Abstract
Gain of function mutations in the androgen receptor (AR) are found in prostate cancer and are implicated in the failure of hormone therapy. Most studies have emphasized the ligand binding domain (LBD) where mutations can create promiscuous receptors, but mutations in the N-terminal transactivation domain (NTD) have also been found. To assess AR alteration as a mechanism of treatment resistance, a mouse model (h/mAR-TRAMP) was used in which the murine AR coding region is replaced by human sequence and prostate cancer initiated by a transgenic oncogene. Mice received either no treatment, androgen-depletion by castration or treatment with antiandrogens, and twenty AR transcripts were sequenced per end-stage tumor. All tumors expressed several mutant alleles, although most mutations were low frequency. Some mutations that occurred multiple times within the population were differentially located dependent on treatment. Mutations in castrated or antiandrogen-treated mice were widely dispersed but with a prominent cluster in the LBD (amino acids 736–771), while changes in intact mice centered near the N-terminal polymorphic glutamine tract. Functional characterization of selected LBD mutant alleles showed diverse effects on AR activity, with about half of the mutations reducing transactivation in vitro. One receptor, AR-R753Q, behaved in a cell- and promoter-dependent manner, even though as a germline mutation it causes androgen insensitivity syndrome. This suggests that alleles that are loss of function during development may still activate a subset of AR targets to become gain of function in tumorigenesis. Mutant ARs may thus utilize multiple mechanisms to evade cancer treatment.
Keywords: androgen receptor, prostate cancer, mouse models, therapy resistance, mutation
Introduction
Somatic mutations are a hallmark of cancer initiation, progression and metastatic disease. During tumorigenesis, genomic instability leads to a mutator phenotype in which it is estimated that each cancer cell may harbor as many as 1,000 mutations (1). While most mutations likely are “passengers” with little effect on selection, some may be “drivers” that provide a growth advantage (2). In the case of prostate cancer, numerous mutations in the androgen receptor (AR) have been identified. Intriguingly, many of them are gain of function (3), but the extent to which they influence disease progression is debated.
Prostate cancer is initially androgen-dependent and responds to treatments that inhibit androgen synthesis and/or antagonize AR action. However, tumors ultimately recur and AR remains not only present but active despite hormone therapy (4). Numerous mechanisms have been proposed for AR activation following androgen depletion, including Ar gene amplification, AR activation by growth factors or altered levels of coregulators. Mutations within the receptor itself have also been found that allow androgen-independent activation, increase sensitivity to low androgen, and alter ligand specificity (5).
A paradigm of how gain of function mutations may allow treatment evasion is AR-T877A, found in the LNCaP cell line and some advanced prostate cancers (6). For wild type AR, hormone binding alters conformation of the ligand binding domain (LBD) to create a coactivator interaction surface, activation function 2 (AF2), which is not formed with bound antiandrogen. The subtle shift in structure due to T877A, proximal to AF2, permits an active conformation with various noncanonical ligands, including the antiandrogen hydroxyflutamide (7). This and other mutations that allow promiscuous AR activation may underlie the phenomenon of flutamide withdrawal syndrome in which tumors regress after antiandrogen treatment is stopped (8). Because ligand is key in AR transactivation, the search for AR mutations in prostate cancer has focused on the LBD (3). However, the DNA binding domain (DBD) and the large N-terminal transactivation domain (NTD) also influence hormonedependent function, and as importantly, may enter into ligand-independent activation (9). Unfortunately, the NTD is often ignored since its high GC content and polymorphic repeats impede sequencing.
The prevalence of AR mutations in prostate cancer and their possible association with specific treatments has been difficult to evaluate because of human genetic heterogeneity, disparate patient treatment, lack of biological samples and the small sizes of most clinical studies. Mouse models circumvent these issues, providing an opportunity to study somatic mutations in cancer. The transgenic adenocarcinoma of mouse prostate (TRAMP) model (10) has been used to compare AR mutations in tumors from intact (untreated) versus castrated (androgen-ablated) mice. Although a small study, location of AR missense mutations in primary tumors varied with hormonal status, with seven of nine mutations from castrated mice occurring in the NTD and all mutations from intact mice confined to the LBD (11). One of the mutants identified, AR-E231G, is sufficient to cause cancer when expressed as a prostate-specific transgene, highlighting the oncogenic potential of AR mutations (12).
To examine whether somatic mutations in prostate cancer correlate with treatment in the context of human AR, we utilized a knock-in mouse in which hAr exon 1 was swapped into the mouse locus to create an AR nearly identical to human (13), bred to TRAMP to initiate prostate cancer. Our objective was two fold: 1) to determine whether somatic mutations in the humanized AR (h/mAR) are random or are selected by treatment and 2) to examine whether different treatments select for distinct mutation clusters in functional AR domains. Sequencing the entire coding region from tumors of mice in varied treatment groups confirmed by an unbiased approach that mutation of AR was common overall but few events achieved high frequency in the tumor cell population. Nevertheless, some mutations reflected treatment and some had context-dependent functions, further delineating the role of AR in cancer progression.
Results
Treatment Affects Tumor Progression in h/mAR-TRAMP Mice
To examine whether somatic mutations correlate with treatment in the context of human AR, the h/mAR model was used, in which homologous recombination replaced the mouse Ar exon 1 with the corresponding human sequence (13). The murine NTD differs from human by 15% in amino acid sequence and lacks the polymorphic glutamine (Q) and glycine (G) tracts that affect AR activity and are implicated in prostate cancer progression (14, 15). Exchanging the NTD creates a hybrid gene within the mouse genomic locus that is 97% identical in coding sequence to hAr. These humanized AR mice were crossed to TRAMP mice that express a prostate-specific SV40 T-antigen oncogenic transgene (10, 16), allowing examination of mutations in human Ar that arise in prostate cancer in genetically homogeneous mice.
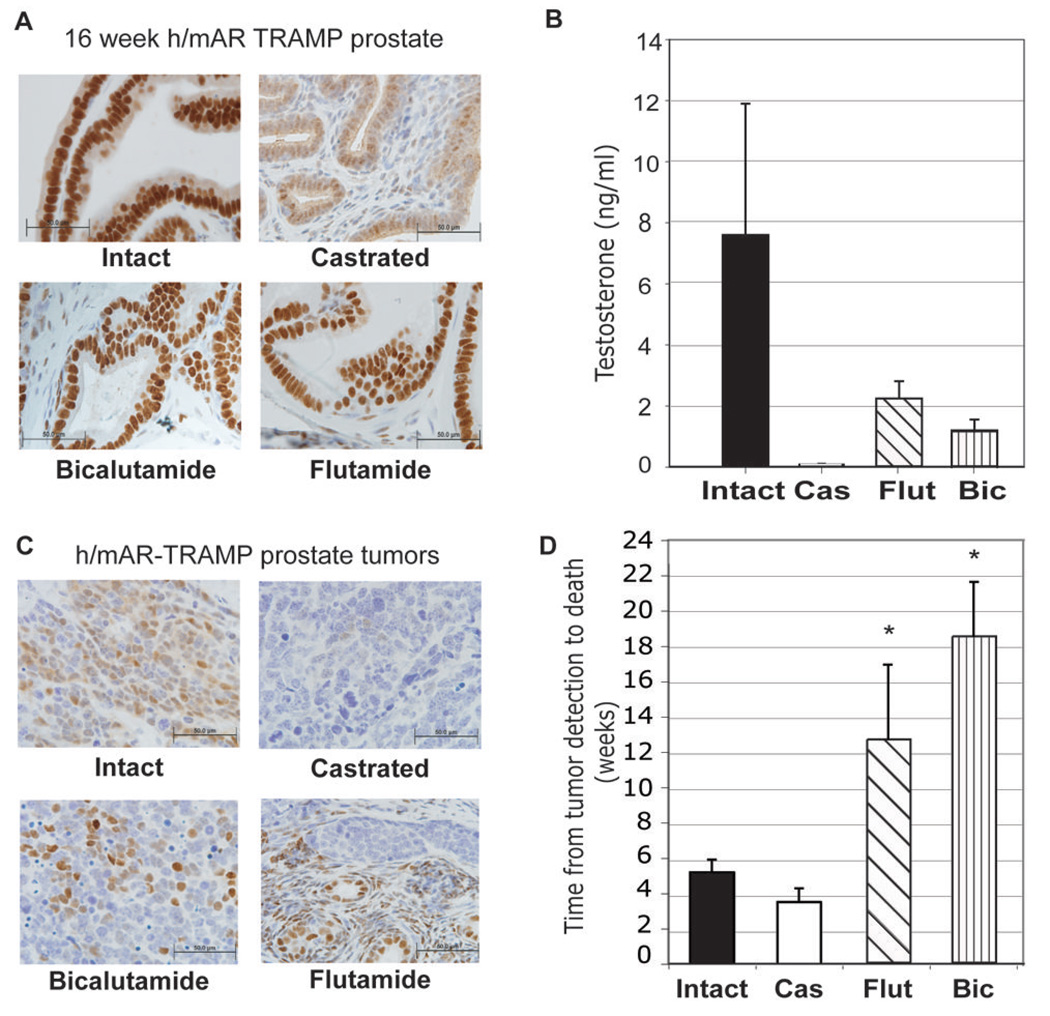
To ensure that the h/mAR-TRAMP mice responded to hormone ablation similarly to wild type mice, short-term effects of androgen blockade were assessed in 12-week-old males treated for 4 weeks by castration or with antiandrogens (flutamide or bicalutamide). Prostatic AR protein detected by immunohistochemistry (IHC) showed nuclear localization in intact mice, while in castrated mice most AR was cytoplasmic indicative of the unliganded state (Fig. 1A). In antiandrogen-treated mice, most AR was nuclear since these antagonists permit nuclear transit and DNA binding but not target gene activation (17). Both antiandrogens reduced serum testosterone (T) to less than 25% of intact levels (Fig. 1B) unlike in men where androgen synthesis inhibitors are necessary to reduce hormone levels. This likely reflects differences in the hypothalamo-pituitary-gonadal axis and adrenal androgen synthesis in mice and men.
