Abstract
The promoter elements and transcription factors necessary for triiodothyronine (T3) induction of hepatic HMG-CoA reductase (HMGR) were investigated by transfecting rat livers with wild type and mutant HMGR promoter-luciferase constructs using in vivo electroporation. Mutations in the sterol response element (SRE), nuclear factor-y (NF-Y) site, and the newly identified upstream transcription factor-2 (USF-2) site essentially abolished the T3 response. Chromatin immunoprecipitation (ChIP) analysis demonstrated that T3 treatment caused a 4-fold increase in in vivo binding of USF-2 to the HMGR promoter. Co-transfection of the wild type HMGR promoter with siRNAs to USF-2, SREBP-2, or NFY nearly abolished the T3 induction, as measured by promoter activity. These data provide in vivo evidence for functional roles for USF-2, SREBP-2, and NF-Y in mediating the T3-induction of hepatic HMGR transcription.
Keywords: Electroporation, Rat model, siRNA knockdown, Chromatin Immunoprecipitation
Introduction
Cholesterol metabolism and serum cholesterol levels are profoundly affected by thyroid hormone (T3) status [1]. Key genes that are involved in regulating cholesterol metabolism include: LDL receptor, cholesterol 7α hydroxylase and HMG-CoA reductase (HMGR) [2]. Each of these genes is regulated at the level of transcription by T3 [2]. Significant advances have been made in defining the mechanisms by which T3 regulates LDL receptor and cholesterol 7α hydroxylase [3; 4]. The promoter elements and transcription factors involved in T3 stimulation have been identified for these genes. In contrast, the elements and transcription factors mediating T3 induction of HMGR have not yet been identified.
Induction of gene transcription by triiodothyronine (T3) is usually mediated by thyroid hormone response elements (TRE) that are often characterized by a direct-repeat-4 (DR4) sequence such as AGGTCAnnnnAGGTCA. Typically in liver, thyroid receptor beta (TRβ1) binds as a heterodimer with retinoic acid receptor (RXR-α) to induce gene transcription in response to T3 [5]. The rat HMG-CoA reductase (HMGR) promoter does not possess a consensus TRE. There is a long lag period of about 36 hr from injection of T3 until induction of HMGR expression, which depends on both RNA and protein synthesis to occur [6]. The absence of a traditional TRE and the late induction of the HMGR promoter are evidence of a potential intermediary in the T3 response [2]. Together, these observations suggest that T3 may act to initially induce the expression of a protein or proteins, which in turn activate transcription of HMGR through an element or elements other than a consensus TRE.
We tested this hypothesis by utilizing in vivo electroporation to introduce HMGR promoter-luciferase constructs directly into the livers of hypothyroid rats treated ±T3. We used this approach to identify promoter elements and transcription factors that operate physiologically to mediate T3 induction of hepatic HMGR. Using numerous mutant promoter constructs, ChIP assays, and siRNA knockdown experiments, we have found that upstream stimulatory factor-2 (USF-2), sterol response element binding protein-2 (SREBP-2), and nuclear factor-y (NF-Y) appear to play significant functional roles in mediating the T3 induction of hepatic HMGR gene expression.
Materials and Methods
Experimental Animals
Hypophysectomized male Sprague-Dawley rats weighing 125 to 150 g were purchased from Harlan (Indianapolis, IN). Hypophysectomized rats received Tekland Iodine Deficient chow and water ad libitum and were housed in a reverse-cycle light-controlled room with a 12-hour light period / 12-hour dark period. The rats were on this diet for at least 20 days prior to experimentation in order to achieve sufficient turnover of T4 and low levels of T3. Rats were given an injection of 1.0 mg/kg T3 72 hours prior to electroporation and an additional injection of 0.25 mg/kg T3 24 hours prior to electroporation. The rats were typically euthanized 24 hours following electroporation, at the mid-dark period when hepatic HMGR expression is at its diurnal high.
Plasmid Construction
The rat HMGR promoter was amplified to −5683 from a genomic BAC clone (CH230-263N2) from the rat genome using the Expand Long Template PCR System (Roche). The −325/+70 HMGR promoter segments in pGL3-Basic (Promega, Madison, WI) were obtained as previously described [7]. A −5123/+70 promoter fragment was obtained by using the primer sequences listed in Table 1. This PCR product contains a NheI site at both ends for cloning. The −3187/+70 and the −2314/+70 promoter fragments were prepared from this large PCR product and cloned into pGL3-Basic vector using the restriction enzymes MluI and SacI, respectively. All clones were confirmed by restriction analysis and DNA sequencing at the Moffitt Molecular Biology Core Facility (Tampa, FL).
