Abstract
Hematopoietic stem-progenitor cells (HSPCs) reside in the bone marrow niche, where interactions with osteoblasts provide essential cues for their proliferation and survival. Here, we use live cell imaging to characterize both the site of contact between osteoblasts and hematopoietic progenitor cells (HPCs) and events at this site that result in downstream signaling responses important for niche maintenance. HPCs made prolonged contact with the osteoblast surface via a specialized membrane domain enriched in prominin 1, CD63 and rhodamine PE. At the contact site, portions of the specialized domain containing these molecules were taken up by the osteoblast and internalized into SARA-positive signaling endosomes. This caused osteoblasts to downregulate Smad signaling and increase production of stromal-derived factor-1 (SDF-1), a chemokine responsible for HSPC homing to bone marrow. These findings identify a mechanism involving intercellular transfer to signaling endosomes for targeted regulation of signaling and remodeling events within an ex vivo osteoblastic niche.
The bone marrow provides the regulatory microenvironment or niche for the proliferation and survival of HSPCs [1], which give rise to all blood and immune cells and repopulate bone marrow following stem cell transplantation. HPCs traffic between the blood circulation and the bone marrow, moving on and off niche sites [1, 2]. Osteoblasts, which reside in the bone marrow, are key participants in providing cues for HPC trafficking, proliferation and survival through the secretion of cell-signaling molecules, including SDF-1 [3]. Osteoblasts also make intimate physical contact with HPCs, modulating their function in response to specific physiological conditions [4, 5]. While contact-dependent communication between HPCs and osteoblasts is critical for establishment and maintenance of progenitor cell proliferation and self-renewal [5-8], the molecular pathways that govern this interaction are largely unknown. Moreover, the downstream events occurring at the HPC/osteoblast contact site remain uncharacterized, despite their major role in signaling and remodeling within the niche.
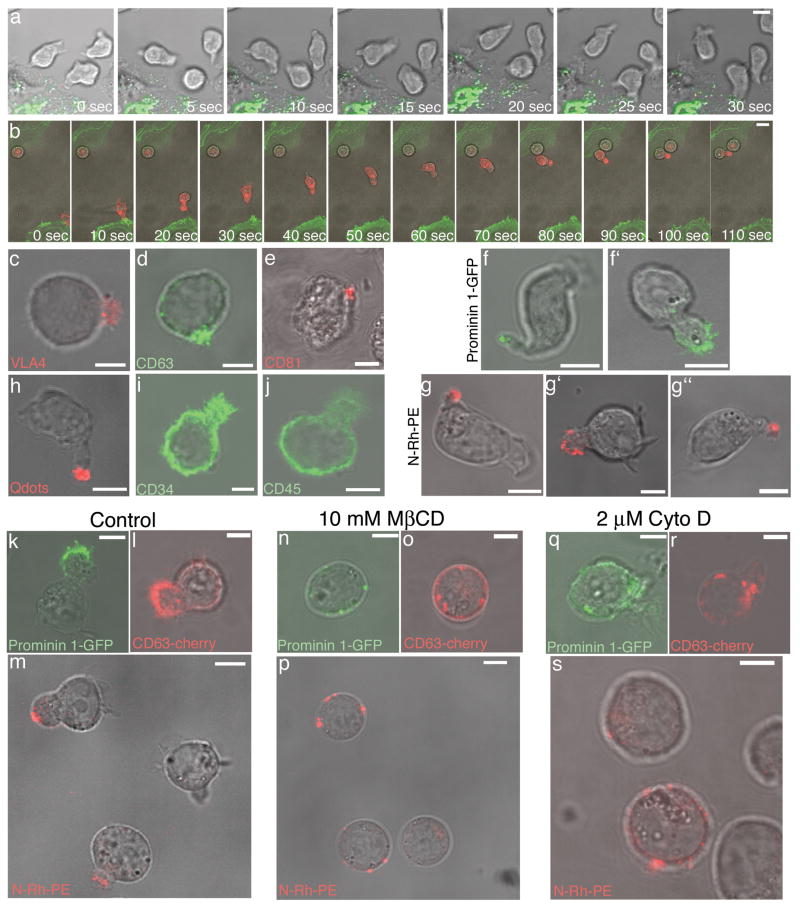
To investigate the morphological and functional characteristics of the HPC-osteoblast interface, we imaged a live-cell, co-culture system consisting of either primary human CD34+ cells or cells from the human KG1a progenitor cell line, co-cultured with the human SaOS2 osteoblastic cell line or primary human osteoblasts. HPCs (i.e., CD34+ and KG1a cells) displayed a rapidly changing morphology and were motile, often with a leading and lagging edge or uropod as previously described (Fig. 1a) (Supp. Mov. 1) [9, 10]. Following movement toward and between osteoblasts, individual HPCs were observed making contact and adhering to osteoblasts (Fig.1b)(Supp. Mov. 2). Given this HPC behavior fit previous descriptions, we began detailed examination of the cell contact interface and its dynamics.
Figure 1.
Organization and maintenance of the HPC plasma membrane. HPC dynamics are revealed by time lapse confocal microscopy of (a) CD34+ cells co-cultured with osteoblasts stably transfected with GalT-YFP (green) or (b) KG1a cells labeled with PKH26 (red) and co-cultured with osteoblasts transiently expressing PH-Akt-GFP (green). Immunofluorescence labeling of KG1a cells with antibodies for (c) VLA4, (d) CD63, and (e) CD81 illustrates an asymmetric distribution of membrane proteins, whereas (i) CD34 and (j) CD45 are distributed more uniformly in the plasma membrane. (h) Live cell imaging of CD34+ cells labeled with cell-labeling QDs (red) (Invitrogen). Live cell microscopy of (f,g) CD34+ cells or (f′,g′) KG1a cells transiently transfected with prominin 1-GFP (green) or 24 h after N-Rh-PE labeling (red). These images are representative of the polarized molecule phenotype observed in > 100 HPCs expressing prominin 1-GFP and > 200 HPCs labeled with N-Rh-PE. Live cell microscopy of (g″) CD34+CD38- cells 24 h after N-Rh-PE labeling. Live cell microscopy of KG1a cells labeled with N-Rh-PE or transiently transfected with CD63-cherry, or prominin 1-GFP and then treated with vehicle control, (k,l,m), 10 mM methyl β cyclodextrin (MβCD) for 30 min at 37°C (n,o,p), or 2 μm cytocholasin D (Cyto D) for 1 h at 37°C (q,r,s). N-Rh-PE was polarized in 92% of the cells treated with vehicle control, 20% of the cells treated with MβCD, and 40% of the cells treated with Cyto D (n > than 100 cells/condition). Following these various drug treatments, KG1a cell viability was greater than 95% as assessed by trypan blue staining (data not shown). Scale bars - 5μm.