Fig. 1. Effects of treatment on the androgen axis in h/mAR-TRAMP mice.
A. Immunohistochemical localization of AR in h/mAR-TRAMP prostates 4 weeks after castration or treatment with the antiandrogens flutamide or bicalutamide. Intact prostate is shown as a control. Bar indicates 50 µm. B. The effect of 4 weeks castration or anti-androgen treatment on serum testosterone levels. C. Localization of AR in representative endstage h/mAR-TRAMP prostate tumors from each treatment group. D. Treatment alters disease length in h/mAR-TRAMP mice. Average length of disease from detection of a palpable tumor to death is plotted for each treatment group. Error bars are the standard error of the mean. * significant difference in length of disease compared to intact by Bonferoni multiple comparison adjustment p<0.01
In end-stage primary tumors from intact, castrate, or antiandrogen-treated mice AR localization was similar to that of short-term treatment, being nuclear in intact and antiandrogentreated mice, but scant in castrates (Fig. 1C). However, AR staining was heterogenous by endstage, in all treatment groups. The time from tumor detection by palpation to death revealed differences in survival with disease in treated compared to intact mice (Fig. 1D). In TRAMP mice, castrates survive only a short time once tumors are detected, suggesting that in this model early reduction in androgen synthesis encourages growth of aggressive androgen-independent tumors (18). Bicalutamide or flutamide treatment after tumor detection significantly extended survival with disease (flutamide versus control, P < 0.008, bicalutamide versus control, P < 0.0001 using a Bonferroni multiple comparisons adjustment).
Analysis of Mutation Frequency in h/mAR-TRAMP Tumors
To detect mutations in Ar mRNA transcripts that may be present in only a subset of tumor cells, a reverse transcription (RT)/amplification/sub-cloning strategy was used (Fig. 2). Sequencing the equivalent of 20 Ar mRNAs per primary tumor (10 from each of two independent (RT) reactions, 760 clones total) identified 994 changes from the reference sequence. 808 of these putative mutations were single base changes. Variation in Q or G tract codon numbers was common and was analyzed separately. Based on the total number of nucleotides sequenced, prostate tumors had an average of 4.0 changes/10,000 bp of Ar coding region. To obtain a baseline mutation rate for the methodology, Ar was sequenced for 17 of these mice from testes where AR but not the T-antigen oncogene is expressed. 33 base substitutions were identified from 230 clones, or about 2.2 changes/10,000 bp. This likely represents the combined error of RT (1/15,000 bp per the manufacturer), the polymerase (1.58/100,000 per the manufacturer) and subcloning and sequencing errors. This error rate may be an overestimate since many of the testis clones included the error-prone Q-tract region (see below). Nevertheless, tumor samples had twice as many sequence changes as non-tumor samples. Of these putative mutations, 54.1% were missense, 8% base deletions, 4% codon deletions, 1.6% base insertions, 4.7% nonsense and 27.4% silent mutations (Table 1). The distribution of mutation types across treatment groups did not differ significantly (Table 1).
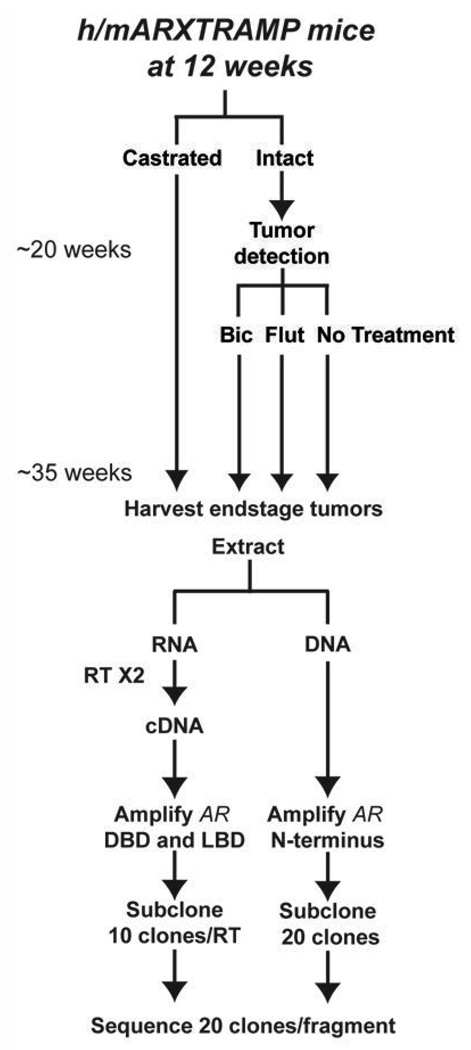
Fig. 2. Treatment Schematic.
h/mAR-TRAMP mice were randomly assigned to treatment groups (9–10 mice/group). At 12 weeks of age, mice were either castrated or left intact. Upon tumor detection by palpation, intact mice either received no treatment or were treated with the antiandrogens bicalutamide (Bic) at 50 µg/g or flutamide (Flut) at 25 µg/g. Endstage tumors were harvested and RNA and DNA extracted. AR was amplified in 4 fragments from cDNA (DBD and LBD) or DNA (NTD). Products were subcloned and 20 clones/ fragment sequenced (10 clones from 2 separate RT reactions for RNA).
Table 1.
Distribution of mutation types within h/mAR-TRAMP treatment groups.
| Treatment Groups | |||||
|---|---|---|---|---|---|
| Intact | Castrated | Flutamide | Bicalutamide | Testis Control | |
| Mutations/10,000 bp | 3.6 | 3.3 | 4.4 | 4.0 | 2.2 |
| Missense (%) | 55.0% | 46.6% | 57.2% | 47.3% | 61.0% |
| Silent (%) | 26.8% | 24.1% | 24.0% | 31.1% | 31.0% |
| Nonsense (%) | 8.2% | 5.1% | 5.3% | 5.8% | 3.0% |
| Codon deletion (%) | 2.3% | 2.8% | 3.2% | 6.6% | 0 |
| Basepair deletion (%) | 7.7% | 7.9% | 10.2% | 7.9% | 6.0% |
| Basepair insertion (%) | 0 | 0 | 0 | 0.8% | 3.0% |
Different cancer types often display a unique mutation spectra favoring one type of basepair change over another (2). Analysis of the h/mAR-TRAMP prostate tumor spectrum revealed a preponderance of mutations at C:G sites that was not seen in the testis. Prostate cancer mutations showed high C:G to T:A transitions (41.0%) and C:G to A:T transversions (21.3%) but low G:C to C:G transversions (1.9%) (data not shown), unlike the reported breast cancer spectrum which has frequent G:C to C:G transversions (2).
The majority of mutations were identified in one or two clones per tumor. Consistent with the previous report on TRAMP mice, this represents a mutation frequency of 5–10% of the cell population (11). However, considering that samples often contain malignant and normal cells and that prostate cancer is multifocal, mutation frequency within the tumor may be higher. As evidence, 24 mutations were present in multiple clones from a single tumor, with four (Q58L, R102X, S324G, A385T) occurring in four clones (20%) each from an independent tumor.