Table 1.
Primer sequences used for cloning, site-directed mutagenesis, and qPCR.
| Target | Sequence |
|---|---|
| Plasmid | |
| −5123/+70 | 5′-CTGAAGCTATGCTAGCAGCTACAGAAATGGAGCGCTCTTCG-3′ |
| 5′-GAGAAGATGCTAGCATCTCAATGGAGGCCACCAAGC-3′ | |
| SDM | |
| SRE | 5′-CCGTGGTGAGAGATGTGTATTGTCCCGTTCTCCGCCCG-3′ |
| 5′-CGGGCGGAGAACGGGACAATACACATCTCTCACCACGG-3′ | |
| USF-2 | 5′-GCTCTTACGCGTCTGCCTTGACAATTCTGAGTTCGGGG-3′ |
| 5′-CCCCGAACTCAGAATTGTCAAGGCAGACGCGTAAGAGC-3′ | |
| SP-1 | 5′-GCGGTGCCCGTTCTCCGAAATTTGTCGAGCAGTGGG-3′ |
| 5′-CCCACTGCTCGACAAATTTCGGAGAACGGGCACCGC-3′ | |
| CRE | 5′-GGGCGACCGTTCGGTCATGCTTCCGTCAGGCTGAGCAG-3′ |
| 5′-CTGCTCAGCCTGACGGAAGCATGACCGAACGGTCGCCC-3′ | |
| qPCR | |
| HMGR | 5′–TGTGGGAACGGTGACACTTA–3′ |
| 5′-CTTCAAATTTTGGGCACTCA-3′ | |
| SREBP-2 | 5′-GGATCATCCAGCAGCCTTTGA-3′ |
| 5′-ACCGGGACCTGCTGCACCTGT-3′ | |
| USF-2 | 5′-CCCAGGATGTGCTTCAAACAGGAA-3′ |
| 5′-TCCTTCTCCGCTCCACTTCATTGT-3′ | |
| NFY-B | 5′-AAGTTCAGAGAGGCCATGAAGGGA-3′ |
| 5′-TCTGCAGTTATTAACCCAGCCGGT-3′ | |
| 18s | 5′-CCATCCAATCGGTAGTAGCG-3′ |
| 5′-GTAACCCGTTGAACCCCATT-3′ | |
| ChIP | |
| USF-2 | 5′-CCACACTCCAACTCTGACACGGT-3′ |
| 5′-CCGAGCCAACCAATGGCTAGT-3′ | |
| SREBP1/2,NFY | 5′-ACTAGCCATTGGTTGGCTCGG-3′ |
| 5′-CGCCAATAAGGAAGGATCGTCCGAT-3′ | |
In Vivo Electroporation
Ten μg of HMGR promoter construct and mutants ligated to the luciferase gene were directly introduced into the livers of rats by electroporation as recently described [8] with minor modifications. pHRL-CMV Renilla vector (Promega, Madison, WI) was co-electroporated at a 1:2000 dilution to control for electroporation efficiency. The total volume of injected DNA was 50 μl. A 5 mm diameter, six-needle array electrode was used to deliver 6 DC electric pulses to the tissue. Each pulse had a potential of 75 V and a duration of 150 ms. Pulses were administered in 150 ms intervals. The voltage applied to the tissue corresponded to an electric field strength of 150 V/cm.
Luciferase Assays
The livers were generally harvested twenty-four hours following electroporation. Once removed, the electroporated regions of the liver were extracted using a 5 mm cork-borer. Approximately 0.1 g of liver was placed in 600 μl of passive lysis buffer (Promega) and homogenized using a tissue disrupter. The lysate was centrifuged at 16,000 g for 5 minutes and the supernatant assayed for luciferase activity using the dual luciferase assay kit from Promega. Luciferase activity was calculated as the average ratio of firefly (reporter) to renilla luciferase for at least 2 injection sites per animal.
