Abstract
Curcumin is the active ingredient of the turmeric powder, a natural spice used for generations in traditional medicines. Curcumin’s broad spectrum of anti-oxidant, anti-carcinogenic, anti-mutagenic, and anti-inflammatory properties makes it particularly interesting for the development of pharmaceutical compounds. Due to curcumin’s various effects on the function of numerous unrelated membrane proteins, it has been suggested that it affects the properties of the bilayer itself. However, a detailed atomic-level study of the interaction of curcumin with membranes has not been attempted. A combination of solid-state NMR and differential scanning calorimetry experiments shows curcumin has a strong effect on membrane structure at low concentrations. Curcumin inserts deep into the membrane in a transbilayer orientation, anchored by hydrogen bonding to the phosphate group of lipids in a manner analogous to cholesterol. Like cholesterol, curcumin induces segmental ordering in the membrane. Analysis of the concentration dependence of the order parameter profile derived from NMR results suggests curcumin forms higher order oligomeric structures in the membrane that span and likely thin the bilayer. Curcumin promotes the formation of the highly curved inverted hexagonal phase which may influence exocytotic and membrane fusion processes within the cell. The experiments outlined here show promise for understanding the action of other drugs such as capsaicin in which drug-induced alterations of membrane structure have strong pharmacological effects.
Turmeric powder prepared from the turmeric plant has been in use for centuries in the traditional medicine of China and India for treating wounds, infections, and other skin problems.1 The active component of turmeric powder, curcumin (Fig. 1 for the structure of curcumin), has a surprising array of anti-oxidant, anti-cancer, anti-mutagenic, antibiotic, anti-viral, anti-fungal, anti-amyloid, anti-diabetic, and anti-inflammatory properties.2–9 Because of its numerous pharmaceutical effects and its inherent non-toxicity, curcumin has been the focus of many recent biochemical investigations, primarily reporting on its medicinal properties and interactions with specific proteins. Despite intense interest in the physiological effects of curcumin, a general mechanism for its action has not been identified.
Figure 1.
Keto-enol form of curcumin, the dominant tatuomer of curcumin. The keto-enol from is stabilized by an intramolecular hydrogen bond, shown here by a dashed red line.
Studies of curcumin have shown that it influences structurally unrelated membrane proteins across several signaling pathways.10 For a small minority of these proteins, specific binding of curcumin to the protein has been detected with a binding constant typically in the nanomolar range. However, for the majority of the proteins affected by curcumin, a curcumin binding site has not been identified. Furthermore, curcumin has been shown to affect these proteins at approximately similar concentrations (micromolar range) despite the lack of a consensus binding motif.11 Since curcumin affects such a large array of unrelated membrane proteins at approximately similar concentrations, it has been proposed that curcumin can regulate the action of membrane proteins indirectly by changing the physical properties of the membrane rather than by the direct binding of curcumin to the protein.11
Integral membrane proteins are embedded in the membrane with hydrophobic segments of the protein in contact with the hydrophobic core of the membrane. A mismatch between the length of the transmembrane segments of the protein and the hydrophobic thickness of the bilayer is energetically unfavorable and can therefore have a strong influence on the conformational equilibria of membrane proteins. Because the elastic moduli of proteins are generally much higher than that of the membrane, the membrane tends to deform to match the hydrophobic and anchoring interactions of the protein. Deformation of the membrane imposes an energetic penalty that can be modified by the addition of lipophilic drugs that alter the physical properties of the membrane such as the hydrophobic thickness of the membrane, the elastic modulus, the lateral pressure profile or other physical parameters. Alterations of membrane protein’s activity in this manner has been detected for several lipophilic drugs and is suspected as being responsible for the unwanted nonspecific side effects for many others.12–16
Ingolfsson et al. tested this hypothesis by measuring the effects of curcumin on the activity of gramicidin channels of varying lengths and amino acid sequences in DOPC (dioleoylphosphatidylcholine) membranes.11 The formation of an active gramicidin channel requires the dimerization of gramicidin in the membrane, which in turn is dependent on the deformation of the bilayer to accommodate the gramicidin dimer. The results of this experiment showed that the inclusion of curcumin increased both gramicidin channel lifetimes and appearance rates, leading to the conclusion that curcumin decreased the energetic costs of the bilayer deformations.11 Further work by the research group of Huang has shown that curcumin can bind to the membrane in two modes: a surface associated mode at low curcumin concentrations and a transmembrane mode at higher concentrations.17, 18 While these studies show that it is possible to alter a membrane protein’s activity by alterations in the physical properties of the membrane, a detailed study of the physical changes in the membrane induced by curcumin has not been performed. In this study, we use a combination of solid-state NMR and differential scanning calorimetry (DSC) experiments to show curcumin induces dramatic changes in the fluidity of the lipid bilayer, consistent with a role for the nonspecific membrane binding of curcumin in the physiological activity of curcumin.
MATERIALS AND METHODS
Curcumin (>94%) was purchased from Sigma and used without further purification. Lipids (DMPC (1,2-Dimyristoyl-sn-glycero-3-phosphocholine), DHPC (1,2-Dihexanoyl-sn-glycero-3-phosphocholine), and DPPC (1,2-Dipalmitoyl-sn-glycero-3-phosphocholine)) were purchased from Avanti Polar Lipids (Alabaster, AL). Curcumin was dissolved in methanol to produce a 1.0 mg/mL stock solution and kept at −20°C in glass vials wrapped in aluminum foil for protection from light-induced degradation.
UV-visible spectroscopy
To prepare large unilamellar vesicles for UV-Vis spectroscopy, the appropriate amount of a stock solution of 20 mg DMPC/mL in chloroform was first dried under nitrogen gas to create a lipid film. The lipid film was then held under vacuum overnight to remove any traces of solvent. The dried lipid film was hydrated with phosphate buffer (pH 7.8) and the solution vortexed until the lipid film was completely dissolved. The lipid solution was then extruded twenty times through a 100 nm membrane to create the lipid vesicles. For the measurement of the breakdown of curcumin, the curcumin concentration for each trial was kept constant at 10 μM and the lipid concentration was varied to create the indicated mole ratios of curcumin. The breakdown of curcumin was measured by the disappearance of the absorbance peak at 424 nm, which is indicative of the fragmentation at the keto-enol group.19 Between scans, the sample cuvettes were removed from the spectrophotometer and placed under aluminum foil to protect them from light degradation.
Differential scanning calorimetry
Samples for DSC experiments were prepared using appropriate ratios of lipid to curcumin from stock solutions of curcumin in methanol and DMPC or DPPC in chloroform. The lipid concentration was held constant at 2.95 mM while the curcumin concentration was varied. Samples were vortexed and then dried under a stream of nitrogen gas. Residual solvent was removed by placing the samples under high vacuum overnight, while the sample was protected from light. After the drying process, sodium phosphate buffer (50 mM NaPi with 150 mM NaCl at pH 7.3) was added to the dry lipid film, followed by vortexing and brief sonication. The resultant multilamellar vesicles (MLVs) were then subjected to repeated freeze thaw cycles and degassed before loading into the calorimeter.
A scanning rate of 1.0°C/min was used for both heating and cooling. Temperature ranges of 5–45°C and 20–60°C were used for DMPC and DPPC, respectively. A base line was recorded with buffer in the reference cell and subtracted from the sample data. The peak maximum in the plot of excess heat capacity vs. temperature was recorded at the phase transition temperature TM, and the transition enthalpy was derived from the area under the excess heat capacity vs. temperature curve during the transition.