Recurring Mutations in h/mAR-TRAMP tumors
To further exclude possible random errors, only codons mutated at least twice were further analyzed. Missense mutations in multiple clones of a single tumor (23 codons) or in multiple tumors (109 codons) occurred at an overall rate of 0.53/10,000 bp. Fig. 3A depicts a Venn diagram of mutations that recurred within or between treatment groups. Only four mutations occurred in all groups (Q58L, G228S, R544G and A764S) three of which were in the NTD. Tumors from flutamide-treated mice bore the most unique mutations (11) whereas those treated with bicalutamide shared most mutations with the flutamide-treated group.
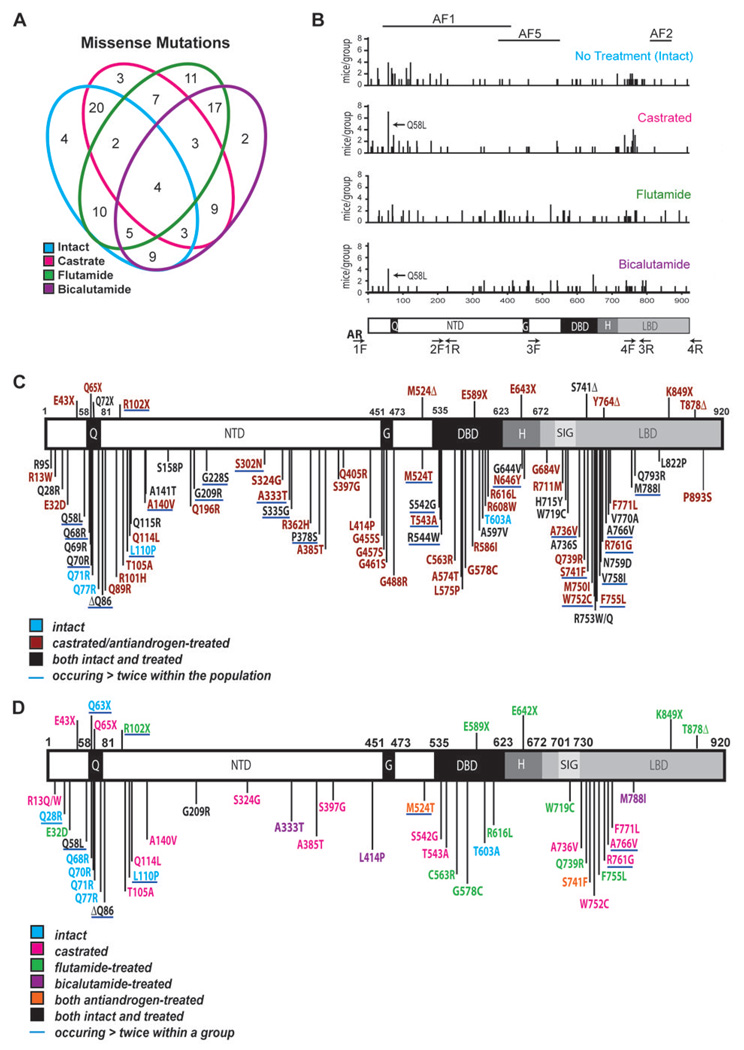
Fig. 3. Analysis of AR mutations from h/mAR-TRAMP tumors.
A. Venn diagram of AR missense mutations recurring in h/mAR-TRAMP tumors grouped by treatment. Numbers represent the mutations shared by overlapping treatment groups. Analysis included only mutations occurring more than once within the entire population. This included all substitutions at the same residue (e.g. R31C and R31H). B. The number of mice per treatment group with missense substitutions in the same residue (Y-axis) positioned along the AR protein (amino acids 1–920; X-axis). The position of the activation functions (AF) 1, 2, and 5 involved in cofactor recruitment are shown above. Arrows indicate Q58L, the most common mutation. AR domains and repeats are shown below. Q: Polyglutamine tract, NTD: N-terminal domain, G: Polyglycine tract, DBD: DNA binding domain, H: Hinge region, LBD: Ligand binding domain. Primer positions are indicated by arrows. Primer 1F is located in the 5’ UTR, 4R in the 3’ UTR and 2R (not shown) in intron 1. Primer pair 2 amplifies the C-terminal half of the NTD including the glycine repeat from genomic DNA. Primer pairs 3 and 4 cross exon/intron boundaries and are used to amplify from cDNA. C, D. Schematics of the amino acid substitutions identified in h/mAR-TRAMP tumors and their relative location within the AR protein domains. C. Amino acid substitutions occurring in more than one mouse within the population or in more than one clone in a single tumor. Mutations underlined in blue occurred in three or more tumors. D. Amino acid substitutions identified in more than one tumor within a treatment group or in more than one clone in a single tumor. Underlined mutations occurred in three or more tumors within a treatment group. Δ: A base deletion within the codon of the specified amino acid causing a frameshift and subsequent premature stop codon.
Missense mutations mapped throughout AR (Fig. 3B) but many clustered within regions mutated in prostate cancer patients, such as a proximal region of the LBD (residues 700–800) (19). Mutations around the Q-tract were found in all groups but most commonly in intact mice. Mutations in three activation function (AF) domains reflected treatment, with the antiandrogen groups carrying many more in the distal NTD region of AF5, while the intact and castrated mice had more mutations in AF1. Overall there were few mutations in AF2, with the least in the castrated group, as might be expected for tumorigenesis under androgen-depleted conditions.
Recurring mutations subdivide into two sets: substitutions at the same codon to different amino acids (e.g., R13Q, R13W), which might represent selection against wild type, and substitutions in one codon to the same residue (e.g., 2 tumors with L110P), perhaps indicating selection for a specific change. 70 codons were mutated to different residues, and 84 codons to the same amino acid (Fig. 3C). Remarkably, only one mutation to different amino acids recurred in testis and none to the same amino acid, suggesting this conservative analysis excluded most of the base changes due to methodology. Each tumor averaged 5 recurring mutations to the same amino acid, with most in amino acids conserved between mouse and man. In the NTD, only 13 mutations recurred in nonconserved residues, of which 10 occurred in or near the Q or G tracts.
Although silent mutations are less likely to alter protein function, they can affect splicing, mRNA stability, or protein folding due to altered codon usage (20). 43 codons carried synonymous changes that recurred within the AR mRNA population, and were distributed throughout the coding region with no apparent differences between treatment groups. Intriguingly, both proline and serine silent mutations were over represented, accounting for 17 codons with synonymous replacements. While the majority of silent mutations went from a more abundant to a less abundant codon, many proline and serine mutations altered rare codons to more frequent ones (Supplemental Tables 1 and 2). Remarkably, silent proline mutations were confined to the N-terminal 172 amino acids, in which 23 prolines occur (with 8 of these codons altered), even though there are 38 subsequent prolines including 8 in a polyproline tract. This hints at an impact of codon usage on AR folding; while silent proline mutations in the proximal portion of the disorganized NTD may be tolerated, subsequent proline codons may be more tightly regulated. Of the synonymous serine mutations, one at S516 from a less to a more abundant codon occurred in two flutamide and three bicalutamide-treated mice. Interestingly, this is a major site of phosphorylation in EGF-stimulated prostate cancer growth (21).
Mutations in or near the NTD polyamino acid tracts
Contraction of the Q tract occurs clinically (22) and expansion and contraction of both Q and G tracts by one or two codons was common in the mouse tumors. While all testis clones, and 96% of those from prostate, had 21 or 22Qs, 2.1% of the tumor clones had 19–20 Qs and 2.3% had 22–31 Qs. The G tract also varied – most clones (84.3%) had the original 23 Gs, but 12.4% lost one and 3.3% lost 2 to 6 Gs. Although there were no large alterations in Q tract length, Q to R substitutions within the tract were common in all groups, occurring in 23 clones from 12 tumors, with none in testis. One such mutation, Q65R, was reported in a clinical sample (19). While a functional significance of disrupting the Q tract with arginine is unknown, disruption by leucine elevates transcriptional activation despite limiting N/C interaction (23).
A stretch of six Qs (amino acids 86–91) of unknown function just beyond the major Qtract showed contraction to five or four Qs in 22 mice from all groups (ΔQ86), but not in testis. The most common mutation in this study, Q58L, occurred next to 4 leucines just before the Q tract in which mutation of the first Q codon expands the L tract from 4 to 5 residues. This was found in 16 mice representing all groups, and was in two or more clones in 4 of these mice, as well as in one clone from a testis control. Interestingly, this tract has expanded from one L in mouse and dog, to three in chimpanzee, and four in man. Whether or not this expansion affects AR activity is unknown.