Serum Preparation and Thyroid Hormone Assay
Blood was collected from animals at time of euthanasia and centrifuged at 16,000 g for 5 min. Supernatant (serum) was collected and used for determination of T3 using the T3 assay from Calbiotech (Spring Valley, CA).
Site-Directed Mutagenesis
Mutant plasmids were produced using the QuikChange kit from Stratagene (Cedar Creek, TX). The plasmid consisted of the double stranded DNA encoding the promoter with point mutations at the site of the SRE, USF-2 site, NF-Y site, CRE, or SP1-like site. Primer sequences used are listed in Table 1. The NF-Y mutant plasmid was previously prepared [7].
Real Time PCR
Total RNA was isolated from rat livers using TRI Reagent (Molecular Research Center, Cincinnati, OH) and resuspended in nuclease-free water. RNA was then DNAse treated using the TURBO DNA-Free Kit (Ambion, Austin, TX). cDNA was prepared using the Reverse Transcription System (Promega) per the manufacturer’s protocol. Primer sequences used are listed in Table 1. mRNA was quantified under the following reaction conditions: 95°C for 5 minutes, followed by 40 cycle of 95°C for 15 seconds, 61°C for 1 minute and melt curve 55°C + 0.5°C each 10 seconds, × 80. All samples were run in duplicate on a Bio-Rad Chromo4 DNAEngine thermal cycler using SYBR green chemistry.
Chromatin Immunoprecipitation
Chromatin was prepared and immunoprecipitated using the ChIP-IT Express kit from Active Motif (Carlsbad, CA). The protocol was modified for liver tissue preparation. All named buffers are included in the kit. Equal portions of liver (100 mg total) from 3 rats per treatment group (±T3) were minced and fixed in 1% formaldehyde for 5 minutes at room temperature. One mL of Glycine Stop-Fix Solution was added to stop fixation and incubated for 5 minutes. Samples were centrifuged for 5 minutes at 720g. The pellets were resuspended in 6 ml scraping solution and centrifuged at 720g for 5 min. The cell pellets were resuspended in 1.5 ml ice-cold Lysis Buffer supplemented with 7.5 μl protease inhibitors (PIC) and 7.5 μl PMSF and incubated on ice for 30 minutes. The cells were homogenized in a Dounce homogenizer for 10 strokes on ice to release the nuclei. The homogenates were then centrifuged at 2400g for 10 min at 4°C to pellet the nuclei. The nuclei were resuspended in 1 ml of Complete Shearing Buffer supplemented with 5 μl PIC and disrupted using an Ultrasonic model W-375 sonicator, 5 times each for 15 seconds on, 30 seconds off, to shear chromatin. Chromatin size was checked by agarose electrophoresis to ensure an average size between 200 and 500 bp. Chromatin was precleared with 35 μl of magnetic beads per 500 μl chromatin by rotating at 4°C for 30 minutes and collected. Eighty μl of chromatin, 3 μg of antibody, and 25 μl of beads were used for each IP reaction. The antibodies used were: SREBP-2 (Cayman Chemical, #10007663), USF-2 (SCBT, sc-861x), SREBP-1 (SCBT, sc-8984x), NF-Y (SCBT, sc-13045x), and RXR-α (SCBT, sc-553x). The negative and positive control antibodies were part of the ChIP-IT Kit. The reactions were rotated at 4°C with antibody for 17 hours. Final DNA samples were analyzed by qPCR in triplicate as described above. The relative level of transcription factor binding was quantified by correcting for amount of input DNA and negative control antibody DNA (background), which were performed in parallel. Relative induction of binding was calculated as the ratio of relative binding of the factor in the Hx +T3 chromatin preparation to the relative binding of the factor in the Hx chromatin preparation. The data are the results of at least 3 independent immunoprecipitation experiments on pooled chromatin samples. PCR products from ChIPs performed with antibodies to USF-2 and NF-Y were also run on a 2% agarose gel to show in vivo binding of these factors. PCR was carried out as described under real-time PCR methods above with the omission of a melt curve.
siRNA Knockdown Study
Following the treatments described previously under Experimental Animals, 5 μg of SREBP-2 siRNA (Dharmacon cat# L-081475-01), or 20 μg of USF-2 siRNA (SABiosciences cat# SIR449799A-C), or NFY siRNA (SABiosciences cat# SIR252807A-C) was co-electroporated with or without 10 μg −325/+70 HMGR promoter construct in a final volume of 50μl using at least 2 sites per animal. The electroporated sites were removed 24 hours later using a 5 mm cork-borer. For NF-Y experiments, samples were also removed after 48 hrs. Luciferase assays were performed as described above. qPCR was performed to measure knock down of endogenous USF-2, SREBP-2, or NFY mRNA.. 5 μg of Dharmacon cat# L-096650-01 siRNA was used as the negative control, as electroporation of this siRNA did not knockdown the target gene.
