Abstract
A mutational analysis of the matrix metalloproteinase (MMP) gene family in human melanoma identified somatic mutations in 23% of melanomas. Five mutations in one of the most commonly mutated genes, MMP8, reduced MMP enzyme activity. Expression of wild-type but not mutant MMP8 in human melanoma cells inhibited growth on soft agar in vitro and tumor formation in vivo, suggesting that wild-type MMP-8 has the ability to inhibit melanoma progression.
MMPs are proteolytic enzymes that degrade components of extracellular matrix and basement membranes1. MMPs have been associated with cancer metastasis2,3, and small molecule inhibitors of MMPs were tested as potential anticancer agents. However, clinical trials using these inhibitors showed no effect and, occasionally, accelerated tumor growth4,5. In contrast to the idea that MMP activity promotes melanoma progression, mouse models suggested that MMPs can have an antitumor role6-8. In particular, an increase in skin tumor incidence was seen in MMP-8-deficient mice6. These findings suggest that an in-depth analysis of the specific role of individual MMPs in particular cancer types is warranted. We systematically addressed these issues by a comprehensive mutational analysis of the MMP gene superfamily in melanoma.
The human MMP family consists of 23 genes. To evaluate whether these are genetically altered in melanoma, we analyzed the coding exons of this gene superfamily in 32 individuals with melanoma (Supplementary Table 1, Supplementary Methods and Supplementary Note online). Exons were PCR-amplified from cancer genomic DNA samples (see primers listed in Supplementary Table 2 online) and sequenced with dye terminator chemistry. To determine whether a given mutation was somatic, we sequenced the gene in genomic DNA from matched normal tissue. From the ∼5.5 Mb of sequence information obtained, we identified eight MMP genes containing somatic mutations (Table 1). Genes found to have one or more nonsynonymous mutations were then screened for mutations in an additional 47 melanomas. Through this approach, we identified 28 somatic mutations in eight genes, affecting 23% of the melanoma tumors analyzed (Table 1 and Supplementary Fig. 1 online).
Table 1. Mutations identified in MMPs.
| Gene | No. of nonsynonymous mutations (% tumors affected)a | Tumor | Exon | Nucleotideb | Amino acidb | Functional domain | NRAS or BRAF mutation |
|---|---|---|---|---|---|---|---|
| MMP2 | 3 (3.8%) | 10T | 2 | C251T | P84L | PGBD | BRAF |
| 76T | 3 | C384T | I128I | Hemopexin | None | ||
| 63T | 9 | A/T (-4) | Splice site | N/A | NRAS | ||
| 52T | 12 | A1865G | D622G | Hemopexin | BRAF | ||
| MMP8 | 6 (6.3%) | 32T | 2 | C149T | S50F/LOH | PGBD | BRAF |
| 13T | 2 | C232T | P78S/LOH | PGBD | None | ||
| 13T | 2 | A261C | K87N/LOH | PGBD | None | ||
| 85T | 2 | G310A | G104R/LOH | Catalytic domain | BRAF | ||
| 84T | 3 | G412C | E138Q/LOH | Catalytic domain | BRAF | ||
| 45T | 7 | G940A | E314K | Hemopexin | BRAF | ||
| MMP11 | 2 (2.5%) | 12T | 2 | G209T | R70L | Pro-rich | NRAS |
| 12T | 2 | G210A | R70R | Pro-rich | NRAS | ||
| 13T | 5 | C704T | A235V | Catalytic domain | None | ||
| MMP14 | 2 (2.5%) | 64T | 4 | G549A | E183E | Peptidase M | BRAF |
| 71T | 7 | C/T (-7) | Splice site | N/A | BRAF | ||
| 17T | 10 | T1664C | V555A | Hemopexin | NRAS | ||
| MMP24 | 4 (3.8%) | 55T | 3 | G488A | W163X | PGBD | None |
| 39T | 4 | G648A | K216K | Peptidase M | None | ||
| 55T | 4 | G718A | G240R | Peptidase M | None | ||
| 8T | 8 | G1519A | D507N | Hemopexin | BRAF | ||
| 13T | 8 | C1565A | P521Q | Hemopexin | None | ||
| MMP26 | 1 (1.3%) | 17T | 5 | C/T (+5) | Splice site | N/A | NRAS |
| MMP27 | 7 (7.6%) | 12T | 1 | G73A | E25K | PGBD | NRAS |
| 32T | 2 | G193A | E65K/LOH | PGBD | BRAF | ||
| 5T | 2 | G317A | W106X/LOH | Catalytic domain | BRAF | ||
| 76T | 3 | C374T | A125V/LOH | Catalytic domain | None | ||
| 52T | 8 | G1047A | W349X | Hemopexin | BRAF | ||
| 81T | 9 | G1218A | M406I | Hemopexin | BRAF | ||
| 81T | 9 | G1219A | D407N | Hemopexin | BRAF | ||
| MMP28 | 3 (2.5%) | 54T | 5 | G728C | G243A | Catalytic domain | BRAF |
| 44T | 6 | A936T | Q312H | Hemopexin | NRAS | ||
| 44T | 6 | G937A | G313R | Hemopexin | NRAS | ||
| 76T | 7 | G1089A | Q363Q | Hemopexin | None |
X, stop codon; LOH, cases wherein the wild-type allele was lost and only the mutant allele remained. Splice site, case wherein the alteration affected ten bases flanking the exon. None, no mutation observed. PGBD, peptidoglycan binding domain. Nine tumors (12T, 13T, 17T, 32T, 44T, 52T, 55T, 76T and 81T) contained more than one somatic alteration.