Preparation of bicelles for solid-state NMR experiments
Bicelles were prepared by mixing 100 mg total of DMPC and DHPC dissolved in chloroform at a DMPC:DHPC molar ratio (q) of 4.5 with an appropriate amount of curcumin dissolved in methanol. The bulk solvent was removed by slowly evaporating the lipid mixture under a stream of nitrogen gas. Residual solvent was removed by placing the sample under high vacuum overnight. The dried sample was then rehydrated by the addition of 200 μL of 45 mM pH 6.0 HEPES buffer containing 150 mM NaCl. The samples were then subjected to repeating heating and cooling cycles above and below the bicelle formation temperature until clear transparent solutions were formed.
Solid-state NMR
All of the experiments were performed on a Chemagnetics/Varian Infinity 400 MHz solid-state NMR spectrometer. Each sample was equilibrated for at least 30 minutes before starting the experiment. 31P NMR spectra were obtained using a spin-echo pulse sequence (90°-τ–180°-τ-acquisition; τ = 125 μs) with a 90° pulse length of 5 μs under a 30 kHz continuous-wave proton decoupling. Chemical shifts were referenced by setting the isotropic chemical shift peak of phosphoric acid to 0 ppm. 14N quadrupole coupling spectra were recorded using a quadrupolar echo pulse sequence (90°-τ–90°-τ-Acquisition; τ = 80 μs) without proton decoupling.
Proton detected local field (PDLF) spectra were recorded as described elsewhere. 20,21 A ramped-cross-polarization (ramp-CP) sequence with a 3 ms contact time, 70 t1 increments, a 5 s recycling delay, and a 25 kHz FLOPSY-8 proton decoupling were used to obtain 2D PDLF spectra. The observed dipolar couplings were converted into order parameters using the relation
| (Eq. 1) |
where Dobs is the observed C-H dipolar coupling, D0 is the dipolar coupling in the absence of motional averaging (~21.5 kHz), θ is the angle between the membrane and the magnetic field (90°), and k is a scaling factor whose value depend on the homonuclear decoupling sequence employed during the t1 period (0.42 for the BLEW-48 sequence). All measurements were performed at 37°C. The samples were equilibrated for 30 minutes at 37°C before acquisition.
RESULTS
Measurement of curcumin degradation in DMPC liposomes by UV-visible spectroscopy
The rate of curcumin degradation in DMPC liposomes was measured using the loss of the characteristic absorption maximum at 424 nm as a marker for curcumin degradation.19 DMPC exhibits a significant stabilizing effect on curcumin at both pH 6.0 and 7.8 (Figure S1). After 2 days, approximately 95% of the curcumin/DMPC sample was intact, indicating curcumin is stable in lipid bilayers at pH 6 on a time scale relevant to solid-state NMR experiments (approximately 12 hours).
Measurement of main and La to HII phase transition temperatures by DSC
Changes in the phase behavior of membranes that occur with the addition of membrane-affecting molecules can be measured with high accuracy using DSC. Alterations in the packing of lipid molecules within the bilayer are reflected by changes in the thermodynamics of the lipid phase transition. In the absence of curcumin, DMPC and DPPC undergo two phase transitions in the range of temperatures studied. The first phase transition, usually termed the pretransition, is a low enthalpy transition reflective of the membrane undergoing a change from the gel phase to the rippled gel phase.22,23 The rippled gel phase is linked to the presence of periodic ripples on the membrane surface, usually with periods in the range of 100–300 Å depending on the lipid.24
A reduction or elimination of the pretransition is common with most membrane ligands and usually indicates the molecule in question is interacting with the lipid headgroups.25 Fig. 2 shows the apparent abolition of the pretransition with the increase of the curcumin concentration in both DMPC and DPPC. This implies that curcumin interacts with the PC headgroup of both lipids, regardless of the difference in acyl chain lengths.
Figure 2. Curcumin lowers the enthalpy and phase transition temperature of DMPC and DPPC bilayers.
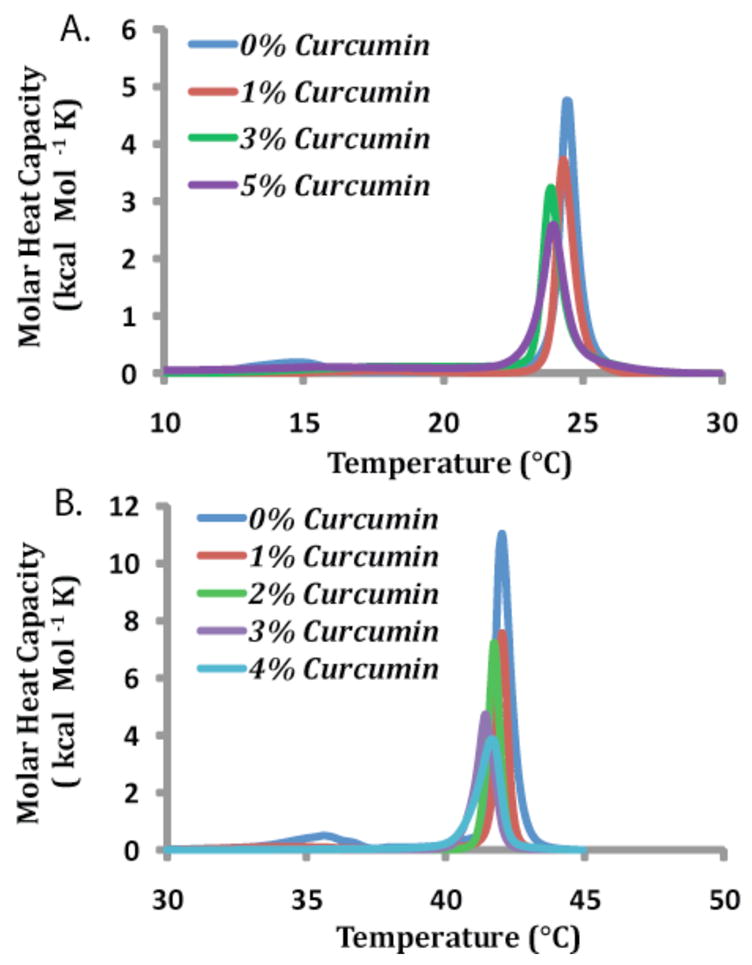
DSC heating scans for (A) DMPC and (B) DPPC with varying concentrations of curcumin show that it lowers the temperature of the DMPC bilayer phase transition (from gel to liquid crystalline phase) and decreases the enthalpy of main phase transition, making the gel phase more fluid and the liquid crystalline phase more rigid. All curcumin values are given as mole percentages with a constant lipid concentration of 2.95 mM.
The main phase transition is associated with the melting of the acyl chains in the hydrophobic core of the membrane from the rigid gel phase to the more fluid liquid crystalline phase. As the curcumin concentration increases, the transition temperature TM, enthalpy, and entropy all generally showed a mild decrease for both DMPC and DPPC as a result of curcumin (Fig. 2 and Table S1). The main phase transition peaks for both DMPC and DPPC were symmetrical, strongly implying that the curcumin was evenly dispersed throughout the bilayers.
DSC was also used to determine the effects of curcumin on membrane curvature by monitoring the phase transition temperature (TH) of the liquid-crystalline (Lα) to inverted hexagonal (HII) phase transition of DiPoPE vesicles.26,27,28 The HII phase is characterized by a high negative curvature. Ligands that either stabilize this curvature or increase the bending elasticity will favor the HII phase and lower TH, conversely those that promote positive curvature or decrease the bending elasticity will increase TH. Our experiments show that DiPoPE has a TH of 45.5°C in the absence of curcumin. The addition of a small amount of curcumin (0.1 mol %) to DiPoPE decreases TH to 41.8 °C, indicating curcumin promotes negative curvature in the bilayer even at low concentrations (Fig. 3). A higher percentage of curcumin (0.5 mol %) decreases TH further to 41.2°C.
