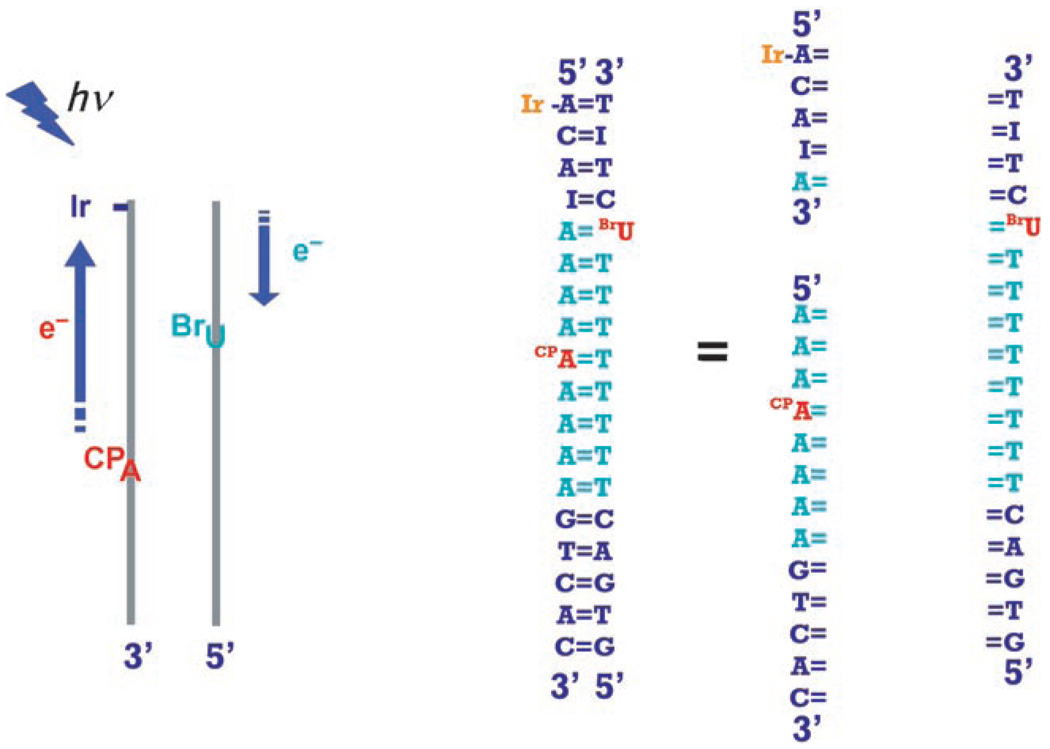
We have designed Ir–DNA conjugates containing two modified bases embedded in an AT tract,[17] a CPA for hole injection, and either a BrU or a CPC as an electron trap (Scheme 2). For synthetic reasons, we required three different strands, one short strand (5-mer) covalently tethered to the IrIII complex, one strand (13-mer) containing a CPA-modified base, and one complementary strand (18-mer) containing either a BrU or CPC. It should be noted that 1) the presence of a nick in the phosphate backbone of the DNA helix does not affect the CT process,[18] 2) the intercalative dppz′ unit stabilizes the short Ir strand in the duplex,[19] and 3) the irreversible ring-opening of CPA precludes back ET.
Scheme 2.
The assembly of three modified DNA single strands annealed to generate the Ir–DNA duplex for the ping-pong reaction.
