Abstract
Much effort has been directed towards understanding the contributions of electrostatics and dynamics to protein function and especially to enzyme catalysis. Unfortunately, these studies have been limited by the absence of direct experimental probes. We have been developing the use of carbon-deuterium bonds as probes of proteins and now report the application of the technique to the enzyme dihydrofolate reductase, which catalyzes a hydride transfer and has served as a paradigm for biological catalysis. We observe that the stretching absorption frequency of (methyl-d3) methionine carbon-deuterium bonds show an approximately linear dependence on solvent dielectric. Solvent and computational studies support the empirical interpretation of the stretching frequency in terms of local polarity. To begin to explore the use of this technique to study enzyme function and mechanism, we report a preliminary analysis of (methyl-d3) methionine residues within dihydrofolate reductase. Specifically, we characterize the IR absorptions at Met16 and Met20, within the catalytically important Met20 loop, and Met42, which is located within the hydrophobic core of the enzyme. The results confirm the sensitivity of the carbon-deuterium bonds to their local protein environment, demonstrate that dihydrofolate reductase is electrostatically and dynamically heterogeneous, and lay the foundation for the direct characterization protein electrostatics and dynamics, and potentially, their contribution to catalysis.
Introduction
The enzyme dihydrofolate reductase (DHFR) catalyzes hydride transfer from the cofactor nicotinamide adenine dinucleotide phosphate (NADPH) to 7,8-dihydrofolate to produce tetrahydrofolate and serves as a paradigm for biological hydride transfer. As with all enzymes,1–4 it is clear that specific electrostatic environments within DHFR contribute to catalysis.5–7 However, it is also clear that enzymes are highly dynamic. For example, the catalytic domain of DHFR is largely comprised of three loops that undergo significant structural reorganization upon ligand binding, and crystallographic B-factors suggest that the different conformations possess different levels of flexibility.8–10 Because the complexes are thought to mimic various stages of the catalytic cycle, it has also been suggested that changes in protein flexibility contribute to catalysis.10 Indeed, in recent years much debate has emerged regarding the role of protein dynamics in enzyme catalysis in general5,11,12 and in DHFR, in particular.6,13–18 How protein dynamics might contribute to catalysis ranges from conformational gating, where conformational changes might even be rate limiting,19,20 as is thought to be the case with DHFR,21–23 to a much more speculative direct coupling of specific protein motions to the reaction coordinate, perhaps similar to the strong and selective Fermi resonance between the stretching and bending motions of C-H moieties.24–26 Unfortunately, the study of protein electrostatics and dynamics has been limited by the challenges associated with the direct characterization of specific protein bonds, including their environment and motion.
In an effort to characterize protein electrostatics and dynamics, much work has been directed towards the development and characterization of spectroscopic probes incorporated within a protein,27–31 and many studies have focused on small infrared (IR) chromophores, such as cyano groups.32,33 Cyano groups absorb in a region of the IR spectrum that is free of protein absorptions (~2200 cm−1), which facilitates both their detection and analysis. In addition, they may be attached to amino acids and incorporated into different parts of a protein. However, cyano groups still introduce structural and electrostatic perturbations, and more problematically, they are likely to engage in artificial cation and/or H-bonding interactions. These interactions are not native to the protein but are likely to dominate the probe’s absorption properties.32,34
An alternative approach that does not rely on the introduction of artificial probes is based on the characterization of native protein absorptions using isotope labeling and difference Fourier transform infrared (FT IR) spectroscopy. This method has been widely used to examine heavy atom vibrations, such as those associated with amide bonds.35 However, the isotope-shifted absorptions remain in a highly congested region of the protein absorption spectrum, mandating the deconvolution of the probe absorption from a much larger background signal. In addition, the interpretation of their frequency and linewidth in terms of electrostatics and/or dynamics is complicated by both extensive coupling and strong and specific molecular interactions such as H-bonding.35
Unlike heavy atom isotopic substitution, substitution of a carbon deuterium (C-D) bond for a CH bond results in an isolated absorption at ~2100 cm−1.36 The unique absorption frequency facilitates both detection and interpretation, as the C-D absorption is adiabatically decoupled from most other protein vibrations. Moreover, isotopic substitution does not introduce any artificial interactions into the protein. Here, we explore the utility of C-D bonds as probes of electrostatics using (methyl-d3) methionine in different solvents. We also present the results of preliminary efforts toward the use of these probes for the characterization of dynamics and electrostatics in DHFR as a function of ligand binding.