The surface distribution of different HPC plasma membrane components was investigated since this might be important for HPC interactions with osteoblasts. The very late antigen-4 (VLA-4) protein, which interacts with VCAM-1 [8], was highly enriched within a specialized membrane domain of HPCs (Fig. 1c) regardless of contact with osteoblasts. A similar asymmetric distribution was seen for CD63 (Fig. 1d) and CD81 (Fig. 1e), which are tetraspanins implicated in integrin regulation [11, 12]. The stem and progenitor cell marker, prominin 1, which localizes to highly curved membranes [13, 14], also showed the polarized pattern (Fig. 1f, f′), as did the fluorescent phosphatidyl ethanolamine analogue, N-Rh-PE (1,2-dipalmitoyl-sn-glycero-3-phophoethanolamine-N-[lissamine rhodamine B sulfonyl]) (Fig. 1g, g′), a cone-shaped lipid that inserts within the plasma membrane [15]. N-Rh-PE labeling of a more enriched population of HSCs, human CD34+CD38- cells, showed a polarized distribution in 97% of the cells (n = 115 cells). Quantum dot (QDs) labeling of HPCs also showed clustering within the membrane domain (Fig. 1h). This presumably occurred by QD clustering along the plasma membrane or potentially by a capping event, rather than by internalized delivery, since QDs could be removed by proteinase K digestion (not shown). The polarized localization of these markers was specific because other surface molecules, CD34 (Fig. 1i) and CD45 (Fig. 1j), were distributed throughout the plasma membrane. Therefore, HPCs are highly polarized and contain a specialized membrane domain with specific protein and lipid components.
To better understand HPC surface polarity and ultimately the importance of asymmetry for events occurring at the HPC-osteoblast interface, cholesterol depletion was performed using the cholesterol-sequestering agent, methyl-β-cyclodextrin. After methyl-β-cyclodextrin treatment, molecules normally enriched in the specialized domain, including prominin1-GFP, CD63-Cherry and N-Rh-PE (Fig. 1k-m), redistributed into punctate clusters across the plasma membrane (Fig. 1n-p). Actin-destabilization using cytochalasin D treatment also disrupted the domain organization, resulting in prominin 1-GFP, CD63-Cherry and N-Rh-PE dispersing throughout the plasma membrane (Fig. 1q-s). Hence, cholesterol and actin-based processes are important for the polarized domain localization of HPC cell surface components.
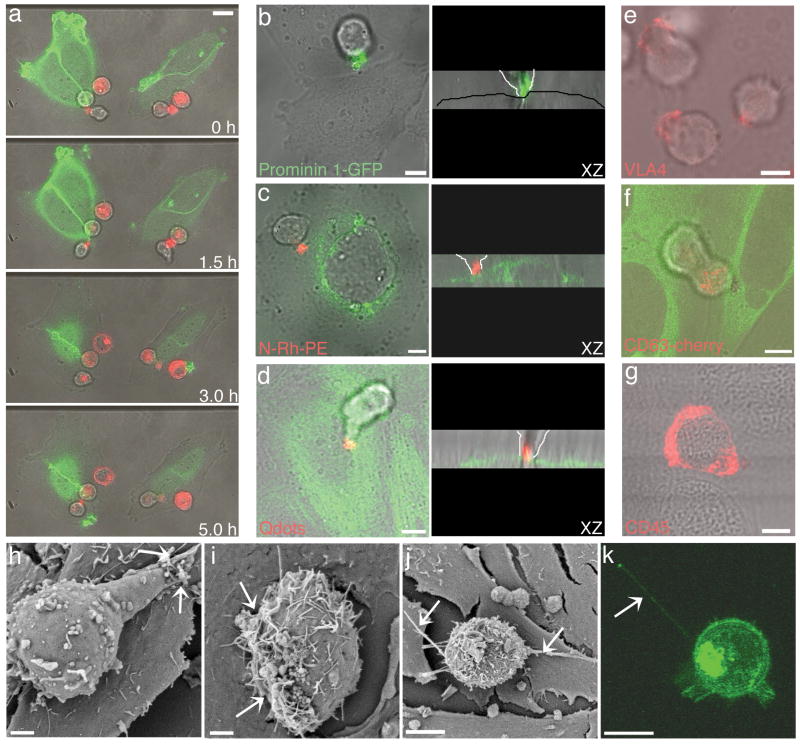
Upon contact with an osteoblast (Fig. 2a, 0h), an HPC (red) often remained attached to the same osteoblast (green) for more than 5 h (Fig. 2a)(Supp. Mov. 2). Confocal Z series imaging revealed that the HPC contact site was often mediated by the uropod, which resided in close association with the osteoblast plasma membrane (Fig. 2b-d, XZ images) [9, 16]. Interestingly, the specialized membrane domain components, including prominin1-GFP (Fig. 2b), N-Rh-PE (Fig. 2c), VLA-4 (Fig. 2e), and CD63-Cherry (Fig. 2f) were all highly enriched at the contact site or uropod, in contrast to CD45 (Fig. 2g), which remained dispersed over the HPC surface. In QD labeled HPCs, the QDs also clustered at the contact site (Fig. 2d). Time lapse imaging of N-Rh-PE labeled CD34+CD38- cells co-cultured with primary human osteoblasts further established the polarized N-Rh-PE domain as the site of cell-cell contact (Supp. Mov. 3).
Figure 2.