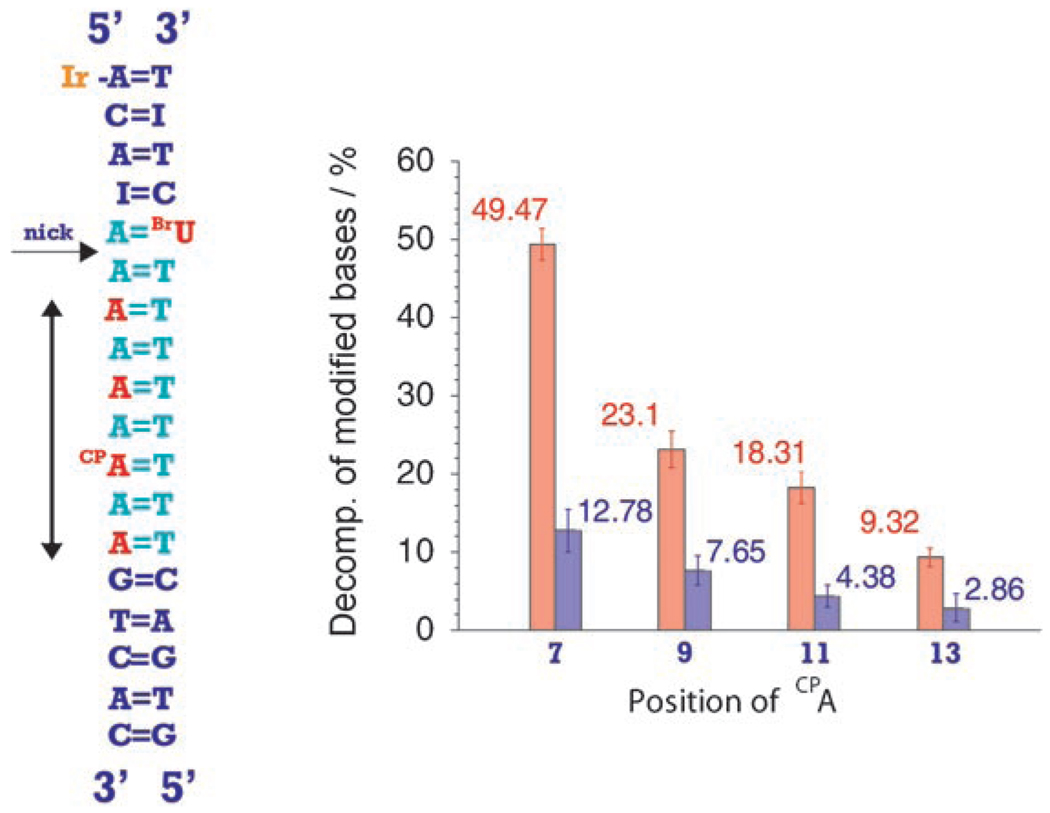
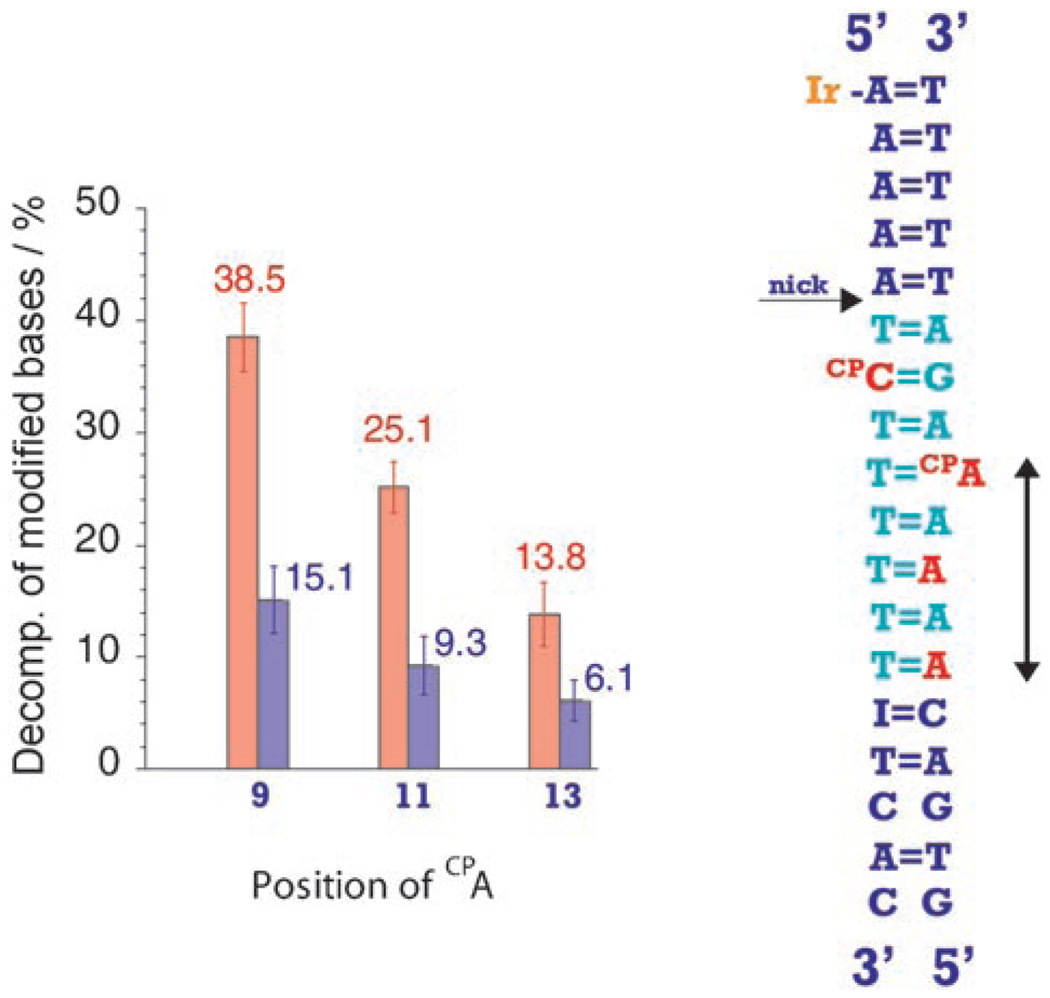
Figure 1 shows the decomposition percentage of CPA and BrU obtained after 30 min irradiation of the DNA assemblies at 365 nm as a function of the CPA position (see the Experimental Section). Clearly, both modified bases undergo a decomposition reaction upon photolysis. No decomposition of BrU is observed without both CPA and tethered IrIII complex in the assembly. Moreover, the extent of BrU decomposition varies with the position of CPA. We therefore see that the BrU decomposition is correlated with CPA ring-opening. We ascribe this correlation to the ping-pong reaction, where the highly oxidizing IrIII excited state[13, 15] is reductively quenched in an intraduplex reaction by the distal CPA (migration of a “ping” electron); the ground-state Ir reductant, formed by the intraduplex flash-quench scheme, is identical to the one generated using an external quencher[13] and is capable of reducing BrU by ET through the DNA stack (migration of a “pong” electron).
Figure 1.
Decomposition percentage for CPA (red) in position 7, 9, 11, or 13 from Ir attachment and BrU (blue) in position 5 from Ir after 30 min irradiation (365 nm).