Materials and Methods
Wild-type and mutant DHFR proteins were expressed in E. coli BL21 (DE3) at 37 °C in the presence of 100 µg/mL ampicillin. Cells were grown in 2 L M9 minimal media containing 1 mM MgSO4, 0.2% dextrose, 400 mg/L amino acids, and 1× Gibco MEM vitamin solution (Invitrogen). Isotopic labeling was accomplished by substitution with 50 mg/L (methyl-d3) methionine. Protein expression was induced at OD600 of 0.6–0.8 with 1 mM IPTG, and cells were grown 4–6 hr before harvesting. Protein purification and sample preparation was performed as described in the Supporting Information. Briefly, cells were lysed by sonication, and DNA was removed from the lysate by streptomycin sulfate precipitation. DHFR was purified first by MTX affinity chromatography, followed by DEAE ion-exchange chromatography. DHFR solutions were exchanged into 50 mM potassium phosphate, pH 7.0, 100 mM KCl, 1 mM DTT by dialysis, filtered, and concentrated to 3.5–4 mM prior to use. DHFR complexes were formed by the addition of concentrated stock solutions of NADP+ and NADPH in H2O and of folate and MTX in 400 mM NaOH. Solutions were adjusted to pH 7.0, and the concentrations of DHFR and all compounds were determined spectrophotometrically.
Samples of protein, (methyl-d3) methionine (Cambridge Isotopes), or BOC-protected (methyl-d3) methionine (prepared as described in Cremeens et al.37) were placed in a liquid sample cell with CaF2 windows and a 25 µm Teflon spacer. FT IR spectra were recorded on a Bruker Equinox 55 spectrometer equipped with a liquid nitrogen cooled MCT detector, as described previously.36,37 The sample chamber was purged with nitrogen (14 psi) for 30 min after opening chamber and during data acquisition. Absorption spectra were obtained from measured interferograms using Bruker OPUS software. A Blackman-Harris function was used for apodization; phase correction was performed with the power spectrum method and phase resolution of 32 cm−1. FT IR measurements were acquired at 4 cm−1 resolution, which is appropriate for condensed phase spectra.38,39 We also determined that there were no significant differences when the spectra were acquired at 2 cm−1 resolution. The resolution parameter for each absorption (ratio of nominal, or instrument resolution to the peak FWHM) was at least 0.5, thus the observed spectra should reflect the true absorption spectra given the intensity of the absorptions.38
For each (methyl-d3) methionine spectrum, 4000 scans were averaged and an unlabeled sample in the same solvent was used as a reference. The spectra were acquired for the free and/or BOC-protected amino acid in H2O, ethanol, methanol, isopropanol, t-butanol, benzyl alcohol, p-dioxane, toluene, as well as in mixtures of H2O and isopropanol, whose dielectric constants have been reported.40 The absorptions were well fit by neither a Gaussian nor a Lorentzian function, so a Voigt function, approximated by a linear combination of a Lorentzian and a Gaussian function,41 was used and yielded an average Lorentzian character of ~20%.
For each (methyl-d3) methionine labeled DHFR (d3Met DHFR) spectrum, three to six sets of 8192 averaged scans of both proteo and deutero-labeled proteins were recorded. Proteo-DHFR spectra were subtracted from the d3Met labeled DHFR spectra in the range 1900–2600 cm−1 using the autosubtract feature of the Bruker OPUS software. A polynomial was fit in the range ~2080–2180 cm−1, excluding a ~30 cm−1 window (2118–2144 cm−1) around the symmetric stretch absorption band, to approximate the background and subtracted from spectra to yield baseline-corrected spectra. The range of data included in the baseline fit, the region of the absorption band excluded from the fit, and the order of polynomial were varied and found not to significantly affect the observed differences in the spectra. The spectra of DHFR containing only d3Met1 was fit to a Voigt function, approximated by a linear combination of a Lorentzian and a Gaussian function,41 which yielded an average 98% Gaussian character. Thus, the spectra in all complexes were fit using a single Gaussian, and these Gaussian functions were used to deconvolute the absorption of d3Met1 from those of d3Met16, d3Met20, or d3Met42 in the spectra of d3Met1/d3Met16, d3Met1/d3Met20 or d3Met1/d3Met42 labeled proteins.
Density functional calculations were performed at the B3LYP/cc-pVTZ level using energy minimized CH3SCD3 in external fields ranging from +0.01 to −0.01 a.u. (1 a.u. = 5.19 × 109 V/cm, Supporting Information) or in the presence of potentially hydrogen-bonding water molecules using the Gaussian 03 program.