Number of nonsynonymous and splice-site mutations observed and percentage of tumors affected for each of the eight genes in the panel of 79 melanoma cancers.
Nucleotide and amino acid change resulting from mutation.
In seven tumors, both alleles of the MMP gene were affected, a characteristic associated with tumor suppressor genes. In addition, 6 of the 28 mutations were nonsense or splice-site alterations, which were predicted to result in aberrant or truncated proteins. Most tumors with MMP gene mutations also contained mutations in NRAS or BRAF. The clinical information associated with melanoma tumors containing MMP mutations is described in Supplementary Table 3 online.
The observed somatic mutations could be either ‘driver’ mutations that have a functional role underlying neoplasia or nonfunctional ‘passenger’ changes. In the eight genes found to be mutated, 28 nonsynonymous (N) and 5 synonymous (S) somatic mutations were identified, yielding a N:S ratio of 28:5, significantly higher than the N:S ratio of 2:1 predicted for nonselected passenger mutations9 (P < 0.026), suggesting that these are driver mutations. The ratio of C>T mutations compared to other nucleotide substitutions had a significant prevalence of C:G>T:A transitions (P < 0.0001) (Supplementary Fig. 2 online), confirming previously reported melanoma signatures10.
MMP8 and MMP27 were the most frequently mutated MMP genes in our samples. Notably, MMP-8 and MMP-27 have an identical protein domain structure1 (Supplementary Fig. 3 online). We chose to focus on MMP8 because it has been previously shown to protect against skin tumor development6,8, and most of its mutations were accompanied by loss of heterozygosity, suggesting that MMP8 is a tumor suppressor gene. To test whether MMP8 is also inactivated through epigenetic events, we screened the sequence surrounding MMP8 for DNA methylation using an EpiTYPER system (Sequenom). We found that local epigenetic changes are not involved in MMP8 inactivation in our melanoma samples (data not shown), consistent with our finding that MMP8 is expressed in melanoma (Supplementary Fig. 4 online).
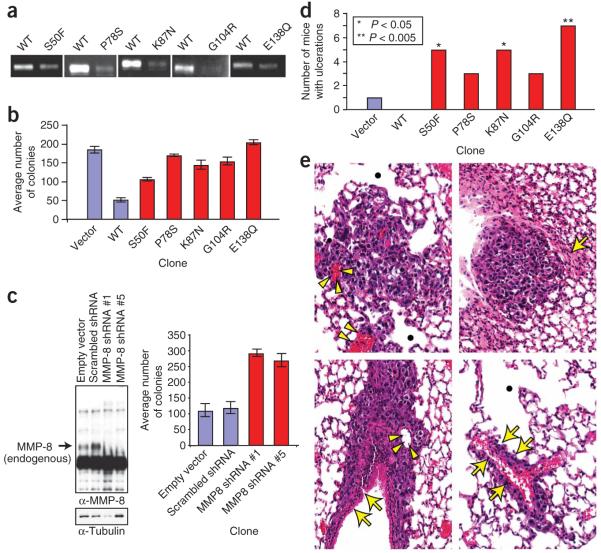
To determine whether the mutations in MMP8 affected the protein’s proteolytic activity, we expressed wild-type MMP-8 and five tumor-derived mutants (S50F, P78S, K87N, G104R and E138Q) in HEK 293T cells. A similar expression level of all MMP-8 constructs was observed. Evaluation of MMP activities in immunoprecipitated MMP-8 complexes by type I collagen substrate gel zymography showed that altered MMP-8 proteins conferred less proteolytic activity toward collagen I than the wild-type protein (Fig. 1a).