AR mutations that may affect AR function
Previously identified as well as novel mutations within functionally significant regions were found in multiple tumors. In the NTD, few of the recurring mutations had been identified previously since few studies have systematically sequenced this region. However, Q114L, found in two castrated mice (one primary tumor and one lymph node metastasis) was noted in a primary prostate tumor (24). This same study also reported L575P, which we found in one bicalutamide- and one flutamide-treated mouse. A novel NTD mutation, M524T, is located in AF5, an area that interacts with p160 coactivators and is involved in ligand independent activation (9). M524T occurred in 3 flutamide- and 2 bicalutamide-treated mice but not in intact or castrated mice. Four of 7 mutations previously identified in patients with flutamide withdrawal symptoms were located in this area, between residues 502 to 535 (24).
Within the LBD, three novel mutations, R711M, H715Y and W719C, were identified within the most highly conserved segment, the signature sequence (Fig. 3C), where other mutations alter ligand specificity (25–27). All three mutations lie on the same surface of helix three, adjacent to AF2, where they may influence protein-protein interactions. Of the remaining LBD mutations, 10 clustered between amino acids 736–771 (Fig. 3C) where they might affect ligand specificity. 8 of these were common, occurring in 3 or more mouse tumors (Fig. 3C). M750I, F755L and V758I, were identified previously in patients (19). In particular, M750I was identified in disease recurring after orchiectomy and bicalutamide treatment (28). Here, M750I was found in two mice, one treated with flutamide and one with bicalutamide.
Three novel mutations in this cluster were of particular interest based on their presence in treatment groups or their location. S741F lies adjacent to W742 where a mutation allows bicalutamide, but not flutamide, to act as an agonist (29). This mutant was identified only in antiandrogen-treated mice, in two each per treatment. W752C might expand the base of the binding pocket to accommodate other ligands, and was found in two clones from a castrated mouse, and one each from a flutamide- and a bicalutamide-treated mouse. R753 contacts androgen and its mutation to Q previously was shown to cause complete androgen insensitivity (CAIS) in man and rat (19), but had not been noted in prostate cancer. R761G was unique to the castrated group, in two primary tumors and a metastasis.
Some AR mutations are treatment group-specific
Substitutions that occurred more than once within a group, either in more than one mouse of a group (29 mutations) or multiple times in a single tumor (25 mutations), revealed treatment effects (Fig. 3D). 14 of these mutations also occurred once in other groups. Few substitutions overlapped between groups, with only Q58L and ΔQ86 ocurring in all groups and only G209R ocurring in both castrated and intact mice. Nine of 11 substitutions recurring in the intact group clustered around the Q tract, with 4 Q to R substitutions within the tract itself. Most recurring mutations in the castrated group (13 of 20) were in the NTD, as noted previously (11), and six occurred more than once within a tumor. Two mutations were found proximal to the first zinc finger of the DBD (S542G and T543A) and 5 clustered in the LBD between residues 736 – 771.
The two antiandrogen treatment groups showed distinct mutation patterns, perhaps due to differences in structure of the ligands. There were many more recurring mutations with flutamide than with bicalutamide, and most (10 of 16) were in the DBD and LBD. One of three substitutions in the DBD, C563R, mutated a zinc-chelating cysteine in the first zinc finger and is likely to be a loss of function. Another, G578C, altered a conserved base in the P-box important for DNA binding specificity (30), and the last, R616L, contacts the phosphate backbone (31). Four missense mutations were located in the LBD where they could affect ligand-dependent activation (see below). The flutamide group also had many more nonsense mutations (5 out of 16) than other groups. The bicalutamide-treated group had few recurring mutations: 3 were unique and 2 overlapped with only the flutamide-treated group.
Transcriptional Activity of Select AR Mutants
Mutations clustered in the proximal portion of the LBD included novel as well as previously reported but uncharacterized mutations. For functional analysis, five LBD mutations (S741F, M750I, W752C, R753Q and R761G), and M524T that was specific to the antiandrogen groups, were introduced into an AR expression vector. Wild type and mutant residues in the LBD cluster are represented in Fig. 4A. All mutant ARs expressed at levels equivalent to wild type when transfected into CV-1 cells (Fig. 4B). M524T migrated more slowly, possibly due to differential protein modification. Treatment with phosphatase did not abolish the M524T size difference indicating it is likely not due to altered phosphorylation (data not shown).
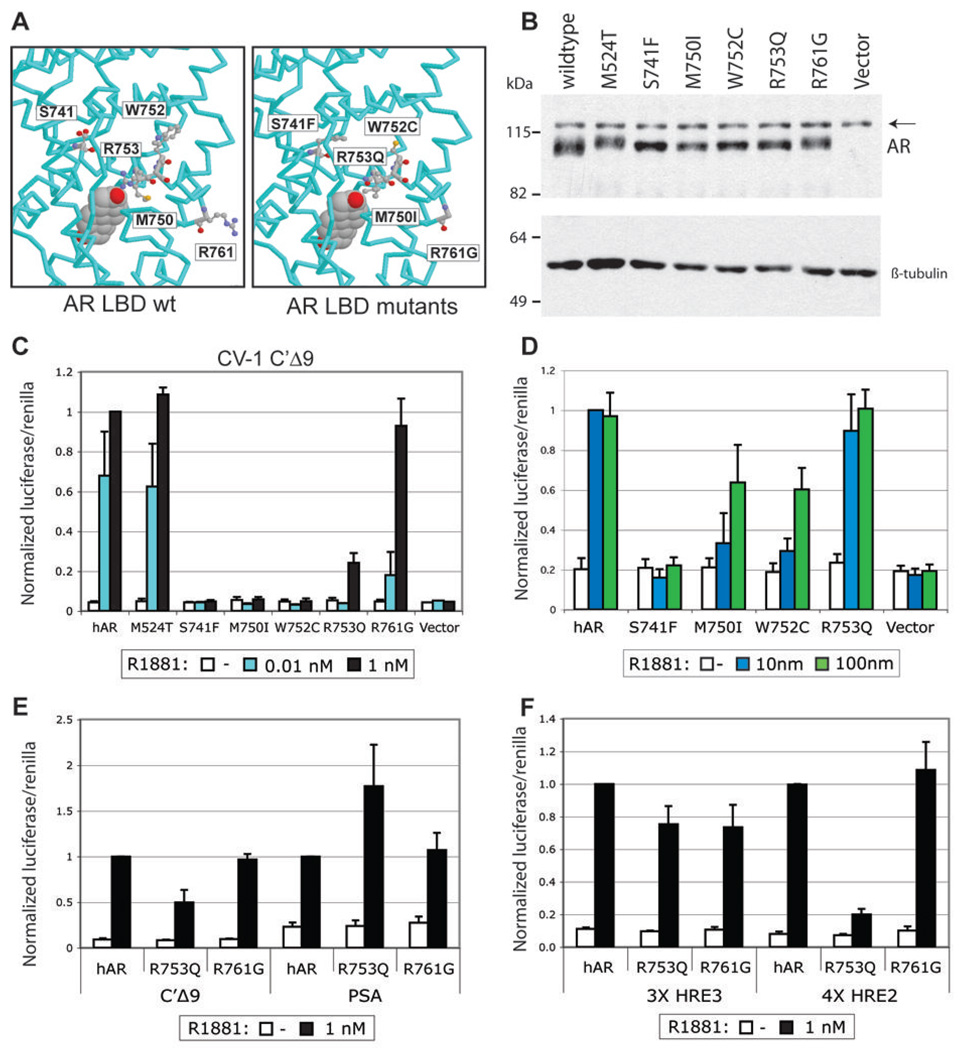
Fig. 4. Functional characterization of mutant ARs from mouse prostate tumors.
A. Protein structure model of the AR LBD representating the five residues (left) and the h/mAR- TRAMP mutations (right) introduced into expression vectors. B. Western blot of 20 µg of whole cell protein lysates from CV-1 cells transfected with 100 ng expression vector containing wild type or mutant AR and treated for 24 hours with 1 nM R1881. Arrow - nonspecific band; β-tubulin was used as a loading control. C. Transactivation of wild type and mutant receptors (4 ng) transfected into CV-1 cells along with 400 ng of the C’ Δ9 luciferase reporter and 100 ng of promotorless renilla. Cells were harvested 24 hours after treatment with the indicated concentration of R1881 and assayed for firefly and renilla luciferase activity. The average normalized values of at least three trials are represented as the fraction of wild type activity at 1 nM R1881. Error bars are the standard error of the mean (SEM). D. Transactivation of wild type and mutant ARs transfected into PC-3 cells with the PSA-luciferase reporter. Cells were harvested as in C. after treatment with 10 or 100 nM R1881. Average normalized values of at least three trials are shown +/− SEM. Values are the fraction of wild type activity at 10 nM R1881. E, F. Transactivation of R753Q in PC-3 cells is promoter-dependent. PC-3 cells were transfected with wild type AR, R753Q or R761G, and reporters with complex enhancers (E) or with tandem repeats of single HREs (F). Normalized values are as in C.