Statistical Analyses
Data is expressed as the mean +/− standard error for a minimum of 4 animals per group. Experimental treatments included hypophysectomized Sprague-Dawley and hypophysectomized Sprague-Dawley rats treated with T3.
Results and Discussion
Functional Analysis of the HMGR Promoter
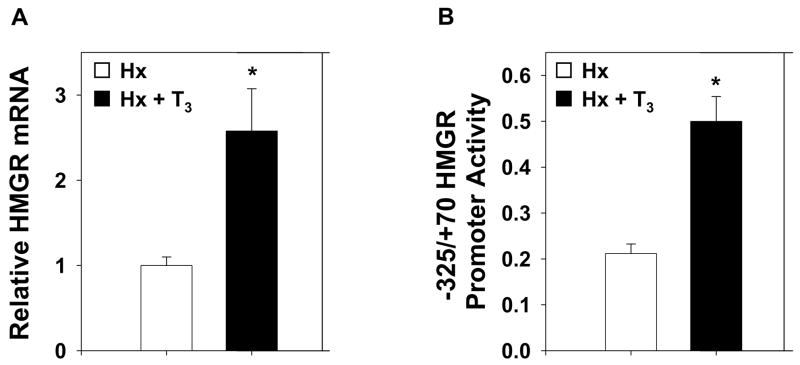
We utilized in vivo electroporation in a hypothyroid (Hx) rat model to analyze the regulatory mechanism of T3 induced hepatic HMGR gene expression. Hx rats were treated ± T3 96 hours before harvest of liver tissue to measure hepatic HMGR gene expression by real-time PCR. HMGR gene expression was induced approximately 2.7-fold by thyroid hormone (Fig. 1A). In order to define the region of the HMGR promoter necessary for this induction, four promoter-luciferase constructs inclusive of the HMGR promoter from −5683, −3190, −2314, or −325 to +70 bp were generated. The plasmids were electroporated into the livers of Hx rats treated ±T3. We found that the −325/+70 region was sufficient to induce maximum thyroid hormone stimulation of HMGR promoter activity (Fig. 1B). With this construct, a 2.5-fold induction of HMGR promoter activity was observed in response to T3, which correlates well with the increase observed in mRNA levels.
Figure 1.
Thyroid Hormone Induces Hepatic HMGR. A, Real-time PCR analysis of total HMGR mRNA isolated from livers of hypophysectomized rats treated ± T3. B, An HMGR promoter construct inclusive of the −325/+70 region was electroporated into the livers of hypophysectomized rats treated ±T3. Data are reported as the mean ± standard error of the mean for each promoter construct. Statistically significant differences are relative to Hx with wild-type construct. For all conditions, n≥4 and *p<0.05.