HPC/osteoblast contact occurs through a specialized HPC membrane domain. (a) Time-lapse microscopy of PKH26 labeled KG1a cells (red) co-cultured with osteoblasts transiently transfected with PH-Akt-GFP (green) reveals long-term adhesion of HPCs to osteoblasts. The HPC/osteoblast contact domain is enriched in various membrane molecules illustrated by (b) prominin 1-GFP transfecton of KG1a cells co-cultured with osteoblasts (representative of > 50 HPC/osteoblast contacts), (c) N-Rh-PE labeled KG1a cells (red) co-cultured with Rab7-GFP expressing osteoblasts (green) (representative of > 100 HPC/osteoblast contacts), and (d) CD34+ cells labeled with cell labeling QDs (red) co-cultured with actin-YFP expressing osteoblasts. In the corresponding XZ images, the osteoblast monlayer is outlined with a black line for the osteoblasts imaged with bright field only, and the progenitor cell contact is outlined with white lines. The enrichment of specific molecules at the cell contact site was also illustrated by (e) VLA-4 immunofluorescence staining of KG1a cells (red) co-cultured with osteoblasts and (f) live cell imaging of a KG1a cell transiently transfected with CD63-cherry (red) and co-cultured with osteoblastic cells stably transfected with tubulin-YFP (green). (g) CD45 immunofluorescence staining of CD34+ cells (red) co-cultured with osteoblastic cells displayed a uniform membrane distribution.(h-j) SEM of CD34+ cells co-cultured with osteoblastic cells for three hours. Arrows indicate the contact sites. (k) 3D projection image of a live KG1a cell transiently transfected with the wtPrP-GFP construct. The arrow indicates the membrane nanotube formed between the KG1a and an osteoblast. All scale bars – 5 μm, except (h,i) scale bars – 2 μm.
Scanning electron microscopy (SEM) of the contact interface revealed the presence of microvilli-like projections on the HPC membrane similar to those previously described (Fig. 2h,i) [16]. The contact zone of the HPC seen in the SEM images comprised a region several microns in diameter, with the microvilli in this zone appearing to almost penetrate the osteoblast at the contact site (Fig. 2i, see arrows). Nanotube-like structures projecting from many HPCs were also observed making contact with osteoblasts, but were distinct from the contact interface (Fig. 2j,k). Thus, the HPC-osteoblast contact site is an elaborate, often microvilli-rich, region containing stem cell markers (i.e., prominin1) and adhesion-signaling molecules (i.e., VLA4, CD63 and CD81).
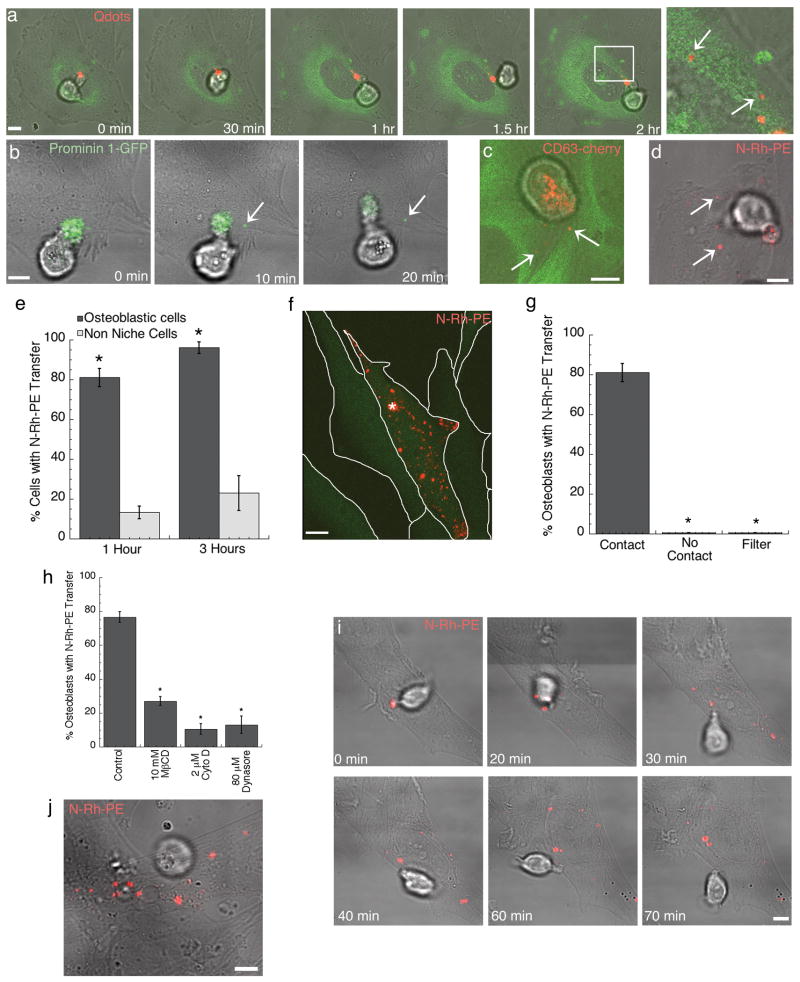
To study the contact site membrane dynamics, we imaged QD-labeled HPCs in contact with osteoblastic cells. Strikingly, QDs transferred from the HPC uropod into the cytoplasm of tubulin-YFP expressing osteoblasts (Fig. 3a) (Supp. Mov. 4). The cytoplasmic localization of the transferred QDs was confirmed by Z series confocal imaging through the osteoblast, which showed that the QDs were in the same plane as the intercellular marker, tubulin-YFP, and not detected on the osteoblast surface (Fig. 3a, zoom). This suggested that intercellular transfer was occurring at the contact site. Imaging of prominin 1-GFP-labeled HPCs in contact with osteoblasts revealed punctate structures enriched in the marker being transferred to the osteoblast over a 20 min contact period (Fig. 3b). Unlike QDs, which reside peripherally on the surface of HPCs and so might be transferred to osteoblasts by a sticking and release process, prominin-1-GFP is embedded in the HPC bilayer and so can only be transferred along with HPC membrane. Co-culturing of CD63-cherry-labeled HPCs with tubulin-YFP osteoblasts also revealed uptake of CD63-cherry into the osteoblast (Fig. 3c) (Supp. Mov. 5). Because both prominin 1-GFP and CD63-cherry proteins were engineered with the fluorescent proteins fused to the carboxy-terminus, which is located in the cytosol, their detection in the osteoblast cells after transfer from HPCs suggested that the entire protein was transferred, not a cleaved fragment. Intercellular transfer of N-Rh-PE into osteoblasts also occurred when N-Rh-PE-labeled HPCs contacted osteoblasts (Fig. 3d). This indicated that both lipid and protein components from the HPC uropod were transferred to osteoblasts.
Figure 3.