Mutant transactivation was tested in CV-1 cells on C’ Δ9-luc, a luciferase reporter driven by the AR-specific enhancer of mouse Slp(32). Three types of response to the synthetic androgen R1881 were exhibited: M524T was as active as wild type, R753Q and R761G were less responsive at lower ligand concentrations, and S741F, M750I and W752C were inactive even at 1 nM R1881 (Fig. 4C). None of the mutants responded to either hydroxyflutamide or bicalutamide at doses of hydroxyflutamide that strongly activate AR-T877A (data not shown).
To compare transactivation in prostate cancer cells, mutants were transfected into ARnegative PC-3 cells along with C’ Δ9-luc or PSAe1p-luc, containing the prostate specific antigen (PSA) upstream enhancer and proximal promoter (33). M524T and R761G had wild type activity with 1 nM R1881 for both reporters (data not shown). S741F, M750I, and W752C that were inactive in CV-1 cells were also weak in PC-3 cells. However, increasing R1881 concentration to 100 nM rescued receptor activity for all but S741F (Fig. 4D). Interestingly, R753Q, which had very low activity in CV-1 cells, was more potent in PC-3 cells, with half the activity of wild type AR on C’ Δ9-luc and as much activity as wild type on PSAe1p-luc (Fig. 4E). The differential response of R753Q could reflect promoter-specific differences in AR response elements or interactions with other factors.
Androgen response elements are either canonical inverted repeats of a TGTTCT half-site that can bind multiple steroid receptors or direct repeats of the half-site that selectively bind AR but confer weak activation (32). Both PSA and Slp enhancers consist of a complex mix of canonical and selective elements, as well as binding sites for other factors. To determine whether the differential activity of AR-R753Q depended on the response element, activity was tested on either canonical (HRE3) or AR-selective (HRE2) elements from the Slp enhancer. While R753Q was able to transactivate the HRE3 reporter at wild type levels, it showed little activity on the AR-selective HRE2 (Fig. 4F). This is intriguing since the equivalent mutation in rat, R734Q, is the germline mutation accounting for CAIS in the tfm rat (34). Characterization of this subset of mutants suggests that some mutations have subtle, but potentially important, affects on AR function that can be cell and promoter context dependent. In particular, loss of function mutations such as those identified in AIS may, in the context of prostate cancer, be gains of function by differentially activating a subset of AR targets.
Discussion
This study queried somatic hAr mutations expressed in h/mAR-TRAMP mouse prostate cancer for evidence of selection due to treatment. Numerous alterations were present at a low frequency, in part reflecting increased mutation during oncogenesis (1). Based on an error rate estimated from testis cDNA, about half of the AR changes in tumors are putative somatic mutations arising during disease. Restricting analysis to recurring mutations highlighted those with a possible selective advantage as evidenced by their variation in position and frequency with treatment.
The number of AR mutations identified was higher than in previous studies, although most mutations were only present in one or two clones of the twenty sequenced per tumor. While accumulation of rare mutations may be a general characteristic of tumor genomes, mutations in AR are more phenotypically evident because the gene is monoallelic. Many of these mutant alleles may confer similar growth advantages, resulting in heterogeneous cell subpopulations within a tumor. Multiple mutations in a single tumor have been reported in hormone-refractory prostate cancer (35). In one case, two mutations were identified in a single transcript and both influenced AR function (36). Because cDNAs in this study were amplified in fragments, it is not possible to determine whether mutations in different segments exist in a single transcript.
Among studies, differences in the number of mutations detected likely reflect the methodology, disease stage, and experimental model. By amplifying and subcloning Ar cDNA, sequence changes are evident in as few as 5% of the transcripts, while genomic DNA, examined in most clinical studies, can only reveal mutations present in a majority of cells. Analyzing the entire Ar coding region showed that about half of the mutations occur in the usually ignored NTD. The prior TRAMP study examined earlier stage tumors and identified fewer mutations per mouse but all were also at low frequency, suggesting that mutation abundance may be influenced by the model (11). The survival benefit of individual mutations may be modest relative to the strong proliferative drive conferred by T-antigen. We have gone on to analyze AR mutations from human prostate cancer metastases using this same approach and have found a similar mutation rate in the AR1. However, more AR mutations from the patient metastases were found in multiple clones per sample, suggesting that end stage primary tumors from TRAMP mice may be more heterogeneous than patient metastases that are more clonal in nature.
Clustering of recurring AR mutations in distinct functional domains highlights the influence of therapy. Mutations are least frequent in the intact mice, in accord with clinical studies that find fewer AR mutations in untreated or androgen-responsive tumors (37, 38). In the previous TRAMP study (11), most mutations in intact mice occurred in the AR C-terminus, in contrast to their NTD location here. This could reflect AR species differences, particularly since mutations in intact h/mAR tumors centered around the Q-tract that is poorly conserved in mice. On the other hand, mutations after castration are found primarily in the NTD similar to the earlier study (11). This may be because all NTD mutations in the castrate group occurred in residues conserved between mouse and man, suggesting a role for these residues in androgenindependent activation.
Mutations in antiandrogen-treated mice overlap little with those in the castrate group, indicating that early depletion of androgen leads to tumor development distinct from that of antagonist treatment. Two mutations, M524T and S741F, occured in multiple mice for both flutamide and bicalutamide treatments, and thus may affect interactions particular to antiandrogen-bound AR. Since M524T had altered electrophoretic mobility, post-translational modification may influence differential AR activity. Numerous protein modification sites are nearby, including ones for Erk2 phosphorylation at S516, RACK1 phosphorylation at Y535, and ligand-dependent SUMOylation at K520 (39–41). The silent mutation at S516 occurring in both flutamide and bicalutamide-treated mice may also affect phosphorylation by an unknown mechanism. Perhaps use of a more abundant codon could increase the rate of protein synthesis and thus alter protein conformation (20). The other shared mutation, S741F, is transcriptionally inactive; the presence of stable cytoplasmic protein suggests ligand binding is defective.
Many more recurring mutations were identified in flutamide- than in bicalutamide-treated mice. Since flutamide is a partial agonist of AR at high concentrations in vitro, lower concentrations such as those found in patients may activate mutant alleles (42). Altering the LBD to achieve activation by the bulkier bicalutamide may be more difficult, but has been reported for AR-W742L (29). Flutamide-treated mice also express several nonsense mutants. Whereas the N-terminal AR-R102X most likely is a loss of function allele, other mutants terminating within the DBD or LBD may produce constitutive receptors (43, 44). Moreover, some truncated transcripts may have paracrine effects, promoting androgen-independent activation of wild type AR in adjacent cells (45). Reduced AR activity may promote growth of late stage tumors that have amassed other mutations, especially when antiandrogen treatment inhibits AR activation. In fact, AR expression is reduced late in disease (46). There may be an early window when AR gain of function mutations are advantageous but become less so as other growth factor pathways predominate.
Certain areas within the AR carried many more recurring mutations indicating potential hotspots for mutation or selection. One cluster identified within the LBD in all treatment groups has also been reported in clinical studies of prostate cancer, with mutations affecting AR function in diverse ways. AR-A749T identified in an untreated metastasis reduces receptor stability (47). Modeling of AR-V758I, found in a patient after orchiectomy and in this study, predicts a distortion that alters ligand specificity (35). Likewise, M745I that leads to CAIS as a germline mutation allows activation of AR by estradiol (48). None of the mutations from this cluster characterized in this study allow AR activation with noncanonical ligands or antiandrogens in transfection. Instead, they exhibit varying degrees of reduced transactivation. Three of the mutations, M750I, W752C, and R753Q, reside at the bottom of the ligand binding pocket within the same alpha helix as the loss of function mutation A749T. All three showed lower activation than wild type at 1 nM R1881 in CV-1 cells, but normal to higher activation at increased ligand levels. This is intriguing since intracrine hormone synthesis may maintain intraprostatic androgen levels in hormone-refractory prostate cancer (49). This might provide sufficient stimulus for these alleles to activate at least a subset of AR targets.