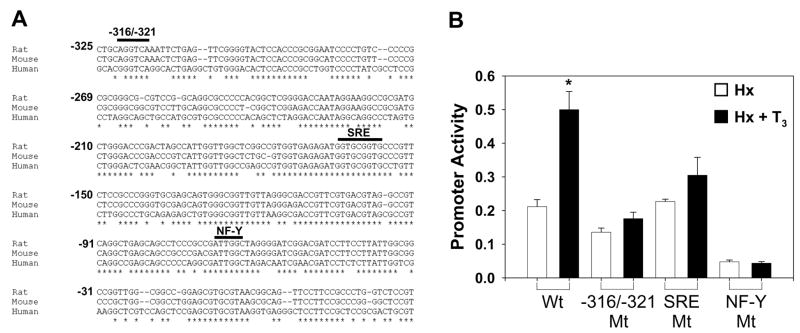
The −325/+70 region of the promoter is highly conserved in mammalian species (Fig. 2A). Specifically, the −316/−321 element, the sterol response element (SRE), and the NF-Y binding sites all lie within regions of very high conservation in the HMGR promoter. The element at −316/−321 is a conserved element that serves as a near-consensus binding sequence for the USF-2 transcription factor [9] as well as other common transcription factors [10; 11; 12]. USF-2, SREBP-2, and NF-Y have previously been shown to function in T3 regulation of genes such as fatty acid synthase, S14, and carnitine palmitoyltransferase-I [13; 14; 15]. Thus, we hypothesized that T3 induction could occur through these sites. We prepared plasmids inclusive of the −325/+70 promoter region (Wt) with point mutations in each of these elements as well as the Sp1-like site at −145/−138 and the cAMP response element at −104/−97. The mutated plasmids were then introduced into the livers of Hx rats treated ± T3 as described previously. Independent mutations in the −316/−321 element, SRE, and NFY binding site resulted in essentially complete loss of the T3 induction (Fig. 2B). These data suggest that all three elements and their respective transcription factors are necessary for T3-mediated induction of HMGR transcription. Mutations in the SP1-like site and CRE did not abolish the T3 response of the HMGR promoter (data not shown).
Figure 2.
−316/−321, SRE, and NF-Y Promoter Elements are Highly Conserved across Species. A, Multiple sequence alignment was performed using the CLUSTALW2 tool provided by EMBL-EBI. HMGR promoter sequences for H. sapiens, R. norvegicus, and M. musculus were obtained from GenBank. Nucleotides are labeled ± from the transcription start site. B, HMGR promoter construct spanning the −325/+70 region (wild-type) as well as constructs with mutations in the −316/−321, SRE, or NF-Y binding sites were electroporated into the livers of hypophysectomized rats treated ±T3 (n≥4). Data are reported as the mean ± standard error of the mean for each promoter construct. Statistically significant differences are relative to Hx with wild-type construct. *p<0.05
Binding of USF-2 to the HMGR Promoter
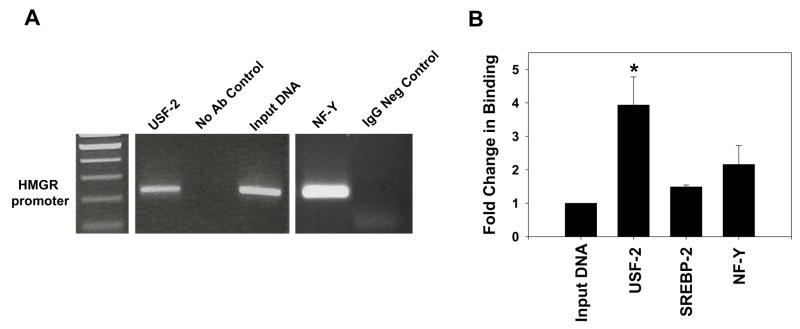
We used the online transcription factor prediction program PROMO to identify a number of potential factors that could bind the −316/−321 sequence [16]. Electrophoretic mobility shift assays (EMSAs) with liver nuclear extract were performed to determine the ability of these factors to bind the −316/−321 element. EMSAs showed antibody supershifts for only USF-2 and RXRα (data not shown). Shifts were not observed for peroxisome proliferator activated receptor-α, TRβ-1, or CAAT enhancer binding protein α/β. To confirm the binding of USF-2 to the HMGR promoter in vivo, ChIP analysis was performed on pooled chromatin samples prepared from the livers of Hx + T3 rats. PCR was performed on the immunoprecipitated DNA and products were run on agarose gels for identification. USF-2 was found to bind the HMGR promoter in vivo (Fig. 3A). This is the first report of in vivo evidence showing USF-2 binding to the HMGR promoter at −316/−321. A previous report has shown this sequence to bind the LRH-1/FTF protein, though we were unable to replicate this binding by EMSA [17]. Chromatin was then prepared in two pools of 3 equal amounts of liver from Hx rats treated ±T3. ChIP assays were performed and immunoprecipitated DNA analyzed using qPCR to measure the induction of transcription factor binding to the HMGR promoter in response to T3. Most notably, USF-2 binding was induced approximately 4-fold in response to T3 (Fig. 3B). It is possible that although USF-2 is bound to the promoter in a basal state, additional USF-2 protein is recruited to the HMGR promoter in response to T3. The USF proteins function in transcriptional activation via a number of pathways including cooperative interaction with other factors [18], homo- and hetero- dimerization and DNA looping [19]. The binding of SREBP-2 or NF-Y was not significantly increased in response to T3 treatment.