Intercellular transfer occurs between HPC and osteoblastic cells in a contact dependent manner. (a) KG1a cell labeled with QDs (red) and co-cultured with osteoblastic cells stably transfected with tubulin-YFP (green) for 2 h before live-cell imaging. The white box indicates the zoomed region and arrows indicate transferred QDs internalized within the osteoblasts. KG1a cells transiently transfected with (b) prominin1-GFP (green) or (c) CD63-cherry (red) and co-cultured with osteoblasts for one hour before live cell confocal imaging. Arrows indicate transferred protein. CD63-cherry transfected cells were co-cultured with osteoblasts stably transfected with tubulin-YFP (green). (d) Live-cell confocal microscopy of N-Rh-PE (red) transfer from KG1a cells to osteoblastic cells. Arrows indicate transferred lipid. (e) The percentage of osteoblastic cells (black, n=120) or HeLa cells (grey, n=75) that acquired N-Rh-PE transfer following contact with an N-Rh-PE labeled HPC after either 1 or 3 h of co-culture. (n, number of HPC/osteoblast or HPC/Hela contacted cells scored over three independent experiments). (f) Live cell imaging of N-Rh-PE (red) labeled CD34+ cell contacting an osteoblastic cell stably transfected with GFP (green). The asterisk indicates the site of CD34+ cell contact and the white lines outline the osteoblasts in the field of view. (g) The percentage of osteoblastic cells that acquired N-Rh-PE transfer following: direct contact with an N-Rh-PE labeled HPC, no contact with a labeled HPC (osteoblasts neighboring HPC contacted cells), or labeled HPC co-culture with osteoblasts through a 0.4μm membrane filter. (n > 100 osteoblasts for each condition). (h) The percentage of osteoblastic cells that acquired N-Rh-PE transfer following contact with a labeled HPC treated with control, 10 mM methyl β cyclodextrin (MβCD), 2 μm cytocholasin D (Cyto D), or 80 μm Dynasore (n > 100 for each condition). (n, number of HPC/osteoblast contacted cells counted over three independent experiments). (i) Live cell confocal microscopy of CD34+ cells labeled with N-Rh-PE and co-cultured with osteoblastic cells. Cells were co-cultured for 1 h before imaging began and intercellular transfer was observed. (j) Live cell confocal microscopy of CD34+CD38- cells labeled with N-Rh-PE and co-cultured with primary human osteoblasts. Scale bars – 5μm.
Efficient intercellular transfer did not occur when HPCs were co-cultured with a non-niche cell type. For example, when N-Rh-PE labeled HPCs were co-cultured with HeLa cells for up to 3 h, only ∼20% of the HeLa cells showed evidence of transfer (Fig. 3e). By contrast, greater than 80% of osteoblastic cells contacting HPCs became labeled within 3 h of co-culturing (Fig. 3e). Since no transferred HPC molecules were observed in neighboring osteoblasts not contacting a labeled HPC during 1 h of co-culture (Fig. 3f,g), efficient molecular transfer only occurred between HPCs and osteoblasts that made direct cell contact. In addition, co-culturing of HPCs and osteoblasts separated by a 0.4 μm membrane filter revealed no molecular transfer from HPCs to osteoblasts during 3 h of co-culture (Fig. 3g). This argued against transfer by bulk cellular release of exosomes, which are small membrane vesicles ∼50-100 nm in diameter [17]. Instead, the observed transfer appeared to be mediated by specific events occurring at the HPC-osteoblast contact site.
To investigate the importance of the specialized membrane domain or uropod in intercellular transfer, this region was disrupted using methyl-β-cyclodextrin or cytochalasin D treatment, as described before. Transfer between a contacting HPC and osteoblast was assessed by live cell imaging of N-Rh-PE-labeled HPCs pretreated with these reagents and then co-cultured with osteoblasts for 1 h. A significantly reduced level of intercellular transfer between contacting cells was observed (Fig. 3h) indicating that efficient intercellular transfer requires contact of the specialized HPC uropod domain with osteoblasts.
HPC uropod domain components were transferred into punctate compartments inside the osteoblast without being noticeably diluted (see Fig. 3b for prominin 1-GFP). This suggested that intercellular transfer did not involve fusion of the HPC uropod with the osteoblast plasma membrane, as transferred components would disperse into the plasma membrane. Instead, transfer appeared to occur by a cytophagocytosis event, in which portions of the HPC uropod domain were engulfed by the osteoblast. This was most convincingly shown in co-cultures of primary CD34+ cells and osteoblasts. There, an entire domain enriched in N-Rh-PE of the primary CD34+ cell could be seen being transferred to the osteoblast over a short transfer period leaving the cell depleted of N-Rh-PE (Fig. 3i) (Supp. Mov. 6). The N-Rh-PE domain transfer was also detected from the more primitive CD34+CD38- cells to primary human osteoblasts (Fig. 3j). Addition of dynasore, an inhibitor of the endocytic effector protein, dynamin [18], to the co-culture system significantly reduced the amount of uptake of HPC components at the contact site (Fig. 3h), suggesting the intercellular transfer mechanism was dynamin dependent.
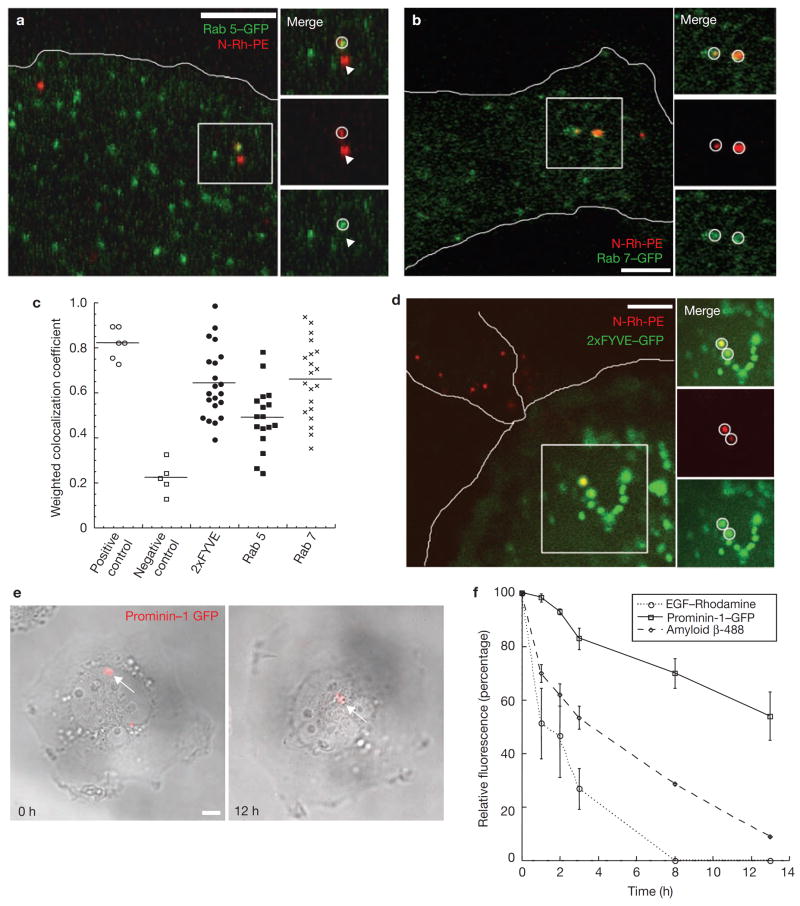
We next followed the fate of transferred material taken up by the osteoblast. After 30 min of uptake, HPC components were seen in various osteoblast endocytic compartments, including Rab5- and Rab7-positive structures (Fig, 4a-c). In addition, HPC components were observed in endocytic structures positive for the 2xFYVE domain [20] (Fig. 4c,d). Imaging of single osteoblasts that had taken up prominin 1-GFP from a contacting HPC (Fig. 4e) showed that the transferred molecules remained in distinct intracellular structures within the osteoblast for up to 12 h post-transfer. In contrast, endocytosed EGF and the soluble amyloid beta protein, which are delivered to lysosomes for degradation [21], were completely destroyed by the osteoblast during a similar time period (Fig. 4f). These results suggested that while transferred molecules could be degraded, a proportion of these molecules avoided destruction within osteoblast lysosomes and were delivered through endocytic intermediates to a longer-lived intracellular compartment.