An indication that differential activation of AR targets might enhance cancer progression is the context-dependent function of R753Q. As a germline mutation, R753Q causes CAIS in patients and in rats (19, 34). Previous characterization of this mutant revealed reduced androgen binding capacity, reduced N-C interaction, and reduced activation in CV-1 cells (34, 50). This study has shown both cell-specific and promoter-dependent effects of R753Q, since activation is near wild type in PC-3 cells and robust for canonical HREs but impaired on AR-selective HREs. These effects may be accounted for at least in part by the altered N-C interaction which may destabilize AR binding on selective elements (51). An inability to activate AR-selective elements due to DBD differences in a knock-in mouse model leads to reduced fertility (52). Although germline AR-R753Q may not activate genes critical in male development, partial function of somatic AR-R753Q may activate a subset of promoters in prostate cancer. This residual activity may favor target genes involved in proliferation over those involved in differentiation. There may even be an optimal oncogenic AR activity that promotes proliferation over differentiation and can be achieved either through a reduction in androgen levels or reduced activity of AR. In support of this idea, Nkx3.1;Pten mutant mice treated with low doses of testosterone (T) develop more aggressive disease than mice treated with either no or normal levels of T (53). Moreover, h/mAR mice with Q tract variant alleles that differ in AR transactivational strength show poorer response to hormone therapy with weaker AR alleles (14).
In conclusion, this study has identified a diverse group of mutations in both the NTD and LBD of the AR that fall within functionally important domains. While different treatments favored certain mutations, all tumors carried numerous overlapping mutations. The presence of so many AR mutations at low levels has implications for treatment, suggesting that, while targeting the AR may slow disease progression, treatment will ultimately select for different cell populations allowing hormone-refractory growth. Furthermore, functional characterization of mutants from this study indicates that partial loss of function mutations that differentially activate AR targets may be selected during tumorigenesis. These partial loss of function alleles may tip the balance between proliferation and differentiation to confer a gain of function advantage in the context of tumor growth.
Materials and Methods
Mice
Female h/mAR mice (13) were mated to TRAMP males on a C57BL/6J background. Short-term treatments of 12-week-old males were castration by surgical orchiectomy or treatment with bicalutamide (Bic) or flutamide (Flu). Bic was compounded with food pellets and administered ad lib for a dose of 50 µg/g based on average food consumption. Flu (25 µg/g) was administered in food pellets or by a 75 mg 60 day slow release subcutaneous pellet (Innovative Research, Sarasota, FL). Age-matched intact mice served as controls. After 4 weeks, animals were sacrificed. AR N20 antibody (Santa Cruz Labs) was used to detect AR in formalin fixed, paraffin embedded prostate tissue. Serum T levels were measured by RIA (DSL, Webster, TX).
For tumor analysis, h/mAR-TRAMP males were randomly assigned to 4 treatment groups (9–10 mice/group): castration at 12 weeks of age, treatment with 50 µg/g Bicalutamide or 25 µg/g Flutamide upon tumor detection, or no treatment. Tumors were monitored by MRI or abdominal palpation and mice were euthanized when moribund as described (14). Primary prostate tumors, metastases and testes were harvested into RNAlater buffer (Ambion, Inc., Austin, TX) or flash frozen in liquid N2. All mouse procedures were approved by the University of Michigan Committee on Use and Care of Animals, in accordance with the NIH guidelines for the Care and Use of Experimental Animals.
RNA and DNA isolation
Tumor RNA was isolated with RNeasy kits (Qiagen Inc, Valencia, CA) and 1 µg was reverse transcribed using Superscript II RT (Invitrogen, Carlsbad, CA) with 0.5 µg oligo(dT) in a 20 µl reaction. Two RT reactions were performed per sample to control for enzyme error. Testis RNA from 17 mice was reverse transcribed and select fragments of AR were amplified and sequenced as controls. DNA was extracted as previously described (54). Briefly, tissue was digested overnight at 37°C in Lysis Buffer (10 mM Tris HCl, 400 mM NaCl, 2 mM Na2EDTA, pH 8.2) with 250 µg/ml proteinase K and 0.5% SDS. Proteins were removed by precipitation with NaCl and the DNA was precipitated with 2 vol 100% ethanol.
Amplification, subcloning and sequencing of the AR coding region
The AR coding region was amplified in 4 fragments, three from RNA (proximal NTD, DBD and LBD) and one from DNA (NTD). PCR primers for h/mAR samples are listed below. PCR amplifications were performed in 25 µl reactions containing 2.5 units Platinum Pfx DNA polymerase (Invitrogen), 2X Buffer and 1X GC enhancer (supplied by the manufacturer), 1.5 mM MgSO4, 0.3 mM dNTPs, 0.5 µM each primer, and 1–3 µl of the RT reaction or 100 ng genomic DNA. PCR conditions optimized for each primer pair were: 94°C for 5 min, 25–35 cycles of 94°C for 30 sec, 55°C (57°C for primer pair 1) for 30 sec, and 68°C for 90 sec, with a final extension step at 68°C for 10 min.
| h/mAR 1 Sense | 5’ TCGGTGGAAGCTACAGACAA 3’ |
| h/mAR 1 Antisnese | 5’ CCGACACTGCCTTACACAAC 3’ |
| h/mAR 2 Sense | 5’ TTCGACCATTTCTGACAACG 3’ |
| h/mAR 2 Antisnese | 5’ TTGGTCAAAAGGAGGCATTT 3’ |
| h/mAR 3 Sense | 5’ AGTGTGGTACCCTGGTGGAG 3’ |
| h/mAR 3 Antisnese | 5’ TTGTGCATGCGGTACTCATT 3’ |
| h/mAR 4 Sense | 5’ CAACTTGCATGTGGATGACC 3’ |
| h/mAR 4 Antisnese | 5’ TCTGGAAAGGGAACAAGGTG 3’ |
Products were visualized on 1% agarose gels and bands were excised and purified with the QiaexII gel extraction kit (Qiagen). 3’-A overhangs were added to the blunt-ended product by incubation with Taq polymerase (Invitrogen) at 70°C for 30 min. Products were ligated into the pGEM-T easy vector (Promega, Madison WI) and transfected into DH5α chemically competent bacterial cells (Invitrogen). DNA from 20 clones /sample (10 clones/RT reaction) was purified with QIAprep Spin Miniprep columns (Qiagen) or with the MacConnell Mini-Prep 24 Machine (MacConnell Research, San Diego, CA) according to the manufacturer’s directions and submitted to the University of Michigan DNA Sequencing Core for analysis.
Sequence Analysis
Sequence was compared to the human AR sequence (Genbank Accession# NM_000044) using Sequencher software (version 4.1, Gene Codes, Ann Arbor, MI). Putative mutations were then checked against the Androgen Receptor Gene Mutations Database (19).
Mutagenesis of AR expression plasmids
Six mutations (M524T, S741F, M750I, W752C, R753Q, and R761G) were introduced into pCMV5-hAR using the Quickchange Site Directed Mutagenesis kit (Stratagene) and the primer pairs below containing each mutation (underlined):
| M524T Sense: | CCACTTGTGTCAAAAGCGAAACGGGCCCCTGGA |
| M524T Antisense: | TCCAGGGGCCCGTTTCGCTTTTGACACAAGTGG |
| S741F Sense: | GCTGTCATTCAGTACTTCTGGATGGGGCTCATG |
| S741F Antisense: | CATGAGCCCCATCCAGAAGTACTGAATGACAGC |
| M750I Sense: | GCTCATGGTGTTTGCCATTGGCTGGCGATC |
| M750I Antisense: | GATCGCCAGCCAATGGCAAACACCATGAGC |
| W752C Sense: | TGTTTGCCATGGGCTGTCGATCCTTCACCAATG |
| W752C Antisense: | CATTGGTGAAGGATCGACAGCCCATGGCAAACA |
| R753Q Sense: | TGTTTGCCATGGGCTGGCAGTCCTTCACCAATGTCAAC |
| R753Q Antisense: | GTTGACATTGGTGAAGGACTGCCAGCCCATGGCAAACA |
| R761G Sense: | CACCAATGTCAACTCCGGGATGCTCTACTTCGC |
| R761G Antisense: | GCGAAGTAGAGCATCCCGGAGTTGACATTGGTG |
The mutant synthesis, DpnI digestion, and transformation were performed as detailed in the manual. DMSO was added to the mutant strand synthesis reaction to prevent CAG and GGN repeat contraction. Plasmids were sequenced to verify the presence of the mutation and retention of the original number of repeats within the polyamino acid tracts.