Figure 3. Curcumin promotes the formation of the negatively curved inverted hexagonal (HII) phase.
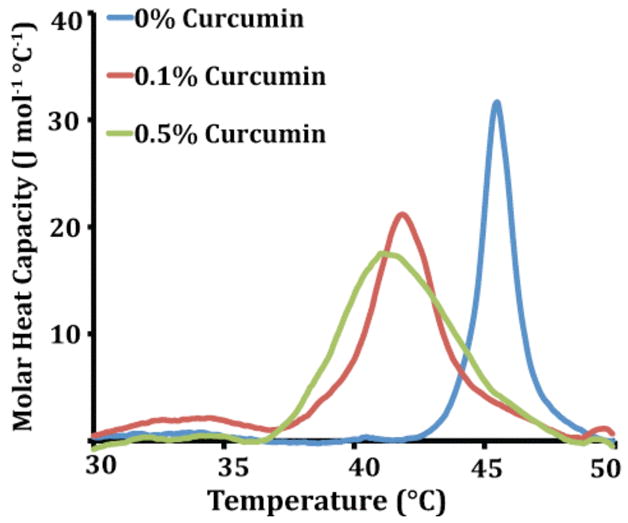
DSC heating scans of DiPoPE containing the listed molar percentage of curcumin.
Nitrogen-14 NMR spectra of magnetically aligned bicelles
The nitrogen atom of the choline group in DMPC can serve as an effective probe for changes in the electrostatic environment of the headgroup region of the membrane.32–33 The quadrupolar splitting of the 14N nucleus is determined by the electric field gradient tensor at the site of the nucleus and in most molecules is quite large. In lipids, however, it is considerably reduced by the near tetrahedral symmetry of the choline headgroup. The electrostatic interactions between the positively charged choline group and the negatively charged phosphate groups reduce this tetrahedral symmetry for lipids in a membrane. Electrostatic interactions of ligands with the phosphate group oppose this interaction and decrease the 14N quadrupolar coupling. Direct electrostatic interactions with the choline group have the opposite effect, inducing a torque on the headgroup that alters the direction of the P-N dipole and increases the quadrupolar coupling. Measurement of 14N quadrupolar couplings is particularly useful as it is generally independent of the order of the acyl chains in the hydrophobic core of the membrane.
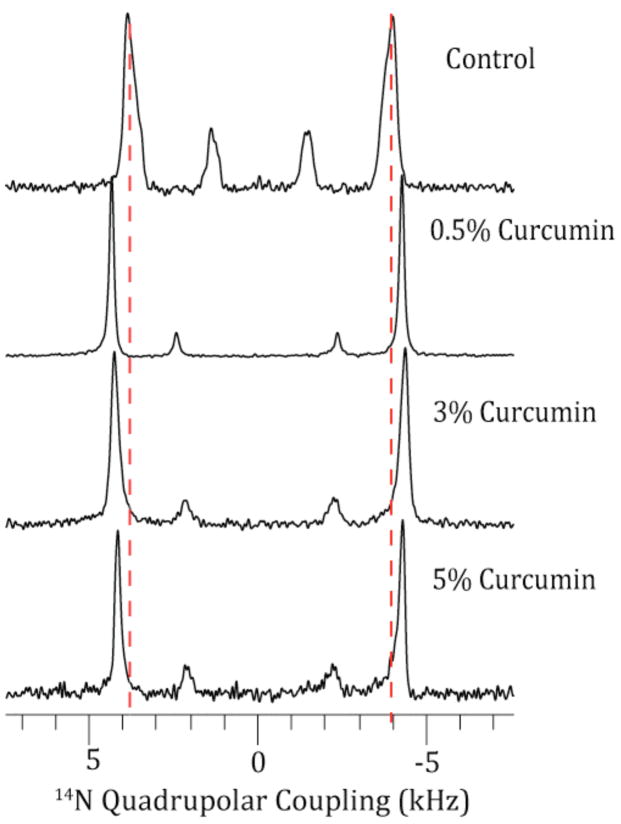
Figure 4 shows the 14N-NMR quadrupole-coupling spectrum for curcumin in magnetically aligned bicelles composed of DMPC and DHPC. The smaller inner peaks are associated with DHPC and the outer peaks with DMPC. The quadrupole coupling associated with DMPC in control bicelles without curcumin was 7.83 kHz. The addition of 0.5 mole % and 3 mole % curcumin increases the 14N quadrupolar coupling to 8.57 and 8.59 kHz respectively. At 5 mole %, the 14N quadrupolar coupling is slightly less (8.42 kHz) than the values at lower percentages of curcumin but still higher than the control sample.
Figure 4. Changes in the 14N quadrupole coupling of magnetically aligned 9:2 DMPC:DHPC bicelles containing the listed molar concentration of curcumin show that curcumin increases the asymmetry around the 14N nucleus in the choline group.
The inner peaks are associated with DHPC and the larger, outer peaks are associated with DMPC. The vertical lines are for guide purposes only.
Determination of site-specific order parameters in bicelles
A 2D separated local field (SLF) type experiment was used for the determination of molecular order parameters in a site-specific manner by the measurement of 13C-1H dipolar couplings. The observed dipolar couplings in 2D SLF spectra are the motional average of the intrinsic C-H dipolar coupling which is a direct function of the C-H bond length. Because the C-H bond length is unlikely to be affected by ligand binding, changes in the dipolar couplings along the lipid molecule upon ligand binding can be used to directly map changes in the mobilities of the different regions of the phospholipid molecules. The proton-detected 2D SLF sequence (called PDLF (Proton Detected Local Field)) used here has the advantage that the dipolar couplings associated with each C and H nucleus are detected independently because the low natural abundance of 13C ensures each proton is in the local field of only one 13C nucleus. This is in contrast to other 2D SLF experiments in which the observed dipolar coupling is in the local field of several protons, leading to a convolution and not a simple superposition of signals.