Figure 4.
Transferred molecules are detected within various endocytic compartments of the osteoblasts. Live cell microscopy of N-Rh-PE (red) labeled KG1a cells co-cultured with osteoblastic cells transiently transfected with (a) Rab 5-GFP (green) or (b) Rab 7-GFP (green). White circles indicate transferred N-Rh-PE localized to a Rab 5-GFP or a Rab 7-GFP positive vesicle. Arrows indicate transferred N-Rh-PE not present in a Rab 5- or Rab 7-GFP vesicle. The osteoblast edge is outlined in white. (c) Histogram of weighted co-localization coefficients. As a positive control for a weighted co-localization coefficient, osteoblasts were transfected with EEA1-YFP and immunostained with an EEA-1 antibody (positive control, n = 6 cells). For a negative control, osteoblasts were transiently transfected with clathrin light chain-YFP and co-cultured with KG1a cells labeled with N-Rh-PE to allow for transfer. Weighted co-localization coefficients were calculated three hours following transfer of N-Rh-PE (negative control, n = 6 cells). All other co-localization coefficients were calculated following 1 h of co-culture (n = 20 cells for each condition). (d) Live cell microscopy of N-Rh-PE (red) labeled KG1a cells co-cultured with osteoblastic cells transiently transfected with 2xFYVE-GFP (green). White circles indicate transferred N-Rh-PE localized to a 2xFYVE-GFP positive vesicle. Both the KG1a cell and the contacted osteoblast are outlined in white. (e) Live-cell confocal imaging of osteoblasts following prominin 1-GFP (red) transfer from a transiently transfected KG1a cell. Before imaging, osteoblasts were washed with fresh medium to remove all KG1a cells, so only transferred prominin 1-GFP was detected. (f) Quantification of the relative fluorescence intensities over the course of 12 h for transferred prominin 1-GFP (solid line, n = 3 cells), internalized soluble amyloid beta-488 (dashed line, n = 6 cells), and internalized rhodamine-EGF (dotted line, n = 6 cells). Scale bars – 5μm.
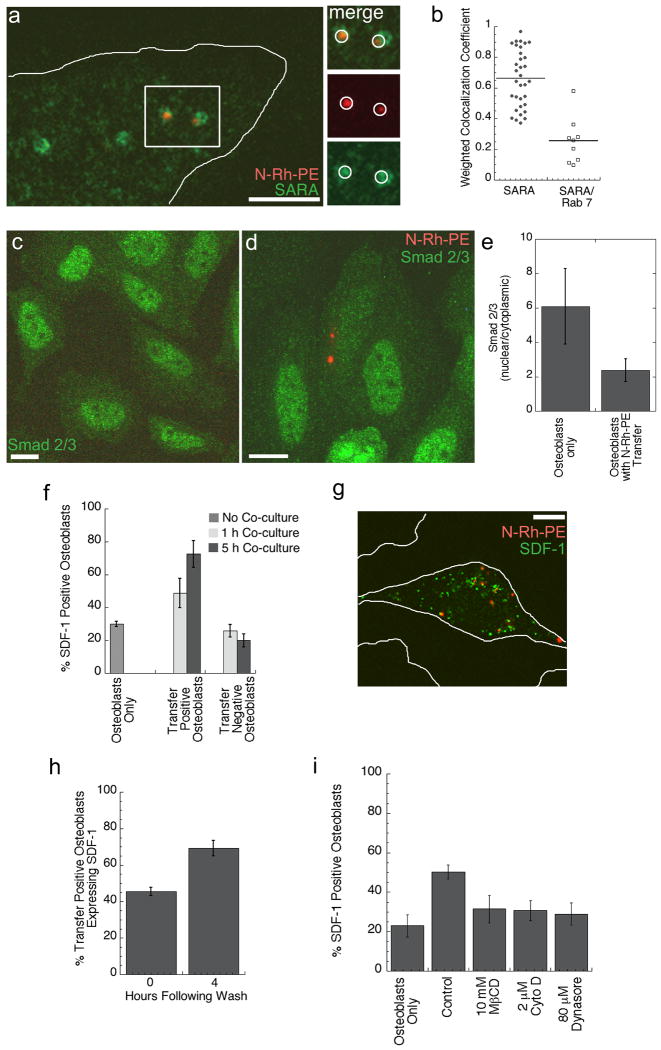
To determine the significance of transferred molecule localization for cell communication between HPC and osteoblast, we further characterized the endocytic structures containing transferred molecules. In particular, we focused on the endocytic structures that were positive for the 2xFYVE domain-GFP, since many FYVE domain-containing proteins are involved in signal transduction [22]. SARA (Smad Anchor for Receptor Activation) is a FYVE domain-containing protein localized to endosomes, which correspond to a signaling compartment specialized for the propagation of extracellular signals such as TGFβ signaling [23, 24]. Upon TGFβ receptor activation, Smads are translocated to the nucleus with the help of the cofactor, SARA, resulting in gene activation. Osteoblasts fixed following N-Rh-PE transfer and stained using SARA-specific antibodies displayed a significant co-localization of N-Rh-PE with SARA-labeled endosomes (Fig. 5a). The histogram of the weighted co-localization coefficients (Fig. 5b) revealed a similar level of N-Rh-PE co-localization with SARA as with Rab 7, although SARA and Rab 7 appeared to represent two different populations of endosomes. These data showed that molecules transferred from the HPC membrane domain can be delivered to a SARA-positive signaling endosome within osteoblasts.