Transactivation assays
CV-1 cells were cultured in DMEM + 10% fetal bovine serum, 1% Glutamax and 1% penicillin/streptomycin. PC-3 cells were cultured in RPMI + 10% fetal bovine serum, 1% Glutamax and 1% penicillin/streptomycin. The day before transfection, cells were seeded at 5 × 104 (CV-1) or 1 × 105 (PC-3) cells/well in a 12-well plate. Four hrs before transfection, media was replaced with DMEM or RPMI + 2.5% charcoal-stripped NuSerum, 1% Glutamax. Cells were transfected using Fugene 6 reagent (Roche) at 3 vol Fugene/µg DNA with 4 ng pCMV5-hAR (wild type or mutant), 400 ng luciferase reporter plasmid and 100 ng promoterless renilla (Promega) for normalization. 24 hrs post-transfection, cells were rinsed in 1X PBS and fed with phenol red-free DMEM or RPMI media + 10% charcoal-stripped Nuserum +/− hormone. Cells were harvested 48 hrs post-transfection. The PSA-luc reporter (33) was a gift from K. Burnstein.
Western blotting
CV-1 cells were seeded at 4 × 105 cells/ 60 mm dish the day before transfection. Four hrs before transfection cells were rinsed in 1X PBS and fed with phenol red-free DMEM + 10% charcoal-stripped NuSerum, 1% Glutamax +/− 1 nM R1881. Cells were transfected with 100 ng receptor (hAR or mutant) and 1.9 µg empty vector (pCMV5) with Fugene 6 (3 µl/µg DNA). 24 hrs after transfection cells were rinsed in ice cold 1X PBS and harvested in 100 µl RIPA buffer + protease inhibitors. Cell lysates were incubated at 4°C for 10 min and then centrifuged at 4°C for 10 min to pellet cell debris. Protein quantification was performed using the Dc Protein Assay (Bio Rad). 20 µg protein lysate was run on an 8% SDS-polyacrylamide gel and transferred onto a nitrocellulose membrane. The blot was probed with antibody N20 to the AR NTD (Santa Cruz, CA) at 1:500 dilution and incubated with HRP-conjugated anti-rabbit IgG at 1:5,000 dilution for 45 min. Bands were detected with ECL western blotting reagents (Pierce, Rockford, IL).
3D structure representations
LBD mutations were examined by homology to the X-ray structure of AR complexed to the FXXLF motif (7) using the SWISS-MODEL automated protein structure homology-modeling server (55). 3-D structures were viewed in Protein Explorer (56).
Statistics
Significance of differences in disease length between treatments was determined using a Bonferoni multiple comparison adjustment (p<0.01).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Norman Greenberg (Fred Hutchinson Cancer Center, Seattle) for advice throughout this study, and Drs. Andy Lieberman and Chris Krebs (University of Michigan) for helpful discussions. This work was supported by grants to D.M.R. (DOD17-02-1-0099, NIDDK-RO1-56356 and NCI-P50 CA69568). O.A.M. was supported by a DOD fellowship W81XWH-05-1-0105, M.P.S. by the Organogenesis Training Grant (NIH-T32-HD075005) and M.A.A. by NCRR/NIH-T32-RR07008. Additional support came from the University of Michigan Cancer Center Support Grant (5 P30 CA46592) and the Michigan Diabetes Research and Training Center (NIH5P60 DK20572).
Footnotes
Steinkamp, M.P., O’Mahony, O.A., Brogley, M., Rehman, H., Dhanasekaran, S., Hofer,’ M., Chinnaiyan, A., Rubin, M., Pienta, K. and Robins, D.M. Androgen receptor mutations in human prostate cancer metastases vary with treatment. (manuscript in preparation)
REFERENCES
- 1.Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci U S A. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi XB, Ma AH, Xia L, Kung HJ, de Vere White RW. Functional analysis of 44 mutant androgen receptors from human prostate cancer. Cancer Res. 2002;62:1496–1502. [PubMed] [Google Scholar]
- 4.Scher HI, Kolvenbag GJ. The antiandrogen withdrawal syndrome in relapsed prostate cancer. Eur. Urol. 1997;31:3–7. doi: 10.1159/000474540. [DOI] [PubMed] [Google Scholar]
- 5.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 6.Taplin ME, Bubley GJ, Shuster TD, et al. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N Engl J Med. 1995;332:1393–1398. doi: 10.1056/NEJM199505253322101. [DOI] [PubMed] [Google Scholar]
- 7.Hur E, Pfaff SJ, Payne ES, Gron H, Buehrer BM, Fletterick RJ. Recognition and accommodation at the androgen receptor coactivator binding interface. PLoS Biol. 2004;2:E274. doi: 10.1371/journal.pbio.0020274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hara T, Miyazaki J, Araki H, et al. Novel mutations of androgen receptor: a possible mechanism of bicalutamide withdrawal syndrome. Cancer Res. 2003;63:149–153. [PubMed] [Google Scholar]
- 9.Shen HC, Coetzee GA. The androgen receptor: unlocking the secrets of its unique transactivation domain. Vitam Horm. 2005;71:301–319. doi: 10.1016/S0083-6729(05)71010-4. [DOI] [PubMed] [Google Scholar]
- 10.Greenberg NM, DeMayo F, Finegold MJ, et al. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci U S A. 1995;92:3439–3443. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han G, Foster BA, Mistry S, et al. Hormone status selects for spontaneous somatic androgen receptor variants that demonstrate specific ligand and cofactor dependent activities in autochthonous prostate cancer. J Biol Chem. 2001;276:11204–11213. doi: 10.1074/jbc.M008207200. [DOI] [PubMed] [Google Scholar]
- 12.Han G, Buchanan G, Ittmann M, et al. Mutation of the androgen receptor causes oncogenic transformation of the prostate. Proc Natl Acad Sci U S A. 2005;102:1151–1156. doi: 10.1073/pnas.0408925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albertelli MA, Scheller A, Brogley M, Robins DM. Replacing the mouse androgen receptor with human alleles demonstrates glutamine tract length-dependent effects on physiology and tumorigenesis in mice. Mol Endocrinol. 2006;20:1248–1260. doi: 10.1210/me.2006-0021. [DOI] [PubMed] [Google Scholar]
- 14.Albertelli MA, O'Mahony OA, Brogley M, et al. Glutamine tract length of human androgen receptors affects hormone-dependent and -independent prostate cancer in mice. Hum Mol Genet. 2008;17:98–110. doi: 10.1093/hmg/ddm287. [DOI] [PubMed] [Google Scholar]
- 15.Robins DM, Albertelli MA, O'Mahony OA. Androgen receptor variants and prostate cancer in humanized AR mice. J Steroid Biochem Mol Biol. 2008;108:230–236. doi: 10.1016/j.jsbmb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gingrich JR, Barrios RJ, Morton RA, et al. Metastatic prostate cancer in a transgenic mouse. Cancer Res. 1996;56:4096–4102. [PubMed] [Google Scholar]
- 17.Marcelli M, Stenoien DL, Szafran AT, et al. Quantifying effects of ligands on androgen receptor nuclear translocation, intranuclear dynamics, and solubility. J Cell Biochem. 2006;98:770–788. doi: 10.1002/jcb.20593. [DOI] [PubMed] [Google Scholar]
- 18.Johnson MA, Iversen P, Schwier P, et al. Castration triggers growth of previously static androgen-independent lesions in the transgenic adenocarcinoma of the mouse prostate (TRAMP) model. Prostate. 2005;62:322–338. doi: 10.1002/pros.20148. [DOI] [PubMed] [Google Scholar]
- 19.Gottlieb B, Beitel LK, Wu JH, Trifiro M. The androgen receptor gene mutations database (ARDB): 2004 update. Hum Mutat. 2004;23:527–533. doi: 10.1002/humu.20044. [DOI] [PubMed] [Google Scholar]
- 20.Kimchi-Sarfaty C, Oh JM, Kim IW, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 21.Ponguta LA, Gregory CW, French FS, Wilson EM. Site-specific Androgen Receptor Serine Phosphorylation Linked to Epidermal Growth Factor-dependent Growth of Castration-recurrent Prostate Cancer. J Biol Chem. 2008;283:20989–21001. doi: 10.1074/jbc.M802392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alvarado C, Beitel LK, Sircar K, Aprikian A, Trifiro M, Gottlieb B. Somatic mosaicism and cancer: a micro-genetic examination into the role of the androgen receptor gene in prostate cancer. Cancer Res. 2005;65:8514–8518. doi: 10.1158/0008-5472.CAN-05-0399. [DOI] [PubMed] [Google Scholar]
- 23.Buchanan G, Yang M, Cheong A, et al. Structural and functional consequences of glutamine tract variation in the androgen receptor. Hum Mol Genet. 2004;13:1677–1692. doi: 10.1093/hmg/ddh181. [DOI] [PubMed] [Google Scholar]