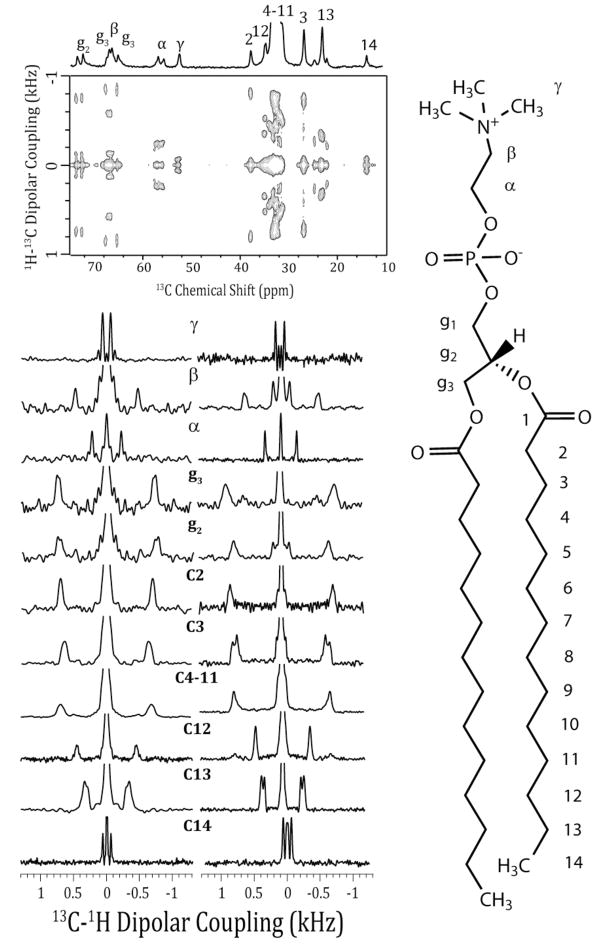
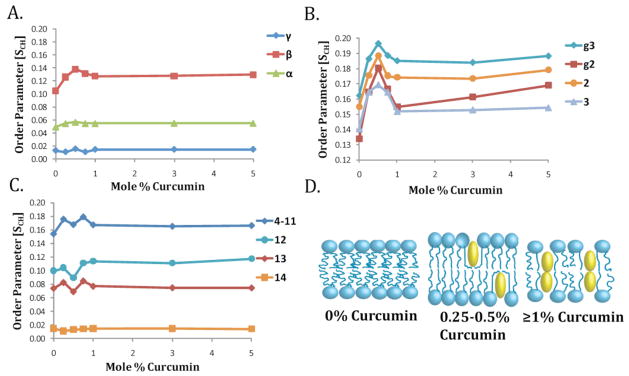
A representative 2D PDLF spectrum and representative C-H dipolar coupling slices are given in Fig. 5 along with the molecular structure and labeling convention used for the DMPC molecule. The calculated order parameter profile for DMPC molecules in bicelles containing 0.25–5 mole % curcumin is given in Fig. 6. The molecular order of the bilayer increased throughout the length of the acyl chain of the lipid in the bilayer upon the incorporation of curcumin. This effect persisted at all concentrations studied from 0.25 to 5 mole %. Increasing the concentration of curcumin from 3 to 5 mol % did not have a significant effect on the order parameter profile, most likely due to the membrane saturation with curcumin. The most prominent ordering effect is in the region of the glycerol backbone from the g3 carbon to carbon 2 (Fig 6B). The mobile carbons at the two ends of the lipid molecule are less affected by the presence of curcumin. Notably, while the β carbon in the headgroup region shows a strong ordering effect upon curcumin binding, the neighboring α carbon does not show an effect upon curcumin binding at all concentrations studied. The acyl chains comprising the hydrophobic core of the membrane are also ordered by curcumin but to somewhat lesser extent than the glycerol region.
Figure 5.
Top: A 1D 13C chemical shift and a 2D PDLF spectrum of magnetically aligned 9:2 DMPC:DHPC bicelles (pH 6.0) in the absence of curcumin. Right: Molecular structure of DMPC showing the numbering convention used. Bottom: C-H dipolar coupling spectral slices extracted from 2D PDLF spectra of magnetically aligned bicelles without (bottom left) and with 5 mole % curcumin (bottom right).
Figure 6. The molecular order parameters determined by 2D PDLF experminents show site-specific ordering of DMPC in DMPC/DHPC bicelles by curcumin.
Curcumin induced overall ordering of the bilayer at all concentrations tested. A. Order parameter profile in the lipid headgroup region (carbons γ -α). The non-monotonic response is indicative of a conformational change of the lipid headgroup in the presence of curcumin. B. Order parameter profile in the lipid headgroup region (carbons g3–3). The ordering effect is strongest in this region. C. Order parameter profile in the hydrophobic core of the membrane (carbons 4–11). The terminal carbons are more disordered than the pure bicelle sample at the lower concentrations of curcumin but are more ordered at higher concentrations (1–5%). D. Cartoon schematic of the physical changes induced by curcumin in the membrane. Curcumin at low molar concentrations is monomeric and oriented with its long axis along the membrane normal. Curcumin is effective at these concentrations in ordering the membrane except for the terminal carbons below the curcumin molecule. At higher concentrations, curcumin oligomerizes in the membrane, reducing its thickness and molecular order relative to the samples with lower concentrations of curcumin.
DISCUSSION
Numerous drugs that have specific action against target membrane proteins at pico- to nanomolar concentrations also have a “secondary pharmacology” at higher concentrations, where the same protein regulates other membrane proteins nonspecifically.16 Nonspecific regulation of embedded membrane proteins can result from changes in the physical properties of the membrane as membrane proteins are energetically coupled to the membrane bilayer.16,29–31 The nonspecific action of lipophillic drugs on non-target membrane proteins has led to significant problems with attrition in drug discovery as pharmacological studies frequently utilize drug concentrations that can alter the properties of the phospholipid bilayer.16,29–31 For this reason, drug leads with a high lipophillicity are frequently considered poor drug candidates regardless of their effectiveness against the target protein.32 However, many natural products maintain bioavailability and effectiveness despite violating classical rules of what determines a drug-like molecule.33 The reasons for this discrepancy are poorly understood. A critical problem in this area is the relatively poor understanding of drug-membrane interactions, particularly at an atomic level.
The preferential cytotoxicity of curcumin to cancer cells is an example of where such an understanding of drug-induced membrane interactions could be crucial. Cancer cells treated with curcumin display some features typical of apoptotic cell death, such as increased membrane permeability, decrease in mitochondrial membrane potential, and transient phosphatidyl serine exposure among other effects.34 However, other features of curcumin induced cytoxicity, such as the very early loss of membrane integrity and the partial reversibility of some of the effects, are not typical of standard apoptosis and point to a direct action of curcumin on the membrane as initial step in the cytotoxic effect of curcumin on cancer cells.34,35 Such apoptosis-independent effects of curcumin on membrane structure have been confirmed at the level of gross morphological changes in erythrocytes.35 Our experiments show at an atomic-level that curcumin physically alters the membrane, restricting the motion of the lipid acyl chains and inducing negative curvature in the membrane.
Curcumin is protected from aqueous degradation when bound to liposomes, suggesting a mechanism for the enhanced clinical efficiency of liposomal curcumin
Curcumin breaks down readily in aqueous solution to ferulic acid, feruloylmethane, vanillin, and acetone.36,37 The instability of curcumin in solution at physiological pH and poor absorption have been implicated in the low bioavailability of curcumin when administered orally.38 Studies have shown that maximum curcumin plasma levels are only in the range of 1.8–11 nM in patients, even when administered at the high level of several grams of curcumin per day.38 The rapid degradation of curcumin in aqueous solution can be alleviated by the use of polymeric or detergent micelles.19,39,40 It has also been reported that liposome-encapsulated curcumin has greater bioavailability and in vivo efficiency.41–47 Our results suggest that the incorporation of curcumin into liposomes strongly enhances the stability of curcumin and may have a strong impact on the demonstrated greater effectiveness of liposomal curcumin.41–47
Curcumin is anchored to the bilayer by a hydrogen bond near the phosphate group in a manner similar to cholesterol
The headgroup region of the lipid (α, β, and γ carbons) was relatively unaffected by the presence of curcumin, as is expected for a largely hydrophobic, electrically neutral molecule. Curcumin did not have a significant effect on the γ-carbons of the choline group, and only a minor effect was seen on the α-carbon (Fig. 6a). However, a significant increase in the observed dipolar coupling was detected for the neighboring β-carbon. This increase is difficult to rationalize as the dynamics of neighboring atoms should change in similar ways.20 However, a conformational change in the phosphocholine headgroup, specifically a movement of the P-N+ dipole away from the water phase of the membrane induced by the O-H dipole of curcumin, will cause a change in θ in Eq. 1 and alter the observed dipole couplings.20, 48, 49
There are two possible interactions of curcumin with the lipid headgroup that can cause this conformational change. The first possibility is an electrostatic interaction with either the polar methoxy or hydroxyl groups of curcumin with the positively charged choline group. Such an interaction will naturally pull the choline group towards the curcumin molecule and alter the conformation of the lipid headgroup. However, the relatively modest increase in the 14N quadrupolar splitting for the curcumin samples in Fig. 4 shows the tetrahedral symmetry of the choline group is not severely disrupted by the presence of curcumin in the bilayer, indicating curcumin is not in proximity to the choline group.