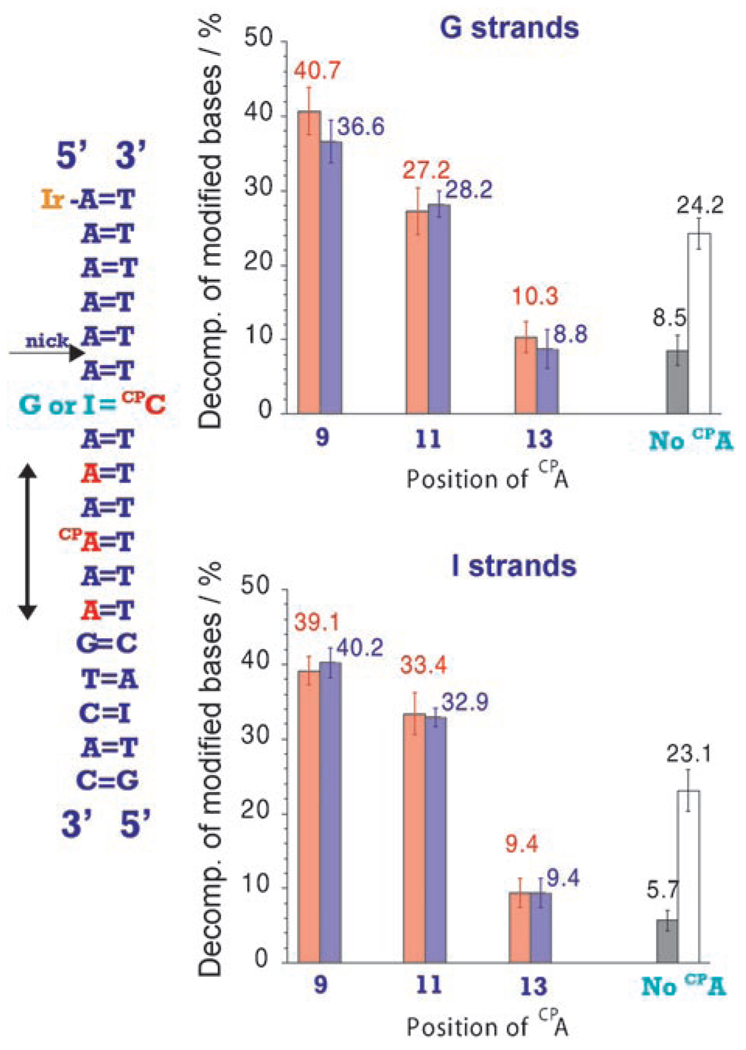
We have further tested the ping-pong reaction using CPC as a reductive probe, and here substantial yields of decomposition of the modified base are observed.[11, 15] While CPC can be either photoreduced or photooxidized by the IrIII complex, it has been shown that this modified base is reductively ring-opened by excited Ir when embedded in a thymine stack.[15] Figure 2 shows the decomposition percentages of CPA and CPC as a function of CPA position in Ir–DNA conjugates, which contain CPA within an A tract and CPC on the complementary strand. Here, a higher relative yield is obtained for the decomposition of the reductive probe compared to that of BrU. In fact, the CPC decomposition is stoichiometric with CPA decomposition. Importantly, in the absence of CPA, little CPC decomposition is obtained; the presence of CPA is required for significant CPC yields of decomposition. Also, no decomposition is observed without the presence of tethered IrIII complex. Furthermore, no change in CPC decomposition is found in assemblies where CPC is base-paired to an inosine (Figure 2); if decomposition of CPC were to occur through photooxidation, an enhancement in yield would be evident without the competitive oxidative guanine sink.[20] Thus CPC ring-opening here results from reduction by DNA-mediated ET and not from a photooxidation process. The ping-pong reaction is, then, highly efficient. Figure 2 shows also the decomposition yields for CPC in the absence of CPA, both without (gray bars) and with (white bars) an external reductive quencher (sodium ascorbate) known to reductively quench the IrIII excited state.[13] The ground-state reduced metallic species, generated through CPA oxidation, can effectively reduce the distal CPC, indeed comparably to an external reductive quencher.
Figure 2.
Decomposition percentage for CPA (red) in position 9, 11, or 13 from Ir attachment and CPC (blue) in position 7 from Ir after 30 min irradiation (365 nm). The CPC is base-paired either to a guanine (left) or to an inosine (right). In both cases, the gray and white bars show the decomposition percentage for CPC without the presence of CPA, with (white) or without (gray) an external reductive quencher (sodium ascorbate, 200 mm).
The sensitivity of DNA CT to DNA structure and dynamics is illustrated also for the ping-pong reaction through variations in the DNA sequence. Despite equal overall energetics, the decomposition efficiency in these assemblies is seen to depend upon the position of the redox trap in the double helix. For instance, exchanging the CPA and CPC and the AT tract from one strand to the other significantly affects the overall yield, especially for reduction (Figure 3). This result is consistent with sequence variations seen earlier for DNA-mediated ET, and more generally for CT in DNA with variations in the intervening base stack.[13, 21]
Figure 3.