- 24.Tilley WD, Buchanan G, Hickey TE, Bentel JM. Mutations in the androgen receptor gene are associated with progression of human prostate cancer to androgen independence. Clin Cancer Res. 1996;2:277–285. [PubMed] [Google Scholar]
- 25.Culig Z, Stober J, Gast A, et al. Activation of two mutant androgen receptors from human prostatic carcinoma by adrenal androgens and metabolic derivatives of testosterone. Cancer Detect Prev. 1996;20:68–75. [PubMed] [Google Scholar]
- 26.Fenton MA, Shuster TD, Fertig AM, et al. Functional characterization of mutant androgen receptors from androgen-independent prostate cancer. Clin Cancer Res. 1997;3:1383–1388. [PubMed] [Google Scholar]
- 27.Zhao X-Y, Malloy PJ, Krishnan AV, et al. Glucocorticoids can promote androgenindependent growth of prostate cancer cells through a mutated androgen receptor. Nature Medicine. 2000;6:703–706. doi: 10.1038/76287. [DOI] [PubMed] [Google Scholar]
- 28.Haapala K, Hyytinen ER, Roiha M, et al. Androgen receptor alterations in prostate cancer relapsed during a combined androgen blockade by orchiectomy and bicalutamide. Lab Invest. 2001;81:1647–1651. doi: 10.1038/labinvest.3780378. [DOI] [PubMed] [Google Scholar]
- 29.Bohl CE, Miller DD, Chen J, Bell CE, Dalton JT. Structural basis for accommodation of nonsteroidal ligands in the androgen receptor. J Biol Chem. 2005;280:37747–37754. doi: 10.1074/jbc.M507464200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen D, Steinberg SV, Rouault E, et al. A G577R mutation in the human AR P box results in selective decreases in DNA binding and in partial androgen insensitivity syndrome. Mol Endocrinol. 2001;15:1790–1802. doi: 10.1210/mend.15.10.0709. [DOI] [PubMed] [Google Scholar]
- 31.Freedman LP. Anatomy of the steroid receptor zinc finger region. Endocr Rev. 1992;13:129–145. doi: 10.1210/edrv-13-2-129. [DOI] [PubMed] [Google Scholar]
- 32.Robins DM. Multiple mechanisms of male-specific gene expression: lessons from the mouse sex-limited protein (Slp) gene. Prog Nucleic Acid Res Mol Biol. 2004;78:1–36. doi: 10.1016/S0079-6603(04)78001-0. [DOI] [PubMed] [Google Scholar]
- 33.Perez-Stable CM, Pozas A, Roos BA. A role for GATA transcription factors in the androgen regulation of the prostate-specific antigen gene enhancer. Mol Cell Endocrinol. 2000;167:43–53. doi: 10.1016/s0303-7207(00)00300-2. [DOI] [PubMed] [Google Scholar]
- 34.Yarbrough WG, Quarmby VE, Simental JA, et al. A single base mutation in the androgen receptor gene causes androgen insensitivity in the testicular feminized rat. J Biol Chem. 1990;265:8893–8900. [PubMed] [Google Scholar]
- 35.Hyytinen ER, Haapala K, Thompson J, et al. Pattern of somatic androgen receptor gene mutations in patients with hormone-refractory prostate cancer. Lab Invest. 2002;82:1591–1598. doi: 10.1097/01.lab.0000038924.67707.75. [DOI] [PubMed] [Google Scholar]
- 36.Monge A, Jagla M, Lapouge G, et al. Unfaithfulness and promiscuity of a mutant androgen receptor in a hormone-refractory prostate cancer. Cell Mol Life Sci. 2006;63:487–497. doi: 10.1007/s00018-005-5471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen G, Wang X, Zhang S, et al. Androgen receptor mutants detected in recurrent prostate cancer exhibit diverse functional characteristics. Prostate. 2005;63:395–406. doi: 10.1002/pros.20191. [DOI] [PubMed] [Google Scholar]
- 38.Marcelli M, Ittmann M, Mariani S, et al. Androgen receptor mutations in prostate cancer. Cancer Res. 2000;60:944–949. [PubMed] [Google Scholar]
- 39.Callewaert L, Verrijdt G, Haelens A, Claessens F. Differential effect of small ubiquitin-like modifier (SUMO)-ylation of the androgen receptor in the control of cooperativity on selective versus canonical response elements. Mol Endocrinol. 2004;18:1438–1449. doi: 10.1210/me.2003-0313. [DOI] [PubMed] [Google Scholar]
- 40.Funderburk SF, Shatkina L, Mink S, Weis Q, Weg-Remers S, Cato AC. Specific N-terminal mutations in the human androgen receptor induce cytotoxicity. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 41.Kraus S, Gioeli D, Vomastek T, Gordon V, Weber MJ. Receptor for activated C kinase 1 (RACK1) and Src regulate the tyrosine phosphorylation and function of the androgen receptor. Cancer Res. 2006;66:11047–11054. doi: 10.1158/0008-5472.CAN-06-0596. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki H, Akakura K, Komiya A, Aida S, Akimoto S, Shimazaki J. Codon 877 mutation in the androgen receptor gene in advanced prostate cancer: relation to antiandrogen withdrawal syndrome. Prostate. 1996;29:153–158. doi: 10.1002/1097-0045(199609)29:3<153::aid-pros2990290303>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 43.Ceraline J, Cruchant MD, Erdmann E, et al. Constitutive activation of the androgen receptor by a point mutation in the hinge region: a new mechanism for androgenindependent growth in prostate cancer. Int J Cancer. 2004;108:152–157. doi: 10.1002/ijc.11404. [DOI] [PubMed] [Google Scholar]
- 44.Gao T, Marcelli M, McPhaul MJ. Transcriptional activation and transient expression of the human androgen receptor. J Steroid Biochem Mol Biol. 1996;59:9–20. doi: 10.1016/s0960-0760(96)00097-0. [DOI] [PubMed] [Google Scholar]
- 45.Lapouge G, Erdmann E, Marcias G, et al. Unexpected paracrine action of prostate cancer cells harboring a new class of androgen receptor mutation--a new paradigm for cooperation among prostate tumor cells. Int J Cancer. 2007;121:1238–1244. doi: 10.1002/ijc.22830. [DOI] [PubMed] [Google Scholar]
- 46.Shah RB, Mehra R, Chinnaiyan AM, et al. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Res. 2004;64:9209–9216. doi: 10.1158/0008-5472.CAN-04-2442. [DOI] [PubMed] [Google Scholar]
- 47.James AJ, Agoulnik IU, Harris JM, et al. A novel androgen receptor mutant, A748T, exhibits hormone concentration-dependent defects in nuclear accumulation and activity despite normal hormone-binding affinity. Mol Endocrinol. 2002;16:2692–2705. doi: 10.1210/me.2001-0281. [DOI] [PubMed] [Google Scholar]
- 48.Bonagura TW, Deng M, Brown TR. A naturally occurring mutation in the human androgen receptor of a subject with complete androgen insensitivity confers binding and transactivation by estradiol. Mol Cell Endocrinol. 2007;263:79–89. doi: 10.1016/j.mce.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 49.Titus MA, Schell MJ, Lih FB, Tomer KB, Mohler JL. Testosterone and dihydrotestosterone tissue levels in recurrent prostate cancer. Clin Cancer Res. 2005;11:4653–4657. doi: 10.1158/1078-0432.CCR-05-0525. [DOI] [PubMed] [Google Scholar]
- 50.Langley E, Kemppainen JA, Wilson EM. Intermolecular NH2-/carboxylterminal interactions in androgen receptor revealed by mutations that cause androgen insensitivity. J. Biol. Chem. 1998;273:92–101. doi: 10.1074/jbc.273.1.92. [DOI] [PubMed] [Google Scholar]
- 51.He B, Lee LW, Minges JT, Wilson EM. Dependence of selective gene activation on the androgen receptor NH2- and COOH-terminal interaction. J Biol Chem. 2002;277:25631–25639. doi: 10.1074/jbc.M202809200. [DOI] [PubMed] [Google Scholar]
- 52.Schauwaers K, De Gendt K, Saunders PT, et al. Loss of androgen receptor binding to selective androgen response elements causes a reproductive phenotype in a knockin mouse model. Proc Natl Acad Sci U S A. 2007;104:4961–4966. doi: 10.1073/pnas.0610814104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Banach-Petrosky W, Jessen WJ, Ouyang X, et al. Prolonged exposure to reduced levels of androgen accelerates prostate cancer progression in Nkx3.1; Pten mutant mice. Cancer Res. 2007;67:9089–9096. doi: 10.1158/0008-5472.CAN-07-2887. [DOI] [PubMed] [Google Scholar]
- 54.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 56.Martz E. Protein Explorer: easy yet powerful macromolecular visualization. Trends Biochem Sci. 2002;27:107–109. doi: 10.1016/s0968-0004(01)02008-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.