A hydrogen bond between the curcumin hydroxyl group and either the phosphate group or the nearby carbonyls of DMPC would cause an analogous conformational change in the choline headgoup. Similar increases in the apparent order parameter for the β-carbon upon the addition of anionic amphiphiles as a result of headgroup tilt have been reported in the literature.48, 49 The increase in the 14N quadrupolar splitting upon addition of curcumin shown in Fig. 4 is in accord with this model, as 14N quadrupolar splittings are known to increase upon the addition of anionic amphiphiles as a result of the distortion of the near tetrahedral symmetry of the choline group.48 Because this mechanism does not involve a direct interaction with the choline group of DMPC, the magnitude of the increase will be modest, in agreement with the observed 14N spectra.
Such an interaction has been directly detected for cholesterol, another amphipathic molecule with a terminal hydroxyl group and a rigid conjugated structure. Unlike curcumin, cholesterol is asymmetric with a definite preference for the end of the molecule with the terminal hydroxyl group to be near the water surface.50 Cholestane, another asymmetric steroid similar to cholesterol except for the absence of the terminal hydroxyl group, lacks this orientational preference.50 Due to its structural similarity to cholesterol, the hydroxyl group of curcumin may exert a similar anchoring effect.
Curcumin has an ordering effect on the lipid membrane and binds to DMPC in a transbilayer orientation
Outside of the headgroup region, and particularly in the hydrophobic core of bilayers, the influence of electrostatics is negligible. Therefore, the measured C-H dipolar splittings from 2D PDLF experiments can be directly correlated with changes in the dynamics of the bilayer in the hydrophobic region.48 Curcumin is a highly conjugated and therefore rigid planar molecule with a length approximately equal to one leaflet of the lipid bilayer. The addition of a rigid molecule to a membrane can create either segmental order or disorder in the membrane depending on the mode of binding Segments of the membrane in contact with the curcumin molecule will possess a higher degree of order compared to a pure bilayer because the motion of the lipid molecule is sterically hindered in this region by the curcumin molecule. Conversely, a segment of the membrane that is not in contact with curcumin will be more likely to be disordered as the displacement of the neighboring lipid molecule creates a void in the regions of the membrane where curcumin is not in direct contact. The additional volume created by this void would allow more motion in this area of the membrane, decreasing the observed order parameter. In general, amphipathic molecules that bind at the surface of the membrane will decrease the order parameters of the acyl chains, while amphipathic molecules inserted perpendicular to the surface of the membrane usually increase acyl chain order.
Curcumin does indeed have a strong effect on the dynamics of the lipid bilayer, altering the acyl chain packing and imparting order to most of the lipid molecule. As expected, the ordering effect induced by curcumin is not uniform throughout the lipid molecule but has several interesting features that reveal the site-specific interactions of curcumin with the bilayer at atomic-level resolution. At all concentrations tested, curcumin increased the overall order of the membrane. The ordering effect was concentrated in the glycerol region of the membrane (carbons g1, g2, g3) with the hydrophobic core of the membrane (carbons 2–14) affected to a somewhat lesser degree. Within the hydrophobic core, the effect of curcumin on the membrane was uneven, with the carbons close to the glycerol region affected more than the carbons towards the middle of the membrane. Interestingly, this ordering effect is absent at the terminus of the acyl chain (Fig. 6C). The ordering profile indicates curcumin inserts into the bilayer deeply, most likely penetrating nearly to the end of the DMPC molecule. An exact depth of penetration could not be obtained, as the resonances for carbons 4–11 are not resolved in the PDLF spectra due to the high similarity of the chemical shifts in this area.
The ordering profile is consistent with a transbilayer orientation of curcumin. The length of an individual curcumin model, based on a rigid planar conformation and standard bond lengths and angles, can be estimated to be about 16 ± 2 Å.51, 52 This length is slightly less than half of the phosphate-to-phosphate length of a DMPC bilayer (35.3 Å),53 but considerably less than half of the total thickness of a DMPC bilayer (44.5 Å),54 supporting the conclusion that curcumin is anchored to the bilayer by a hydrogen bond to the phosphate group and not the choline headgroup. In a transbilayer orientation, curcumin could therefore be expected to have an ordering effect to nearly the end of the acyl chains as shown in (Fig. 6). In a surface associated orientation, curcumin would disorder most of the acyl chain region in disagreement with the PDLF data.
The effect of curcumin on the phase transitions in both DMPC and DPPC is also in agreement with significant interactions with the hydrophobic core of the membrane. Curcumin only slightly depresses the temperature of the main gel to liquid crystalline phase transition (Tm) of DMPC and DPPC and has a minimal effect at low concentrations on the enthalpy of the phase transition (Table 1). These characteristics are consistent with a ligand that maintains van der Waal contacts with the acyl chains and does not strongly disrupt the packing of the gel phase.55, 56 The DSC profile is similar to cholesterol at equivalent concentrations, which is known to bind in a transbilayer orientation.57 In DPPC these changes are more significant, reflecting the longer length of the acyl chains and the greater hydrophobic mismatch between curcumin and DPPC.
Several other lines of evidence are also consistent with a transbilayer orientation. The deep penetration of curcumin into the bilayer consistent with a transbilayer orientation is suggested by the significant degree of protection of curcumin from aqueous degradation when bound to the membrane (Fig. S1).19, 40 Curcumin is degraded in aqueous solution by a reaction at the keto-enol group.36, 58 The greatly decreased rate of degradation upon binding to the membrane indicates the keto-enol group of curcumin is sufficiently protected from solvent. In a surface associated state, the keto-enol group can be expected to be located in the glycerol region of the lipid if it is anchored to the membrane at the phosphate group as mentioned above. The glycerol region of the lipid is exposed to solvent, hence it can be expected that curcumin bound to the membrane in a surface-associated state will be subject to rapid degradation. The large blue shift in the fluorescence of micelle-bound curcumin also suggests a strongly hydrophobic binding environment.59, 60
Curcumin in DOPC bilayers can adopt both surface-associated and transbilayer orientations, with the surface-associated state prevalent at lower concentrations (< 3%).17, 18 DOPC bilayers are thicker than DMPC bilayers (a 36.9 Ǻ phosphate to phosphate distance for DOPC61 compared to 35.3 Å for DMPC53) and the unfavorable energetics associated with the hydrophobic mismatch between curcumin and DOPC may stabilize the surface-associated state of curcumin. A comparison of the factors influencing the orientation of curcumin would be interesting for further studies, as the orientation of curcumin within the membrane is likely to have both an effect on its antioxidant activity and on the liposome’s protective effect from curcumin degradation.62, 63
The interaction of curcumin with the membrane impacts its ability to protect the body from harmful free radicals in several ways. First, the exact localization of curcumin in the membrane will affect the availability of curcumin to oxidants and other antioxidants.62 In the transbilayer orientation detected here for curcumin in DMPC bilayers, one phenoxy- group resides at the membrane/water interface while the other phenoxy- group and the enol group is buried within the hydrophobic core of the membrane.