Figure 3.
USF-2 Binding to the HMGR Promoter is Increased 4-fold in Response to T3. A, ChIP assays were performed on pooled chromatin prepared from equal portions of liver collected from 3 Hx + T3 rats. Sheared chromatin was immunoprecipitated using the antibodies indicated. Immunoprecipitated DNA was analyzed by PCR and products (220bp) were run on a 2% agarose gel. B, Two chromatin pools were prepared from equal portions of liver collected from 3 rats in each treatment group (±T3). ChIP was performed for the transcription factors listed. Immunoprecipitated DNA was analyzed by qPCR in triplicate. Fold-change in binding was calculated for at least three separate ChIP experiments as the ratio of Hx + T3 binding to Hx binding. Statistically significant differences are relative to the total input DNA set at 1. *p<0.05
siRNA Knockdown of USF-2, SREBP-2, and NFY Abolishes T3 Response
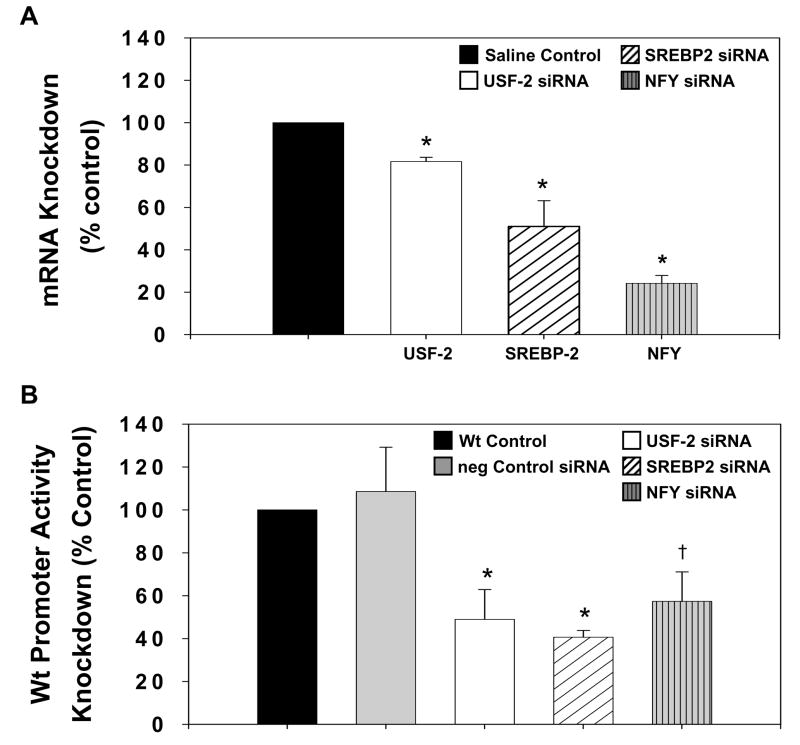
In order to better understand the functional necessity of the USF-2, NF-Y, and SREBP-2 transcription factors in the T3 induction of HMGR transcription, we performed in vivo siRNA knockdown assays. We hypothesized that knockdown of these necessary factors would mimic the results of our previous mutations in the HMGR promoter elements and abolish the T3 response as measured by HMGR promoter activity. siRNA to USF-2, SREBP-2, or NF-Y was electroporated into the livers of Hx + T3 rats. Efficacy of siRNA knockdown was measured by qPCR analysis of endogenous mRNA isolated from individual electroporation sites 24 hours following electroporation. siRNA resulted in maximal knockdown of endogenous USF to 82% of control and SREBP-2 to 51% of control (Fig. 4A) within 24 hours. siRNA to NF-Y required a greater period of time to achieve optimal knockdown, with knockdown to 79% of control achieved at 24 hours and to 24% of control at 48 hours. Having generated a considerable knockdown of endogenous mRNA, we then co-electroporated the Wt HMGR plasmid with siRNA to each factor. siRNA to USF-2 and SREBP-2 generated knockdown of HMGR promoter activity to 49% or 44% of control, respectively and essentially abolished the T3 response (Fig. 4B). This knockdown is consistent with the difference in Wt HMGR promoter activity observed in the Hx rat compared to rats treated with T3 (Fig. 1B). Electroporation of NF-Y siRNA resulted in knockdown of HMGR promoter activity that was highest after 48 hours. The knockdown at 24 hours was to 75% of control compared to 57% of control at 48 hours (Fig. 4B). It is pertinent to note that quantification of endogenous mRNA knockdown (Fig. 4A) may include hepatocytes that were not transfected with siRNA due to the potential collection of extraneous tissue. This may result in masking of the knockdown effect. In contrast, knockdown of HMGR promoter activity (Fig. 4B) is measured by luciferase assay, which only quantifies successfully transfected cells. Thus, discrepancies in relative knockdown between endogenous mRNA and promoter activity are expected. These in vivo knockdown assays indicate functional roles for USF-2, SREBP-2, and NFY in T3 activation of HMGR.