Decomposition percentage for CPA (red) in position 9, 11, or 13 from Ir attachment and CPC (blue) in position 7 from Ir after 30 min irradiation (365 nm). Both the AT tract and the CPA and CPC have been flipped compared to sequences presented in Figure 2.
Thus, for the first time, DNA-mediated HT and ET have been triggered consecutively within the same DNA duplex by irradiation of a unique charge injector. The ping-pong reaction likely involves hole migration primarily through the purine strand with electron migration facilitated by stacked pyrimidines. Critically, the analogous parameters govern both HT and ET through the DNA base-pair stack.
Experimental Section
All DNA oligonucleotides were synthesized using standard phosphoramidite chemistry. Strands containing a CPA or CPC were synthesized by placing an O6-phenylinosine or 4-thiouridine at the target CPA or CPC site, respectively. After 16 h incubation at 60°C in 6m aqueous cyclopropylamine leading to simultaneous substitution reaction, cleavage from the solid support, and deprotection, the strands were purified using reversed-phase HPLC and characterized by MALDI mass spectrometry. [Ir(ppy)2(dppz)′]+ was synthesized and covalently tethered to DNA oligonucleotides according to previously described methods.[13, 15] Ir–DNA conjugates were prepared by combining equimolar amounts (1:1:1) of the desired DNA single strands. After annealing (solution was heated to 90°C for 5 min, then slowly cooled down to 15°C over a period of 3 h), all of the resulting duplexes showed melting temperatures above ambient temperature.
Aliquots (30 µL) of the Ir–DNA conjugates (10 µm in 50 mm TrisHCl, pH 7.0) were irradiated for 30 min at 365 nm (Hg/Xe lamp, 1000W, with monochromator). Subsequent cleavage into deoxynucleosides upon digestion with phosphodiesterase I and alkaline phosphatase was carried out and the results analyzed by HPLC. The percentage of base decomposition, that is, the amount of decomposed CPA, CPC, or BrU, was determined by subtracting the ratio of the area under the peak of the undecomposed base in an irradiated sample over that in a non-irradiated sample from unity, with adenine or inosine as an internal HPLC standard. Irradiation experiments were repeated three times and the results averaged.
Footnotes
This work was supported by the NIH (GM49216).
Contributor Information
Benjamin Elias, Chimie Organique et Médicinale, Université catholique de Louvain (Belgium)
Joseph C. Genereux, Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, CA 91125 (USA)
Jacqueline K. Barton, Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, CA 91125 (USA)
References
- 1.a) Carell T, Behrens C, Gierlich J. Org. Biomol. Chem. 2003;1:2221. doi: 10.1039/b303754a. [DOI] [PubMed] [Google Scholar]; b) O’Neill MA, Barton JK. In: Charge Transfer in DNA: From Mechanism to Application. Wagenknecht HA, editor. New York: Wiley; 2005. p. 27. [Google Scholar]; c) Conwell EM. Proc. Natl. Acad. Sci. USA. 2005;102:8795. doi: 10.1073/pnas.0501406102. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Giese B. Top. Curr. Chem. 2004;236:27. [Google Scholar]