Second, the rate of lipid peroxidation is influenced to a strong degree by the physical state of the membrane in a way that is not completely understood.64 Curcumin is shown here to increase the order parameter of the lipid acyl chain, which is likely to have the effect of decreasing lateral diffusion within the membrane. The decreased rates of diffusion will simultaneously decrease the rate of both the propagation and termination steps of the peroxidation chain reaction.65 While it is unfortunately not possible to definitively predict how rigidification of the membrane by curcumin will impact a given free radical reaction due to the opposing nature of these reactions, it is a factor that must be considered in understanding radical reactions within the membrane.66
Finally, curcumin may counteract the destabilizing influence of peroxyl radicals on the membrane, as has been proposed for vitamin E.67, 68 Acyl chain hydroperoxides formed during free radical reactions are more hydrophilic than unmodified acyl chains and therefore migrate to the surface.65 This motion cannot be easily accommodated in a tightly packed membrane and will disrupt the integrity of the cellular membrane. By rigidifying the membrane, curcumin may counteract this effect.
Curcumin likely forms transbilayer oligomers at higher concentrations
Overall, curcumin induces order in the DMPC bilayer. Surprisingly, the sample with lower concentrations of curcumin (up to 1% curcumin) had a higher effect on the overall order of the membrane (Fig. 6). Increasing the amount of curcumin concentration to 1% led to a significant decrease in the ordering of the glycerol region and the beginning of the acyl chain (carbons 2 and 3, Fig. 6B).
This trend may be explained by the formation of transbilayer oligomers of curcumin. Both the transbilayer and surface associated binding modes have unsatisfied hydrogen bonds that can be satisfied by the formation of oligomeric curcumin complexes.69 The hydrophobic mismatch between the curcumin oligomer and the lengthier DMPC bilayer would cause the bilayer to thin to match the length of the rigid curcumin complex (Fig. 6D). Bilayer thinning would in turn tend to disorder the acyl chain region and counteract the ordering effect induced by curcumin, in agreement with the lower molecular order parameters observed at higher curcumin concentrations. Cholesterol, whose structural similarity to curcumin in some respects has been noted above, also forms transbilayer dimers, although the interaction is not driven by the formation of hydrogen bonds.57, 70, 71
Lateral phase separation to form curcumin-rich and curcumin-poor domains above a critical curcumin concentration would also tend to reduce the ordering effect of curcumin on the membrane, as each curcumin molecule would be in contact with fewer lipids. Domain formation has been seen for the related tetrahydroxycurcumin molecule in DPPC membranes at high (>15%) tetrahydroxycurcumin concentrations.72 However, the phase transition of DMPC is symmetric at the concentrations used in this study (Fig. 2), which is a very strong indication of homogenous mixing of curcumin throughout the bilayer.
Curcumin stabilizes the formation of non-bilayer structures
Curcumin promotes the formation of the inverted hexagonal (HII) phase in DiPOPE (Fig. 3). Phosphotidylethanolamine (PE) is wedge-shaped with a small headgroup compared to most other lipids. The shape of PE lipids imposes a steric restriction on the packing of PE lipids in the flat liquid crystalline (Lα) phase due to the void volume in the headgroup region. The poor packing in the Lα phase manifests itself as a negative (concave) curvature strain, which can be relieved at higher temperatures by rolling the bilayer up into inverted cylinders with the lipid tails facing outward (the inverted hexagonal (HII) phase).73 Ligand binding alters the energetics of the Lα - HII phase transition in a manner largely dependent on steric considerations. Ligands that insert deep into the bilayer increase the effective size of the acyl chain region and therefore increase the negative strain, while binding to the headgroup region has the opposite effect.
Stabilization of the HII phase is indirect but strong evidence that curcumin-induced negative curvature strain on the bilayer.74 At first glance, this result may seem paradoxical in light of curcumin’s known ability to promote gramicidin channel formation. As mentioned above, the formation of gramicidin channel has an energetic penalty associated with deforming the bilayer to accommodate the short gramicidin channels.11 The binding of ligands to the membrane can change the energetics of this process by either altering the thickness of the bilayer, the curvature of the membrane, or the bilayer elasticity.16, 75 This suggests the effects on gramicidin channel function and other mechanosensitive channels with a positive hydrophobic mismatch are dominated by curcumin-induced changes in the bilayer elasticity, as hypothesized by Ingolfsson et al.11, and not by changes in bilayer curvature. A similar finding has been observed for capsaicin,14, 76 which resembles half of the curcumin structure, and polyunsaturated fatty acids.77–79 It should also be noted that small concentrations of cholesterol increase bilayer elasticity,80 contrary to the well-known rigidifying effect seen at higher concentrations.
The promotion of non-lamellar phases can have strong effects on biochemical processes involving lipids. Non-lamellar lipid phases with a high negative curvature are involved in membrane fusion, exocytosis, and membrane blebbing.81, 82 The promotion of negative curvature by curcumin may have a direct effect on apoptosis by increasing the permeabilizing activity of the apoptotic protein tBid.83 This may also explain in part the effect of curcumin analogues, such as dimethoxycurcumin, which exhibit significant pharmaceutical effects in spite of decreased antioxidant activity compared to curcumin.84
CONCLUSIONS
Solid-state NMR on magnetically aligned bicelles shows curcumin alters membrane structure at low concentrations in a manner analogous to cholesterol. Like cholesterol, curcumin binds DMPC in a transbilayer orientation at low concentrations where its rigid molecular structure induces segmental ordering of the lipid membrane. Unlike cholesterol, curcumin is able to form hydrogen bonds at both ends of the molecule. At higher concentrations, curcumin forms transbilayer oligomeric structures in a process most likely driven by hydrogen bond formation between the two phenolic groups. Curcumin also promotes the formation of negatively curved non-lamellar bilayer phases and may therefore have an effect on processes dependent on curved non-lamellar bilayer intermediates such as membrane fusion. The results of this study show the value of solid-state NMR and aligned bicelles for the exact atomic-level resolution of membrane-drug interactions.
Supplementary Material
Acknowledgments
This research was supported by funds from NIH (AI054515 and RR023597 to A.R.). Dong-Kuk Lee was partly supported by a Korea Research Foundation Grant funded (MOEHRD, Basic Research Promotion Funds, KRF-2006-331-CC00153).
Footnotes
SUPPORTING INFORMATION AVAILABLE: Experimental data reporting the kinetics of curcumin degradation in DMPC vesicles at pH 6 and 7.8 and a list of the thermodynamic parameters for the main phase transition of DMPC and DPPC with curcumin incorporated.