Figure 4.
siRNA Knockdown Demonstrates the Functional roles of USF-2, SREBP-2, and NFY on Hepatic HMGR Promoter Activity. A, Total RNA was isolated from the livers of hypophysectomized rats treated with T3 and electroporated with 5μg SREBP-2, 20 μg USF-2 or NFY siRNA, or saline control (n≥3). qPCR was performed to measure relative endogenous mRNA levels of each factor in response to the respective siRNA. Saline electroporation was set to 100% as the control for each gene. siRNA knockdowns are presented as a percentage of this control. B, An HMGR promoter construct spanning the −325/+70 region (wild-type) and either 5 μg SREBP-2 or 20 μg USF-2 or NFY siRNA were co-electroporated into the livers of hypophysectomized rats treated with T3. Wild-type promoter activity was set to 100% as the control. siRNA knockdowns are presented as a percentage of this control. Data are reported as the mean ± standard error of the mean for each treatment. Statistically significant differences are relative to control (n≥3). * p<0.05, † p<0.10
In conclusion, the −325/+70 region of the HMGR promoter appears sufficient for the full T3 response. The newly described element at −316/−321 was shown to bind USF-2 in vivo. Binding of USF-2 to this element was markedly increased by T3 treatment. The transcription factors USF-2, SREBP-2, and NF-Y are necessary for the response, as mutations in the elements binding these factors markedly diminished the T3 response. Furthermore, knockdown of USF-2, SREBP-2, or NFY mRNA was able to nearly eliminate the T3 response, which suggests functional roles for all of these factors in regulating hepatic HMGR promoter activity. Even through T3 did not appear to cause significant increases in binding of SREBP-2 or NF-Y to the HMGR promoter, the decreases observed after siRNA treatment indicate that these factors are also required. Together, these data provide a working model for T3 induction of HMGR gene expression.
Acknowledgments
This work was supported by a grant from the National Institutes of Diabetes and Digestive and Kidney Diseases (R01DK075414) and does not necessarily represent the official views of the NIDDK or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Illingworth DR, McClung MR, Connor WE, Alaupovic P. Familial hypercholesterolaemia and primary hypothyroidism: coexistence of both disorders in a young woman with severe hypercholesterolaemia. Clin Endocrinol (Oxf) 1981;14:145–52. doi: 10.1111/j.1365-2265.1981.tb00609.x. [DOI] [PubMed] [Google Scholar]
- 2.Weitzel JM, Hamann S, Jauk M, Lacey M, Filbry A, Radtke C, Iwen KA, Kutz S, Harneit A, Lizardi PM, Seitz HJ. Hepatic gene expression patterns in thyroid hormone-treated hypothyroid rats. J Mol Endocrinol. 2003;31:291–303. doi: 10.1677/jme.0.0310291. [DOI] [PubMed] [Google Scholar]
- 3.Lopez D, Ness GC. Characterization of the rat LDL receptor 5′-flanking region. Biochim Biophys Acta. 2006;1761:492–500. doi: 10.1016/j.bbalip.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Shin DJ, Plateroti M, Samarut J, Osborne TF. Two uniquely arranged thyroid hormone response elements in the far upstream 5′ flanking region confer direct thyroid hormone regulation to the murine cholesterol 7alpha hydroxylase gene. Nucleic Acids Res. 2006;34:3853–61. doi: 10.1093/nar/gkl506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harvey CB, Williams GR. Mechanism of thyroid hormone action. Thyroid. 2002;12:441–6. doi: 10.1089/105072502760143791. [DOI] [PubMed] [Google Scholar]