- 2.a) Bandyopadhyay A, Ray AK, Sharma AK, Khondaker SI. Nanotechnology. 2006;17:227. [Google Scholar]; b) Porath D, Cuniberti G, Di Felice R. Top. Curr. Chem. 2004;237:183. [Google Scholar]; c) Clever GH, Kaul C, Carell T. Angew. Chem. 2007;119:6340. doi: 10.1002/anie.200701185. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2007;46:6226. [Google Scholar]; d) Guo X, Gorodetsky AA, Hone J, Barton JK, Nuckolls C. Nat. Nanotechnol. 2008;3:163. doi: 10.1038/nnano.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Drummond TG, Hill MG, Barton JK. Nat. Biotechnol. 2003;21:1192. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]; b) Yang Y, Wang Z, Yang M, Li J, Zheng F, Shen G, Yu R. Anal. Chim. Acta. 2007;584:268. doi: 10.1016/j.aca.2006.11.055. [DOI] [PubMed] [Google Scholar]; c) Wang X, Ozkan CS. Nano Lett. 2008;8:308. doi: 10.1021/nl071180e. [DOI] [PubMed] [Google Scholar]
- 4.a) Boon EM, Ceres DM, Hill MG, Drummond TD, Barton JK. Nat. Biotechnol. 2000;18:1096. doi: 10.1038/80301. [DOI] [PubMed] [Google Scholar]; b) Boon EM, Salas JW, Barton JK. Nat. Biotechnol. 2002;20:282. doi: 10.1038/nbt0302-282. [DOI] [PubMed] [Google Scholar]; c) Boal AK, Barton JK. Bioconjugate Chem. 2005;16:312. doi: 10.1021/bc0497362. [DOI] [PubMed] [Google Scholar]
- 5.Merino EJ, Boal AK, Barton JK. Curr. Opin. Chem. Biol. 2008;12:229. doi: 10.1016/j.cbpa.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Wan C, Fiebig T, Kelley SO, Treadway CR, Barton JK, Zewail AH. Proc. Natl. Acad. Sci. USA. 1999;96:6014. doi: 10.1073/pnas.96.11.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Takada T, Kawai K, Cai X, Sigimoto A, Fujitsuka M, Majima T. J. Am. Chem. Soc. 2004;126:1125. doi: 10.1021/ja035730w. [DOI] [PubMed] [Google Scholar]; c) Lewis FD, Zhu H, Daublain P, Cohen B, Wasielewski MR. Angew. Chem. 2006;118:8150. doi: 10.1002/anie.200603455. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2006;45:7982. [Google Scholar]
- 7.a) Hall DB, Holmlin RE, Barton JK. Nature. 1996;382:731. doi: 10.1038/382731a0. [DOI] [PubMed] [Google Scholar]; b) Ghosh A, Joy A, Schuster GB, Douki T, Cadet J. Org. Biomol. Chem. 2008;6:916. doi: 10.1039/b717437c. [DOI] [PubMed] [Google Scholar]; c) Nakatani K, Saito I. Top. Curr. Chem. 2004;236:163. [Google Scholar]; d) Nunez ME, Hall DB, Barton JK. Chem. Biol. 1999;6:85. doi: 10.1016/S1074-5521(99)80005-2. [DOI] [PubMed] [Google Scholar]
- 8.a) Gorodetsky AA, Barton JK. Langmuir. 2006;22:7917. doi: 10.1021/la0611054. [DOI] [PubMed] [Google Scholar]; b) Kelley SO, Hill MG, Barton JK. Angew. Chem. 1999;111:991. doi: 10.1002/(SICI)1521-3773(19990401)38:7<941::AID-ANIE941>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 1999;38:941. [Google Scholar]; c) Drummond TG, Hill MG, Barton JK. J. Am. Chem. Soc. 2004;126:15010. doi: 10.1021/ja044910i. [DOI] [PubMed] [Google Scholar]
- 9.a) Nakatani K, Dohno C, Saito I. J. Am. Chem. Soc. 2001;123:9681. doi: 10.1021/ja010479a. [DOI] [PubMed] [Google Scholar]; b) Musa OM, Horner JH, Shahin H, Newcomb M. J. Am. Chem. Soc. 1996;118:3862. [Google Scholar]; c) O’Neill MA, Dohno C, Barton JK. J. Am. Chem. Soc. 2004;126:1316. doi: 10.1021/ja037802p. [DOI] [PubMed] [Google Scholar]; d) Dohno C, Ogawa A, Nakatani K, Saito I. J. Am. Chem. Soc. 2003;125:10154. doi: 10.1021/ja035887o. [DOI] [PubMed] [Google Scholar]