References
- 1.Hatcher H, Planalp R, Cho J, Tortia FM, Torti SV. Cell Mol Life Sci. 2008;65:1631–1652. doi: 10.1007/s00018-008-7452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joe B, Vijaykumar M, Lokesh BR. Crit Rev Food Sci Nutr. 2004;44:97–111. doi: 10.1080/10408690490424702. [DOI] [PubMed] [Google Scholar]
- 3.Hafner-Bratkovic I, Gaspersic J, Smid LM, Bresjanac M, Jerala R. J Neurochem. 2008;104:1553–1564. doi: 10.1111/j.1471-4159.2007.05105.x. [DOI] [PubMed] [Google Scholar]
- 4.Ono K, Hasegawa K, Naiki H, Yamada M. J Neurosci Res. 2004;75:742–750. doi: 10.1002/jnr.20025. [DOI] [PubMed] [Google Scholar]
- 5.Reinke AA, Gestwicki JE. Chem Biol Drug Des. 2007;70:206–215. doi: 10.1111/j.1747-0285.2007.00557.x. [DOI] [PubMed] [Google Scholar]
- 6.De Clercq E. Med Res Rev. 2000;20:323–349. doi: 10.1002/1098-1128(200009)20:5<323::aid-med1>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 7.Goel A, Kunnumakkara AB, Aggarwal BB. Biochem Pharmacol. 2008;75:787–809. doi: 10.1016/j.bcp.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 8.Strimpakos AS, Sharma RA. Antioxid Redox Signal. 2008;10:511–545. doi: 10.1089/ars.2007.1769. [DOI] [PubMed] [Google Scholar]
- 9.Jagetia GC, Aggarwal BB. J Clin Immunol. 2007;27:19–35. doi: 10.1007/s10875-006-9066-7. [DOI] [PubMed] [Google Scholar]
- 10.Bilmen JG, Khan SZ, Javed MUH, Michelangeli F. Eur J Biochem. 2001;268:6318–6327. doi: 10.1046/j.0014-2956.2001.02589.x. [DOI] [PubMed] [Google Scholar]
- 11.Ingolfsson HI, Koeppe RE, Andersen OS. Biochemistry. 2007;46:10384–10391. doi: 10.1021/bi701013n. [DOI] [PubMed] [Google Scholar]
- 12.Oezdirekcan S, Nyholm TKM, Raja M, Rijkers DTS, Liskamp RMJ, Killian JA. Biophys J. 2008;94:1315–1325. doi: 10.1529/biophysj.106.101782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adachi S, Nagao T, Ingolfsson HI, Maxfield FR, Andersen OS, Kopelovich L, Weinstein IB. Cancer Res. 2007;67:6493–6501. doi: 10.1158/0008-5472.CAN-07-0411. [DOI] [PubMed] [Google Scholar]
- 14.Lundbaek JA, Birn P, Tape SE, Toombes GES, Sogaard R, Koeppe RE, Gruner SM, Hansen AJ, Andersen OS. Mol Pharmacol. 2005;68:680–689. doi: 10.1124/mol.105.013573. [DOI] [PubMed] [Google Scholar]
- 15.Hwang TC, Koeppe RE, Andersen OS. Biochemistry. 2003;42:13646–13658. doi: 10.1021/bi034887y. [DOI] [PubMed] [Google Scholar]
- 16.Lundbaek JA. J Gen Physiol. 2008;131:421–429. doi: 10.1085/jgp.200709948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hung WC, Chen FY, Lee CC, Sun Y, Lee MT, Huang HW. Biophys J. 2008;94:4331–4338. doi: 10.1529/biophysj.107.126888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y, Lee CC, Hung WC, Chen FY, Lee MT, Huang HW. Biophys J. 2008;95:2318–24. doi: 10.1529/biophysj.108.133736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leung MHA, Colangelo H, Kee TW. Langmuir. 2008;24:5672–5675. doi: 10.1021/la800780w. [DOI] [PubMed] [Google Scholar]
- 20.Dvinskikh SV, Durr UHN, Yamamoto K, Ramamoorthy A. J Am Chem Soc. 2007;129:794–802. doi: 10.1021/ja065536k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dvinskikh S, Durr U, Yamamoto K, Ramamoorthy A. J Am Chem Soc. 2006;128:6326–6327. doi: 10.1021/ja061153a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rand RP, Chapman D, Larsson K. Biophys J. 1975;15:1117–1124. doi: 10.1016/S0006-3495(75)85888-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suurkuusk J, Lentz BR, Barenholz Y, Biltonen RL, Thompson TE. Biochemistry. 1976;15:1393–1401. doi: 10.1021/bi00652a007. [DOI] [PubMed] [Google Scholar]
- 24.Janiak MJ, Small DM, Shipley GG. Biochemistry. 1976;15:4575–4580. doi: 10.1021/bi00666a005. [DOI] [PubMed] [Google Scholar]
- 25.Gardikis K, Hatziantoniou S, Viras K, Wagner M, Demetzos C. Int J Pharm. 2006;318:118–123. doi: 10.1016/j.ijpharm.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 26.Israelachvili JN, Marcelja S, Horn RG. Q Rev Biophys. 1980;13:121–200. doi: 10.1017/s0033583500001645. [DOI] [PubMed] [Google Scholar]
- 27.Cullis PR, Dekruijff B. Biochim Biophys Acta. 1979;559:399–420. doi: 10.1016/0304-4157(79)90012-1. [DOI] [PubMed] [Google Scholar]
- 28.Gruner SM. J Phys Chem. 1989;93:7562–7570. [Google Scholar]
- 29.Andersen OS, Koeppe RE. Annu Rev Biophys Biomol Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 30.Lundbaek JA, Birn P, Girshman J, Hansen AJ, Andersen OS. Biochemistry. 1996;35:3825–3830. doi: 10.1021/bi952250b. [DOI] [PubMed] [Google Scholar]
- 31.Lundbaek JA, Birn P, Hansen AJ, Sogaard R, Nielsen C, Girshman J, Bruno MJ, Tape SE, Egebjerg J, Greathouse DV, Mattice GL, Koeppe RE, Andersen OS. J Gen Physiol. 2004;123:599–621. doi: 10.1085/jgp.200308996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leeson PD, Springthorpe B. Nat Rev Drug Discovery. 2007;6:881–890. doi: 10.1038/nrd2445. [DOI] [PubMed] [Google Scholar]
- 33.Ganesan A. Curr Opin Chem Biol. 2008;12:306–317. doi: 10.1016/j.cbpa.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 34.Jaruga E, Salvioli S, Dobrucki J, Chrul S, Bandorowicz-Pikula J, Sikora E, Franceschi C, Cossarizza A, Bartosz G. FEBS Lett. 1998;433:287–293. doi: 10.1016/s0014-5793(98)00919-3. [DOI] [PubMed] [Google Scholar]
- 35.Jaruga E, Sokal A, Chrul S, Bartosz G. Exp Cell Res. 1998;245:303–312. doi: 10.1006/excr.1998.4225. [DOI] [PubMed] [Google Scholar]
- 36.Tonnesen HH, Karlsen J. Z Lebensm-Unters-Forsch. 1985;180:132–134. doi: 10.1007/BF01027775. [DOI] [PubMed] [Google Scholar]
- 37.Tonnesen HH, Karlsen J. Z Lebensm-Unters-Forsch. 1985;180:402–404. doi: 10.1007/BF01027775. [DOI] [PubMed] [Google Scholar]
- 38.Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB. Mol Pharm. 2007;4:807–818. doi: 10.1021/mp700113r. [DOI] [PubMed] [Google Scholar]
- 39.Ma ZS, Haddadi A, Molavi O, Lavasanifar A, Lai R, Samuel J. J Biomed Mater Res A. 2008;86A:300–310. doi: 10.1002/jbm.a.31584. [DOI] [PubMed] [Google Scholar]
- 40.Tonnesen HH. Pharmazie. 2002;57:820–824. [PubMed] [Google Scholar]
- 41.Wang D, Veena MS, Stevenson K, Tang C, Ho B, Suh JD, Duarte VM, Faull KF, Mehta K, Srivatsan ES, Wang MB. Clin Cancer Res. 2008;14:6228–6236. doi: 10.1158/1078-0432.CCR-07-5177. [DOI] [PubMed] [Google Scholar]