Figure 1.
Effects of MMP8 mutations on enzymic activity, growth on soft agar and growth in vivo. (a) Collagen I zymography was used to assess activity of wild-type and altered MMP-8 proteins. Conditioned media from HEK 293T cells expressing wild-type MMP-8 or altered MMP-8 with the indicated amino-acid substitutions were immunoprecipitated and subjected to type I collagen substrate gel zymography. Each panel represents a separate experiment. (b) Anchorage-independent proliferation of Mel-STR cell clones expressing the indicated constructs was assessed by measuring colony growth in soft agar. Graph indicates number of colonies observed after two weeks of growth. (c) SK-Mel-2 cells were infected with either control shRNAs or two different shRNA constructs targeted against MMP8 (#1 and #5) and selected to form stable pooled clones. Left panel: conditioned media of the clones were immunoblotted with the indicated antibodies. Right panel: anchorage-independent proliferation of infected SK-Mel-2 cell clones expressing the indicated constructs was assessed as described in b. (d) NOD/SCID mice were subcutaneously injected with Mel-STR cell clones expressing the indicated constructs and examined on a bi-weekly basis (n=8; *P < 0.05 to **P < 0.005; Fisher’s exact test.) (e) Representative hematoxylin and eosin-stained images of histopathological pulmonary sections from mice injected with MMP-8 mutant clones. Top left: parenchymal metastasis which probably originated in the blood vessels (arrow heads); emphysematous areas are indentified with a black dot. Top right: pleural metastasis island associated with focal parenchymal collapse or atelectasia (arrow). Bottom left: parenchymal metastasis growing along a bronchial structure (arrows). Bottom right: perivascular metastasis (arrows) with surrounding emphysematous areas. All slides are at ×130. α-Tubulin, antibody to tubulin.
To determine whether MMP-8 has tumor suppressor activity that is inactivated by tumor-derived mutations, we created stable pooled clones of human melanocyte Mel-STR cells11 expressing similar levels of wild-type or mutant MMP8 (Supplementary Fig. 5 online). MMP-8 activity of these clones was identical to that of HEK 293T cells (Fig. 1a). These clones were used for subsequent studies.
Expression of wild-type or mutant MMP-8 did not affect the growth rate of Mel-STR cells in tissue culture (Supplementary Fig. 6 online). In contrast, expression of wild-type but not mutant MMP-8 substantially inhibited colony growth on soft agar (Fig. 1b). To test whether inhibition of MMP8 would have the same effect on cell growth on soft agar as MMP8 mutation, we used short hairpin RNA (shRNA) to stably knockdown MMP-8 protein levels in SK-Mel-2 cells, a melanoma cell line that expresses wild-type MMP-8. We examined the effect of two different shRNA constructs against MMP8: both resulted in an increase in the ability of SK-Mel-2 cells to grow on soft agar compared to control shRNA constructs (Fig. 1c).
As a previous study has reported that wild-type MMP-8 reduces cell migration8, we investigated whether cell migration was altered in melanoma lines expressing the various MMP-8 mutants. Cells expressing wild-type MMP-8 had a reduced migration capacity compared to cells expressing mutant MMP-8 (Supplementary Fig. 7a,b online). In contrast, when MMP-8 was knocked down in SK-Mel-2 cells, there was a significant (P < 0.0001, t-test) reduction in the ability of the cells to migrate (Supplementary Fig. 7c,d).