- 10.a) Steenken S, Telo JP, Novais LP, Candeias LP. J. Am. Chem. Soc. 1992;114:4701. [Google Scholar]; b) Seidel CAM, Schultz A, Sauer MHM. J. Phys. Chem. 1996;100:5541. [Google Scholar]
- 11.a) Lu W, Vicic DA, Barton JK. Inorg. Chem. 2005;44:7970. doi: 10.1021/ic051124l. [DOI] [PubMed] [Google Scholar]; b) Shao F, O’Neill MA, Barton JK. Proc. Natl. Acad. Sci. USA. 2004;101:17914. doi: 10.1073/pnas.0408128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Manetto A, Breeger S, Chatgilialoglu C, Carell T. Angew. Chem. 2006;118:325. doi: 10.1002/anie.200502551. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2006;45:318. [Google Scholar]; b) Cook GP, Greenberg MM. J. Am. Chem. Soc. 1996;118:10025. [Google Scholar]; c) Ito T, Rokita SE. J. Am. Chem. Soc. 2003;125:11480. doi: 10.1021/ja035952u. [DOI] [PubMed] [Google Scholar]
- 13.Elias B, Shao F, Barton JK. J. Am. Chem. Soc. 2008;130:1152. doi: 10.1021/ja710358p. [DOI] [PubMed] [Google Scholar]
- 14.Valis L, Wang Q, Raytchev M, Buchvarov I, Wagenknecht HA, Fiebig T. Proc. Natl. Acad. Sci. USA. 2006;103:10192. doi: 10.1073/pnas.0600957103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Shao F, Elias B, Lu W, Barton JK. Inorg. Chem. 2007;46:10187. doi: 10.1021/ic7014012. [DOI] [PubMed] [Google Scholar]; b) Shao F, Barton JK. J. Am. Chem. Soc. 2007;129:14733. doi: 10.1021/ja0752437. [DOI] [PubMed] [Google Scholar]
- 16.a) Chang IJ, Gray HB, Winkler JR. J. Am. Chem. Soc. 1991;113:7056. [Google Scholar]; b) Stemp EDA, Arkin MR, Barton JK. J. Am. Chem. Soc. 1997;119:2921. [Google Scholar]
- 17.The A tract was chosen because of its resistance to inherent charge trapping. See Kawai K, Osakada Y, Fujitsuka M, Majima T. J. Phys. Chem. B. 2007;111:2322. doi: 10.1021/jp0661847. Augustyn KE, Genereux JC, Barton JK. Angew. Chem. 2007;119:5833. doi: 10.1002/anie.200701522. Angew. Chem. Int. Ed. 2007;46:5731.
- 18.Liu T, Barton JK. J. Am. Chem. Soc. 2005;127:10160. doi: 10.1021/ja053025c. [DOI] [PubMed] [Google Scholar]
- 19.Duplexes show higher melting temperature in the Ir band (365 nm) than in the DNA band (260 nm) by 9–10°C. This indicates that the complex prevents the two end strands from separating. This observation is characteristic of a DNA complex containing an intercalated planar ligand and has already been observed in other systems. See Kitamura Y, Ihara T, Okada K, Tsujimura Y, Shirasaka Y, Tazaki M, Jyo A. Chem. Commun. 2005:4523. doi: 10.1039/b505632b. Ossipov D, Pradeepkumar PI, Holmer M, Chattopadhyaya J. J. Am. Chem. Soc. 2001;123:3551. doi: 10.1021/ja003985t.
- 20.Inosine is structurally similar to guanosine but has a higher oxidation potential. Therefore, if CPC is oxidized, a higher extent of decomposition is expected when CPC is base-paired to inosine rather than to guanosine, as less competition for hole formation is expected. No decomposition enhancement with inosine versus guanine indicates that a different process other than hole formation must be involved. See Refs. [11] and [13] for further discussion.
- 21.Shao F, Augustyn KE, Barton JK. J. Am. Chem. Soc. 2005;127:17445. doi: 10.1021/ja0563399. [DOI] [PubMed] [Google Scholar]