- 42.Mishra VK, Mohammad G, Mishra SK. Dig J Nanomater Biostruct. 2008;3:163–169. [Google Scholar]
- 43.Thangapazham RL, Puri A, Tele S, Blumenthal R, Maheshwari RK. Int J Oncol. 2008;32:1119–1123. [PMC free article] [PubMed] [Google Scholar]
- 44.Li L, Ahmed B, Mehta K, Kurzrock R. Mol Cancer Ther. 2007;6:1276–1282. doi: 10.1158/1535-7163.MCT-06-0556. [DOI] [PubMed] [Google Scholar]
- 45.Li L, Braiteh FS, Kurzrock R. Cancer. 2005;104:1322–1331. doi: 10.1002/cncr.21300. [DOI] [PubMed] [Google Scholar]
- 46.Bruzell EM, Morisbak E, Tonnesen HH. Photochem Photobiol Sci. 2005;4:523–530. doi: 10.1039/b503397g. [DOI] [PubMed] [Google Scholar]
- 47.Tonnesen HH, Kristensen S, Grinberg LN. Int J Pharm. 1994;110:161–167. [Google Scholar]
- 48.Scherer PG, Seelig J. Biochemistry. 1989;28:7720–7728. doi: 10.1021/bi00445a030. [DOI] [PubMed] [Google Scholar]
- 49.Porcelli F, Buck B, Lee DK, Hallock KJ, Ramamoorthy A, Veglia G. J Biol Chem. 2004;279:45815–45823. doi: 10.1074/jbc.M405454200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giordani C, Wakai C, Yoshida K, Okamura E, Matubayasi N, Nakahara M. J Phys Chem B. 2008;112:2622–2628. doi: 10.1021/jp0760713. [DOI] [PubMed] [Google Scholar]
- 51.Tonnesen HH, Karlsen J, Mostad A. Acta Chem Scand B. 1982;36:475–479. [Google Scholar]
- 52.Parimita SP, Ramshankar YV, Suresh S, Row TNG. Acta Crystallogr E. 2007;63:O860–O862. [Google Scholar]
- 53.Kucerka N, Kiselev MA, Balgavy P. Eur Biophys J Biophys Lett. 2004;33:328–334. doi: 10.1007/s00249-003-0349-0. [DOI] [PubMed] [Google Scholar]
- 54.Harroun TA, Heller WT, Weiss TM, Yang L, Huang HW. Biophys J. 1999;76:3176–3185. doi: 10.1016/S0006-3495(99)77469-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McElhaney RN. Biochim Biophys Acta. 1986;864:361–421. doi: 10.1016/0304-4157(86)90004-3. [DOI] [PubMed] [Google Scholar]
- 56.Henzler-Wildman KA, Martinez GV, Brown MF, Ramamoorthy A. Biochemistry. 2004;43:8459–8469. doi: 10.1021/bi036284s. [DOI] [PubMed] [Google Scholar]
- 57.Harris JS, Epps DE, Davio SR, Kezdy FJ. Biochemistry. 1995;34:3851–3857. doi: 10.1021/bi00011a043. [DOI] [PubMed] [Google Scholar]
- 58.Wang YJ, Pan MH, Cheng AL, Lin LI, Ho YS, Hsieh CY, Lin JK. J Pharm Biomed Anal. 1997;15:1867–1876. doi: 10.1016/s0731-7085(96)02024-9. [DOI] [PubMed] [Google Scholar]
- 59.Chignell CF, Bilski P, Reszka KJ, Motten AG, Sik RH, Dahl TA. Photochem Photobiol. 1994;59:295–302. doi: 10.1111/j.1751-1097.1994.tb05037.x. [DOI] [PubMed] [Google Scholar]
- 60.Began G, Sudharshan E, Sankar KU, Rao AGA. J Agric Food Chem. 1999;47:4992–4997. doi: 10.1021/jf9900837. [DOI] [PubMed] [Google Scholar]
- 61.Armen RS, Uitto OD, Feller SE. Biophys J. 1998;75:734–744. doi: 10.1016/S0006-3495(98)77563-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jovanovic SV, Boone CW, Steenken S, Trinoga M, Kaskey RB. J Am Chem Soc. 2001;123:3064–3068. doi: 10.1021/ja003823x. [DOI] [PubMed] [Google Scholar]
- 63.Ohara K, Mizukami W, Tokunaga A, Nagaoka S, Uno H, Mukai K. Bull Chem Soc Jpn. 2005;78:615–621. [Google Scholar]
- 64.Dix TA, Aikens J. Chem Res Toxicol. 1993;6:2–18. doi: 10.1021/tx00031a001. [DOI] [PubMed] [Google Scholar]
- 65.Schnitzer E, Pinchuk I, Lichtenberg D. Eur Biophys J Biophys Lett. 2007;36:499–515. doi: 10.1007/s00249-007-0146-2. [DOI] [PubMed] [Google Scholar]
- 66.Cubillos M, Lissi E, Abuin E. J Chil Chem Soc. 2006;51:825–828. [Google Scholar]
- 67.Atkinson J, Epand RF, Epand RM. Free Radic Biol Med. 2008;44:739–764. doi: 10.1016/j.freeradbiomed.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 68.Wang XY, Quinn PJ. Mol Memb Biol. 2000;17:143–156. doi: 10.1080/09687680010000311. [DOI] [PubMed] [Google Scholar]
- 69.Tonnesen HH, Arrieta AF, Lerner D. Pharmazie. 1995;50:689–693. [Google Scholar]
- 70.Rukmini R, Rawat SS, Biswas SC, Chattopadhyay A. Biophys J. 2001;81:2122–2134. doi: 10.1016/S0006-3495(01)75860-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mukherjee S, Chattopadhyay A. Biochemistry. 1996;35:1311–1322. doi: 10.1021/bi951953q. [DOI] [PubMed] [Google Scholar]
- 72.Gardikis K, Hatziantoniou S, Viras K, Demetzos C. Thermochim Acta. 2006;447:1–4. [Google Scholar]
- 73.Epand RM. Biochim Biophys Acta-Rev Biomembr. 1998;1376:353–368. doi: 10.1016/s0304-4157(98)00015-x. [DOI] [PubMed] [Google Scholar]
- 74.Epand RM, Epand RF. Biophys J. 1994;66:1450–1456. doi: 10.1016/S0006-3495(94)80935-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lundbaek JA. J Phys-Condes Matter. 2006;18:S1305–S1344. doi: 10.1088/0953-8984/18/28/S13. [DOI] [PubMed] [Google Scholar]
- 76.Aranda FJ, Villalain J, Gomezfernandez JC. Biochim Biophys Acta-Biomembr. 1995;1234:225–234. doi: 10.1016/0005-2736(94)00293-x. [DOI] [PubMed] [Google Scholar]
- 77.Boland LM, Drzewiecki MM. Cell Biochem Biophys. 2008;52:59–84. doi: 10.1007/s12013-008-9027-2. [DOI] [PubMed] [Google Scholar]
- 78.Bruno MJ, Koeppe RE, Andersen OS. Proc Natl Acad Sci U S A. 2007;104:9638–9643. doi: 10.1073/pnas.0701015104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sogaard R, Werge TM, Bertelsen C, Lundbye C, Madsen KL, Nielsen CH, Lundbaek JA. Biochemistry. 2006;45:13118–13129. doi: 10.1021/bi060734+. [DOI] [PubMed] [Google Scholar]
- 80.Lemmich J, Mortensen K, Ipsen JH, Honger T, Bauer R, Mouritsen OG. Eur Biophys J. 1997;25:293–304. doi: 10.1007/s002490050041. [DOI] [PubMed] [Google Scholar]
- 81.Bezrukov SM. Curr Opin Colloid Interface Sci. 2000;5:237–243. [Google Scholar]
- 82.Cevc G, Richardsen H. Adv Drug Deliv Rev. 1999;38:207–232. doi: 10.1016/s0169-409x(99)00030-7. [DOI] [PubMed] [Google Scholar]
- 83.Epand RF, Martinou JC, Fornallaz-Mulhauser M, Hughes DW, Epand RM. J Biol Chem. 2002;277:32632–32639. doi: 10.1074/jbc.M202396200. [DOI] [PubMed] [Google Scholar]
- 84.Priyadarsini KI, Maity DK, Naik GH, Kumar MS, Unnikrishnan MK, Satav JG, Mohan H. Free Radic Biol Med. 2003;35:475–484. doi: 10.1016/s0891-5849(03)00325-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.