To determine whether the MMP8 mutations resulted in altered growth in vivo, Mel-STR clones expressing empty vector, wild-type or mutant MMP-8 were administered to NOD/SCID mice by subcutaneous injection. Twenty days after injection, none of the mice with cells expressing wild-type MMP-8 had ulcerating lesions, whereas most of the tumors expressing the five mutant MMP-8 proteins presented with ulcerations (Fig. 1d). Ulceration is a measure of aggression in melanoma and is used clinically as a determinant of melanoma stage12. To determine whether MMP-8 expression affected metastasis, we examined sections of paraffin-embedded lungs by hematoxylin/eosin (H&E) staining. No lung metastases were found in the mice injected with wild-type clones, and only one of the mice injected with clones expressing empty vector had micrometastases. In contrast, numerous perivascular, peribronchial and subpleural micrometastases with evidence of tumor invasion were observed in some of the mice receiving clones expressing MMP-8 mutants (Fig. 1e). Because of severe skin ulceration, mice had to be euthanized before macroscopic metastases were evident. It is probable that a larger number of mice injected with mutant clones would have developed metastases had they been euthanized at a later time point. Thus, a correlation between increased ulceration and micrometastasis formation was seen (Supplementary Table 4 online). Although the underlying mechanism for the metastatic phenotype seen in the mutant cells is unclear, it is consistent with murine MMP-8 conferring a reduced invasive ability8. In contrast to the above results, no significant difference was observed in the primary tumor growth rate. This is consistent with the similar growth rate of all the clones in vitro.
The combination of genetic, biochemical and cellular data presented here suggests that MMP8 is a tumor suppressor in human melanoma. This conclusion is consistent with the previously reported antitumor properties of MMP-8 and other proteases5-7,13,14. Indeed, knockdown of MMP-8 phenocopies mutant MMP-8 overexpression when growth on soft agar is evaluated. However, knockdown of MMP-8 does not phenocopy mutant overexpression when cell migration is assessed. This suggests that, in some cases, the MMP-8 mutants may have an assay-specific gain-of-function or dominant-negative effect. This scenario has precedent and has been described for p53 (ref. 15).
To our knowledge, this study represents the first systematic mutational analysis of the MMP family in any human cancer type. Despite decades of research on MMPs, this is the first study that demonstrates that a large portion of melanomas have previously uncharacterized somatic mutations in MMP genes that affect MMP activity. These findings emphasize the need to test the role of each MMP in an individual manner and to precisely define its functional role in cancer. This may allow the development of individualized therapy on the basis of the mutant MMP present in specific tumors.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Xu, J. Polite, I. Cardenas-Navia, D. Leja and I. Ginty for technical assistance. We thank R. Weinberg (Massachusetts Institute of Technology) for the Mel-STR cells, and W. Gahl (National Institutes of Health) and R. Alani (Johns Hopkins University) for normal melanocyte RNA. We also thank L. Matrisian, P. Meltzer, T. Waldman, G. Merlino, T. Hornyak and T. Bugge for their comments on the manuscript. Funded by the National Human Genome Research Institute and National Institute on Aging.
References
- 1.Brinckerhoff CE, Matrisian LM. Nat. Rev. Mol. Cell Biol. 2002;3:207–214. doi: 10.1038/nrm763. [DOI] [PubMed] [Google Scholar]
- 2.Egeblad M, Werb Z. Nat. Rev. Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 3.Freije JM, et al. Adv. Exp. Med. Biol. 2003;532:91–107. doi: 10.1007/978-1-4615-0081-0_9. [DOI] [PubMed] [Google Scholar]
- 4.Coussens LM, Fingleton B, Matrisian LM. Science. 295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 5.Lopez-Otin C, Matrisian LM. Nat. Rev. Cancer. 2007;7:800–808. doi: 10.1038/nrc2228. [DOI] [PubMed] [Google Scholar]
- 6.Balbin M, et al. Nat. Genet. 2003;35:252–257. doi: 10.1038/ng1249. [DOI] [PubMed] [Google Scholar]
- 7.Montel V, et al. Cancer Res. 2004;64:1687–1694. doi: 10.1158/0008-5472.can-03-2047. [DOI] [PubMed] [Google Scholar]
- 8.Gutierrez-Fernandez A, et al. Cancer Res. 2008;68:2755–2763. doi: 10.1158/0008-5472.CAN-07-5154. [DOI] [PubMed] [Google Scholar]
- 9.Sjoblom T, et al. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 10.Greenman C, et al. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta PB, et al. Nat. Genet. 2005;37:1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.American Joint Committee on Cancer . Cancer Staging Handbook. 6th edn Lippincott Williams & Wilkin; New York: 2004. p. 469. [Google Scholar]
- 13.Gorrin-Rivas MJ, et al. Clin. Cancer Res. 2000;6:1647–1654. [PubMed] [Google Scholar]
- 14.Acuff HB, et al. Cancer Res. 2006;66:7968–7975. doi: 10.1158/0008-5472.CAN-05-4279. [DOI] [PubMed] [Google Scholar]
- 15.Ko LJ, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.