Figure 5.
Intercellular transfer correlates with decreased Smad 2/3 activation and an increased production of SDF-1 by the osteoblast. (a) N-Rh-PE (red) labeled KG1a cells were co-cultured for one hour with osteoblasts, which were then fixed and immunostained for SARA (green). Transferred N-Rh-PE could be detected within SARA positive vesicles in the osteoblast as indicated by the white circles. (b) Histogram of weighted co-localization coefficients for N-Rh-PE with a SARA positive endosome (n = 33 cells) and for SARA with Rab 7-GFP positive endosomes (n = 9 cells) indicates that transferred N-Rh-PE is detected within SARA positive endosomes, which are distinct from a Rab 7-GFP compartment. (c) Osteoblasts fixed and stained for Smad2/3 (green) have a high level of nuclear expression indicating activate Smad2/3. Following a transfer event, osteoblasts with detectable transfer (d) have a reduced nuclear localization of Smad2/3. The nuclear to cytoplasmic ratios of Smad2/3 signal intensity is quantified in (e). n > 50 osteoblasts +/- transfer. (f) Quantification of the percent of SDF-1 expressing osteoblasts without co-culture, following 1 h of co-culture, and 5 h of co-culture. (n > 100 osteoblasts or +/- transfer scored for 3 independent experiments). (g) SDF-1 immunofluorescence of a transfer positive osteoblast. White lines outline the osteoblasts in the field of view. (h) Osteoblasts were co-cultured with N-Rh-PE labeled KG1a cells for 1 h and then washed to remove all KG1a cells. SDF-1 expression was detected by immunofluorescence immediately following KG1a cell removal and then 4 h following KG1a cell removal. (n > 100 transfer positive osteoblasts for 3 independent experiments). (i) To evaluate whether transfer rather than KG1a cell contact mediated the increase in SDF-1 expression, KG1a/osteoblast co-cultures were treated with 10 mM methyl β cyclodextrin (MβCD), 2 μm cytocholasin D (Cyto D), or 80 μm Dynasore to allow for KG1a cell contact with osteoblasts, but not transfer. Drug treatment, which reduced N-Rh-PE transfer (Fig. 3h), but allowed for cell contact, resulted in a decreased percent of SDF-1 expressing osteoblasts when compared to control co-cultures. (n > 600 osteoblasts for 3 independent experiments)
SARA endosomes are known to signal by virtue of Smad activation [25, 26]. Therefore, Smad signaling was tested in the presence of transferred molecules in the osteoblasts. Osteoblasts without a transfer event, or osteoblasts cultured without HPCs, displayed a high nuclear localization of Smad2/3, suggesting activated Smad signaling (Fig. 5c). In contrast, osteoblasts with detectable N-Rh-PE transfer primarily had a cytoplasmic localization of Smad2/3 (Fig. 5d), consistent with a reduction in activated Smad2/3 signaling. Quantification of the nuclear-cytoplasmic intensity ratio of Smad2/3 confirmed that Smad nuclear localization, and therefore TGFβ signaling, was diminished in osteoblasts following a N-Rh-PE transfer event (Fig. 5e).
Previous studies have suggested that TGFβ1 signaling can be inhibitory to osteoblast maintenance of HPCs by downregulating a specific chemokine, SDF-1 [27, 28], produced by osteoblasts. SDF-1, also referred to as CXCL12, is known to modulate progenitor cell migration and adhesion [3, 29]. Immunofluorescence showed that approximately 30% of the osteoblasts in culture expressed detectable SDF-1 (Fig. 5f). This finding is consistent with the osteoblasts having activated TGFβ1 signaling and the observed high levels of nuclear Smad2/3 (Fig. 5c,d). We hypothesized that if the intercellular transfer event is reducing TGFβ1 signaling, we may observe a subsequent increase in SDF-1 expressing osteoblasts specifically following intercellular transfer. After 1 h of co-culture, 45% of the osteoblasts containing transferred material expressed SDF-1 (Fig. 5f). Figure 5g shows an example of an SDF-1 expressing osteoblast (green) after uptake of N-Rh-PE (red) from an HPC. When the time of co-culture was increased to 5 h, almost 75% of the osteoblasts with a transfer event were expressing SDF-1 (Fig. 5f). Therefore, after prolonged co-culture, osteoblasts positive for transferred N-Rh-PE were also positive for SDF-1 expression.
To discriminate between induction of SDF-1 expression by the transfer event or by continued KG1a cell contact, we washed the KG1a cells from the osteoblastic monolayer after 1 h of co-culture. After HPC removal, the osteoblasts were cultured an additional 4 h to evaluate SDF-1 expression. Once again, approximately 75% of osteoblasts with a transfer event were positive for SDF-1 expression (Fig. 5h), therefore prolonged HPC contact with osteoblasts does not lead to the increased SDF-1 detected. To address the possibility that HPC contact with osteoblasts rather than transfer resulted in the increased SDF-1, we treated the HPC-osteoblast co-cultures for 1 h with 10 mM methyl β cyclodextrin (MβCD), 2 μm cytocholasin D (Cyto D), or 80 μm Dynasore, which we showed in Fig. 3h reduced N-Rh-PE transfer. Drug treatment, which allows for osteoblast contact but reduces transfer, resulted in a decreased percent of SDF-1 expressing osteoblasts when compared to control co-cultures. These data suggest that intercellular transfer, rather than cell contact, leads to changes in protein expression within osteoblastic cells, more specifically to an increased production of SDF-1.
In this paper, we identify events following HPC-osteoblast contact that may play important roles in the signaling and remodeling events within the bone marrow niche. Specifically, we show that portions of the HPC uropod membrane at the contact site are actively endocytosed by osteoblasts and delivered to SARA-positive, signaling endosomes. In response to intercellular transfer, osteoblasts exhibit decreased Smad signaling and greater production of the bone marrow chemokine, SDF-1. This may occur because the transferred material within SARA-endosomes sequesters SARA away from its general co-factor function in Smad activation, allowing the osteoblast to produce more SDF-1. These events could greatly influence the overall trafficking, proliferation and survival of hematopoietic stem-progenitor cells in the bone marrow niche by permitting signaling events to be initiated and propagated at the single cell level. Furthermore, the intercellular transfer and downstream endosomal signaling described in this study could be significant for cell-cell communication not only in the bone marrow, but also in other intimate cellular microenvironments [30-32].