Results and Discussion
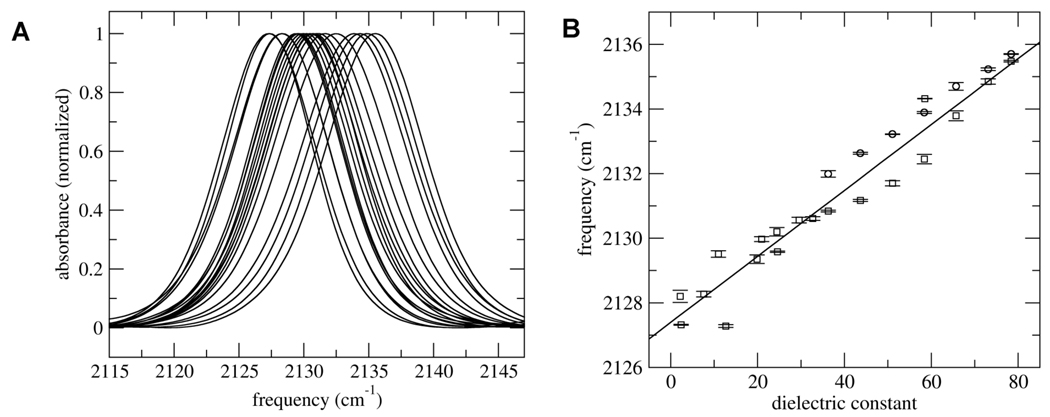
To examine C-D bonds as probes of their environment, we first examined the C-D stretching frequency of free or BOC-protected (methyl-d3) methionine in different mixtures of water and isopropanol, as well as in methanol, ethanol, t-butanol, benzyl alcohol, tetrahydrofuran, p-dioxane, and toluene. Each spectrum showed a strong absorption around 2130 cm−1 and a lower frequency, weaker absorption around 2240 cm−1, which based on previous studies are assigned as the symmetric and overlapping asymmetric stretches of the deuterated methyl group. The symmetric absorption was well fit using a linear combination of a Gaussian and a Lorentzian function to approximate a Voigt function,41 with a center frequency that varied monotonically in the water-isopropanol mixtures between 2136 cm−1 (100% water) and 2129 cm−1 (100% isopropanol) and down to 2127 cm−1 with the organic solvents (Figure 1).
Figure 1.
(A) Superposition of representative spectra of BOC-protected (methyl-d3) methionine symmetric stretch in different solvent examined. (B) Frequency of (methyl-d3) methionine symmetric stretch as a function of solvent dielectric constant. The circle and square symbols correspond to the free and BOC-protected amino acid, respectively. See text for details.
Because dielectric constants are known for all of the solvents and solvent mixtures employed, we first examined the observed C-D absorption frequency as a function this parameter. An approximately linear dependence of the C-D stretching frequency on solvent dielectric was immediately apparent. Correspondingly, the data are not well fit by models that predict a non-linear relationship between frequency and the dielectric constant or refractive index of the solvent, including the KBM equation, the modified KBM equation, or the Buckingham equation42,43 (Supporting Information). In addition, no correlation was found with Gutmann’s solvent acceptor numbers,44 as is found for other IR probes, such as cyano groups32,34 A linear least square fit was performed using the program Origin (squared correlation coefficient = 0.94 for all of the data and 0.98 when limited to just the free amino acid) and a t-test at the 95% confidence level shows the linear correlation to be significant (t value of 35 compared to a critical value of ~245).