- 6.Simonet WS, Ness GC. Transcriptional and posttranscriptional regulation of rat hepatic 3-hydroxy-3-methylglutaryl-coenzyme A reductase by thyroid hormones. J Biol Chem. 1988;263:12448–53. [PubMed] [Google Scholar]
- 7.Lagor WR, de Groh ED, Ness GC. Diabetes alters the occupancy of the hepatic 3-hydroxy-3-methylglutaryl-CoA reductase promoter. J Biol Chem. 2005;280:36601–8. doi: 10.1074/jbc.M504346200. [DOI] [PubMed] [Google Scholar]
- 8.Lagor WR, Heller R, de Groh ED, Ness GC. Functional analysis of the hepatic HMG-CoA reductase promoter by in vivo electroporation. Exp Biol Med (Maywood) 2007;232:353–61. [PubMed] [Google Scholar]
- 9.Corre S, Galibert MD. USF as a key regulatory element of gene expression. Med Sci (Paris) 2006;22:62–7. doi: 10.1051/medsci/200622162. [DOI] [PubMed] [Google Scholar]
- 10.Murad H, Collet P, Brunner E, Schohn H, Becuwe P, Devignes MD, Dauca M, Domenjoud L. Immunoselection and characterization of a human genomic PPAR binding fragment located within POTE genes. Biochimie. 2007;89:329–36. doi: 10.1016/j.biochi.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z, Chen K, Shih JC, Teng CT. Estrogen-related receptors-stimulated monoamine oxidase B promoter activity is down-regulated by estrogen receptors. Mol Endocrinol. 2006;20:1547–61. doi: 10.1210/me.2005-0252. [DOI] [PubMed] [Google Scholar]
- 12.Malo MS, Pushpakaran P, Hodin RA. A ‘Swinging Cradle’ model for in vitro classification of different types of response elements of a nuclear receptor. Biochem Biophys Res Commun. 2005;337:490–7. doi: 10.1016/j.bbrc.2005.09.080. [DOI] [PubMed] [Google Scholar]
- 13.Jackson-Hayes L, Song S, Lavrentyev EN, Jansen MS, Hillgartner FB, Tian L, Wood PA, Cook GA, Park EA. A thyroid hormone response unit formed between the promoter and first intron of the carnitine palmitoyltransferase-Ialpha gene mediates the liver-specific induction by thyroid hormone. J Biol Chem. 2003;278:7964–72. doi: 10.1074/jbc.M211062200. [DOI] [PubMed] [Google Scholar]
- 14.Jump DB, Badin MV, Thelen A. The CCAAT box binding factor, NF-Y, is required for thyroid hormone regulation of rat liver S14 gene transcription. J Biol Chem. 1997;272:27778–86. doi: 10.1074/jbc.272.44.27778. [DOI] [PubMed] [Google Scholar]
- 15.Yin L, Zhang Y, Hillgartner FB. Sterol regulatory element-binding protein-1 interacts with the nuclear thyroid hormone receptor to enhance acetyl-CoA carboxylase-alpha transcription in hepatocytes. J Biol Chem. 2002;277:19554–65. doi: 10.1074/jbc.M111771200. [DOI] [PubMed] [Google Scholar]
- 16.Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–4. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 17.Datta S, Wang L, Moore DD, Osborne TF. Regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase promoter by nuclear receptors liver receptor homologue-1 and small heterodimer partner: a mechanism for differential regulation of cholesterol synthesis and uptake. J Biol Chem. 2006;281:807–12. doi: 10.1074/jbc.M511050200. [DOI] [PubMed] [Google Scholar]
- 18.Liu M, Whetstine JR, Payton SG, Ge Y, Flatley RM, Matherly LH. Roles of USF, Ikaros and Sp proteins in the transcriptional regulation of the human reduced folate carrier B promoter. Biochem J. 2004;383:249–57. doi: 10.1042/BJ20040414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferre-D’Amare AR, Pognonec P, Roeder RG, Burley SK. Structure and function of the b/HLH/Z domain of USF. Embo J. 1994;13:180–9. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]