Methods
Cells and Transfection
The KG1a cell line and the SaOS2 cell line (from the American Type Culture Collection; ATCC, Manassas, VA) were cultured in RPMI 1640 supplemented with 10% FBS, sodium pyruvate, non-essential amino acids, sodium bicarbonate, and MEM vitamins (Invitrogen, Carlsbad, CA). Primary human osteoblast (hOST cells) were purchased from Lonza (Lonza Group Ltd, Switzerland) and cultured according to manufacturers protocol. Human CD34+ cells were collected from normal healthy volunteers who gave informed consent in accordance with the Declaration of Helsinki and National Institutes of Health Institutional Review Board–approved protocols. Donors received 5 days of G-CSF (Filgrastim; Amgen, Thousand Oaks, CA) 10mcg/Kg given as a single daily subcutaneous injection with leukapheresis initiated on the morning of day 5. Large volume (15L) leukapheresis procedures were performed with a model CS-3000 Plus continuous-flow apheresis device (Fenwal Division, Baxter, Deerfield, IL). The apheresis products were enriched for CD34+ cells by immunomagnetic beads affinity elution with a magnetic cell selection system (Isolex 300I, Nexell Therapeutics, Irvine, CA). In some experiments, human CD34+CD38- cells were enriched by FACS with CD34-PE (Becton Dickinson, San Jose,CA) and CD38-PECy7 (Becton Dickinson) antibodies using a FACS Aria II instrument (Becton Dickinson).
Transfection
KG1a and CD34+ cells were nucleofected following the manufacturer protocol (Amaxa, Lonza Group Ltd, Switzerland) with CD63-cherry or with prominin 1-GFP. SaOS2 cells were transfected with PH-Akt-GFP, 2xFYVE-GFP, clathrin light chain-YFP, Rab5-GFP, or Rab7-GFP using the Fugene 6 Reagent according to the manufacturers protocol (Roche, Switzerland). Previously, actin-YFP, tubulin-GFP and pEGFP SaOS2 stable cell lines were generated by G418 selection.
HPC labeling
Cells were labeled with N-Rh-PE by incubating with 5 μM N-Rh-PE (Avanti Polar Lipids, Alabaster, AL) diluted in cold 1 × Hanks buffer (Invitrogen, Carlsbad, CA) for 1 h on ice. Cells were then washed 3 × with cold Hanks buffer before plating back into growth medium. N-Rh-PE labeled cells were used in co-culture experiments approximately 24 h after labeling. HPCs were labeled with Cell Labeling QDs as described by the manufacturer (Invitrogen, Carlsbad, CA) except the cell and QD incubation time was extended to overnight. PKH26 labeling was performed according to manufacturers protocol (Sigma, St. Louis, MO).
Live-cell imaging
SaOS2 cells were plated on collagen I (BD Biosciences, San Jose, CA) precoated four or eight-well chambered coverglasses (Nunc; Thermo Fisher Scientific, Rochester, NY). For transient transfections, SaOS2 cells were transfected in the chambered coverglasses 24 h before co-culturing and imaging. Cells were imaged at 37°C in either CO2 independent medium supplemented with 10% FBS (Invitrogen, Carlsbad, CA) or with mineral oil overlayed upon the culture medium to reduce gas exchange. Cells were imaged by Laser Scanning Confocal Microscopy with Zeiss LSM 510 systems (Zeiss, Germany) using excitation wavelengths of 488 or 543 nm and a 63x oil immersion objective (NA=1.2). Image analysis was performed using the Zeiss LSM 510 software or Image J (NIH, Bethesda, MD). Brightness and contrast were change in some brightfield images to increase cell visibility (Adobe Photoshop).
For intercellular transfer experiments, HPCs and osteoblasts were co-cultured for 1 h on the microscope before imaging. Following co-culture, Z series confocal imaging was performed through HPC/osteoblast contacted cells to identify transferred material.
The weighted co-localization coefficients were calculated using the Zeiss LSM software (Zeiss, Germany). Briefly, the weighted co-localizaton coefficients represent the number of red pixels (N-Rh-PE) that co-localize with green pixels (Rab5, Rab7, 2xFYVE, SARA) divided by the total number of red pixels.
Fixation and Immunostaining
HPC and osteoblast co-cultures were fixed with 4% paraformaldehyde for 15 min at room temperature followed by a 1xPBS wash. Cells were blocked and permeablized in 1xPBS with 10mg/ml of BSA and 0.3% tween-20. Cells were stained at 4°C for approximately 15 h with anti-cd45 (Calbiochem, Gibbstown, NJ), anti-AC133 (prominin-1, Miltenyi Biotec, Germany), anti-CD49d (Calbiochem, Gibbstown, NJ), anti-SARA (Santa Cruz Biotechnology, Santa Cruz, CA), anti-Smad2/3 (BD Biosciences, San Jose, CA) and anti-SDF-1 (Santa Cruz Biotechnology, Santa Cruz, CA). Cells were washed gently with 1xPBS followed by incubation with Alexa-488 goat anti-mouse antibody (Invitrogen, Carlsbad, CA) or Cy3-goat anti-rabbit antibody (Jackson ImmunoResearch, West Grove, PA) for 1 h at room temperature. Cells were washed following secondary antibody with 1xPBS and mounted in Flouromount-G (Southern Biotech, Birmingham, AL) for imaging.
Scanning Electron Microscopy
Cells were rinsed twice with PBS and fixed with 2% glutaraldehyde in 0.1 mol/l Na-cacodylate buffer (with 0.1 mol/l sucrose) at pH 7.4 for 12 h. They were subsequently treated with 1% tannic acid in 0.15 mol/l Na-cacodylate at pH 7.4 for 1 h and post-fixed with 1% osmium tetroxide in 0.1 mol/l Na-cacodylate at pH 7.4 for 1 h. Samples were dehydrated in a graded ethanol series.
Statistical Analysis
Error bars are shown as standard deviations and Student t-tests were used as a statistical test for all data.
Supplemental Movies
Acknowledgments
We are grateful to Victoria Cogger (University of Sydney, ANZAC Research Institute) for her assistance with scanning electron microscopy. We would also like to thank Denis Corbeil (University of Dresden) for providing the prominin 1-GFP construct, and Suliana Manley and George Patterson (NIH) for critical reading of the manuscript. This project was supported by the Intramural Research Program of the US National Institute of Child Health and Human Development, National Institutes of Health.
References
- 1.Adams GB, Scadden DT. The hematopoietic stem cell in its place. Nat Immunol. 2006;7(4):333–7. doi: 10.1038/ni1331. [DOI] [PubMed] [Google Scholar]
- 2.Wright DE, et al. Physiological migration of hematopoietic stem and progenitor cells. Science. 2001;294(5548):1933–6. doi: 10.1126/science.1064081. [DOI] [PubMed] [Google Scholar]
- 3.Mendez-Ferrer S, et al. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452(07186):442–8. doi: 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- 4.Jung Y, et al. Cell-to-cell contact is critical for the survival of hematopoietic progenitor cells on osteoblasts. Cytokine. 2005;32(3-4):155–62. doi: 10.1016/j.cyto.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425(6960):836–41. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 6.Arai F, et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118(2):149–61. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Calvi LM, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425(6960):841–6. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 8.Wilson A, Trumpp A. Bone-marrow haematopoietic-stem-cell niches. Nat Rev Immunol. 2006;6(2):93–106. doi: 10.1038/nri1779. [DOI] [PubMed] [Google Scholar]
- 9.Wagner W, et al. Hematopoietic progenitor cells and cellular microenvironment: behavioral and molecular changes upon interaction. Stem Cells. 2005;23(8):1180–91. doi: 10.1634/stemcells.2004-0361. [DOI] [PubMed] [Google Scholar]
- 10.Giebel B, et al. Segregation of lipid raft markers including CD133 in polarized human hematopoietic stem and progenitor cells. Blood. 2004;104(8):2332–8. doi: 10.1182/blood-2004-02-0511. [DOI] [PubMed] [Google Scholar]
- 11.Feigelson SW, et al. The CD81 tetraspanin facilitates instantaneous leukocyte VLA-4 adhesion strengthening to vascular cell adhesion molecule 1 (VCAM-1) under shear flow. J Biol Chem. 2003;278(51):51203–12. doi: 10.1074/jbc.M303601200. [DOI] [PubMed] [Google Scholar]
- 12.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6(10):801–11. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 13.Bauer N, et al. New Insights into the Cell Biology of Hematopoietic Progenitors by Studying Prominin-1 (CD133) Cells Tissues Organs. 2007 Dec 21; doi: 10.1159/000112847. Published online. [DOI] [PubMed] [Google Scholar]
- 14.Roper K, Corbeil D, Huttner WB. Retention of prominin in microvilli reveals distinct cholesterol-based lipid micro-domains in the apical plasma membrane. Nat Cell Biol. 2000;2(9):582–92. doi: 10.1038/35023524. [DOI] [PubMed] [Google Scholar]
- 15.Willem J, et al. A non-exchangeable fluorescent phospholipid analog as a membrane traffic marker of the endocytic pathway. Eur J Cell Biol. 1990;53(1):173–84. [PubMed] [Google Scholar]
- 16.Freund D, et al. Polarization of human hematopoietic progenitors during contact with multipotent mesenchymal stromal cells: effects on proliferation and clonogenicity. Stem Cells Dev. 2006;15(6):815–29. doi: 10.1089/scd.2006.15.815. [DOI] [PubMed] [Google Scholar]
- 17.van Niel G, et al. Exosomes: a common pathway for a specialized function. J Biochem. 2006;140(1):13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- 18.Damke H, et al. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol. 1994;127(4):915–34. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macia E, et al. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell. 2006;10(6):839–50. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Wurmser AE, Gary JD, Emr SD. Phosphoinositide 3-kinases and their FYVE domain-containing effectors as regulators of vacuolar/lysosomal membrane trafficking pathways. J Biol Chem. 1999;274(14):9129–32. doi: 10.1074/jbc.274.14.9129. [DOI] [PubMed] [Google Scholar]
- 21.Valdez G, et al. Trk-signaling endosomes are generated by Rac-dependent macroendocytosis. Proc Natl Acad Sci U S A. 2007;104(30):12270–5. doi: 10.1073/pnas.0702819104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corvera S. Signal transduction: stuck with FYVE domains. Sci STKE. 2000;2000(37):PE1. doi: 10.1126/stke.2000.37.pe1. [DOI] [PubMed] [Google Scholar]
- 23.Bokel C, et al. Sara endosomes and the maintenance of Dpp signaling levels across mitosis. Science. 2006;314(5802):1135–9. doi: 10.1126/science.1132524. [DOI] [PubMed] [Google Scholar]
- 24.Di Guglielmo GM, et al. Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat Cell Biol. 2003;5(5):410–21. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- 25.Runyan CE, Schnaper HW, Poncelet AC. The role of internalization in transforming growth factor beta1-induced Smad2 association with Smad anchor for receptor activation (SARA) and Smad2-dependent signaling in human mesangial cells. J Biol Chem. 2005;280(9):8300–8. doi: 10.1074/jbc.M407939200. [DOI] [PubMed] [Google Scholar]
- 26.Hayes S, Chawla A, Corvera S. TGF beta receptor internalization into EEA1-enriched early endosomes: role in signaling to Smad2. J Cell Biol. 2002;158(7):1239–49. doi: 10.1083/jcb.200204088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung Y, et al. Regulation of SDF-1 (CXCL12) production by osteoblasts; a possible mechanism for stem cell homing. Bone. 2006;38(4):497–508. doi: 10.1016/j.bone.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 28.Wright N, et al. Transforming growth factor-beta1 down-regulates expression of chemokine stromal cell-derived factor-1: functional consequences in cell migration and adhesion. Blood. 2003;102(6):1978–84. doi: 10.1182/blood-2002-10-3190. [DOI] [PubMed] [Google Scholar]
- 29.Dar A, et al. Chemokine receptor CXCR4-dependent internalization and resecretion of functional chemokine SDF-1 by bone marrow endothelial and stromal cells. Nat Immunol. 2005;6(10):1038–46. doi: 10.1038/ni1251. [DOI] [PubMed] [Google Scholar]
- 30.Al-Nedawi K, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10(5):619–24. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 31.Davis DM. Intercellular transfer of cell-surface proteins is common and can affect many stages of an immune response. Nat Rev Immunol. 2007;7(3):238–43. doi: 10.1038/nri2020. [DOI] [PubMed] [Google Scholar]
- 32.Williams GS, et al. Membranous structures transfer cell surface proteins across NK cell immune synapses. Traffic. 2007;8(9):1190–204. doi: 10.1111/j.1600-0854.2007.00603.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.