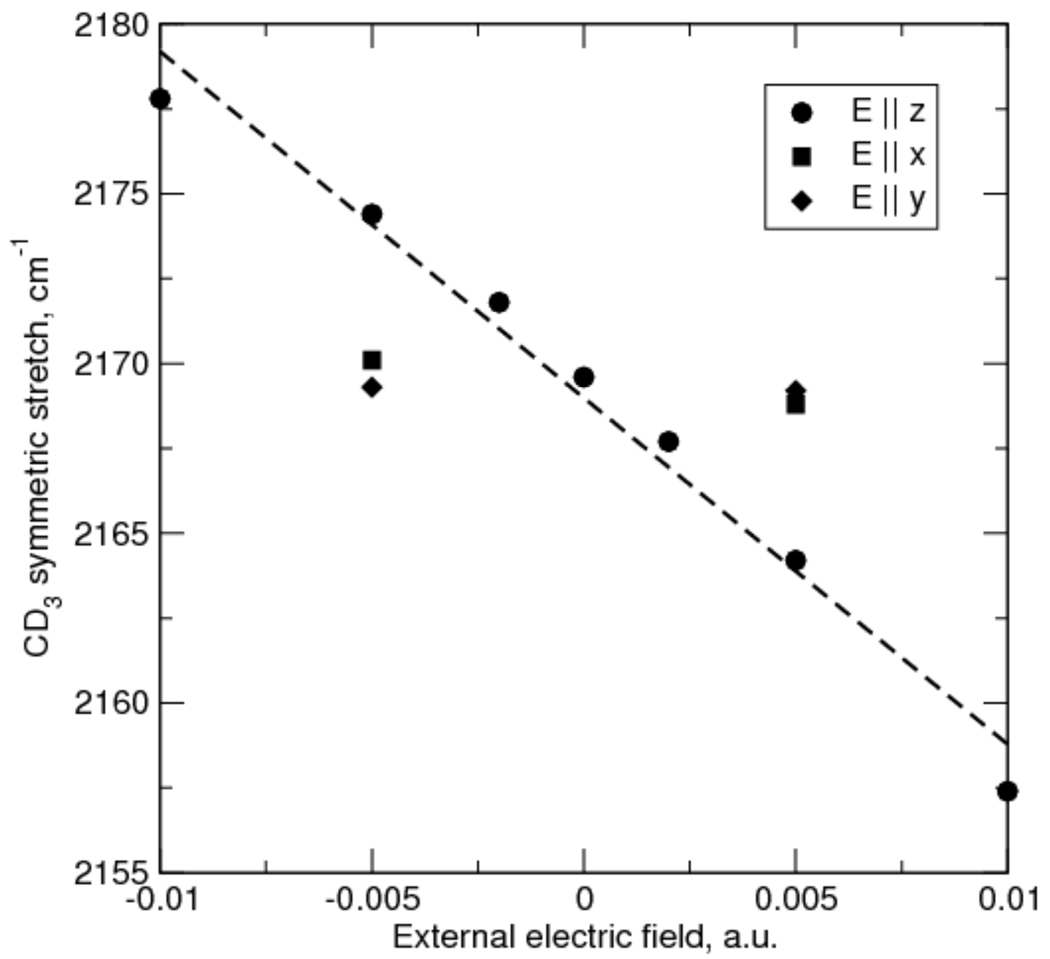
Comparison of the free and BOC-protected data (circle and square symbols in Figure 1B, respectively) suggests that the hydrophobic BOC group makes a contribution to the observed C-D frequency, likely by packing with the labeled methyl group and providing a more hydrophobic environment. In order to further explore the potential origins of the observed frequency shifts we explored the potential contribution of the reaction field. The reaction field, which is the electric field experienced by the solute due to its effect on ordering solvent dipoles, should increase monotonically with increasing solvent dielectric, until it approaches a high dielectric limit46. To characterize how the reaction field affects the C-D absorption frequency, we performed density functional calculations (at the B3LYP/cc-pVTZ level) on CH3SCD3. When the field direction is parallel to the symmetry axis of the CD3 group the frequency of the symmetric stretch is a an approximately linear function of the field (squared correlation coefficient = 0.98), with a slope of −1020 cm−1/a.u (Figure 2). External fields perpendicular to the CD3 symmetry axis yield frequency shifts that are one to two orders of magnitude smaller, suggesting that the component of the field along the symmetry axis is a major determinant of the frequency.
Figure 2.
Computed frequencies (B3LYP/cc-pVTZ) for the symmetric CD3 stretch in CH3SCD3 as a function of external field, E. The z direction is along the S-C bond, y is perpendicular to the CSC plane, and×is perpendicular to y and z. The dashed line is a linear regression on the points for E ‖z.
As the dielectric constant increases, the reaction field should approach an asymptotic value: for a dipole in a spherical cavity, the reaction field is proportional to (2ε-2)/(2εε+1)46, which plateaus at high values of the dielectric constant, ε. Thus, the reaction field alone is insufficient to explain the observed frequency shifts, which continue to increase even at high dielectric constant, and other factors must also contribute. In addition to the electric field, the observed frequencies may be sensitive to hyperconjugative interactions between the C-D σ-bonds and the d-orbitals of the adjacent sulfur atom, and the electron density at the sulfur center is expected to be sensitive to any solvation. While thioethers are generally believed to be very poor H-bond acceptors,47 the methionine sulfur atom has been shown to act as an H-bond acceptor in the human retinoic acid receptor48 and α-lytic protease.49 Thus, to explore the potential contribution of H-bonding to the observed frequencies, we performed additional density functional calculations (again at the B3LYP/cc-pVTZ level) on CH3SCD3 in the presence of waters of solvation. The symmetric-stretch mode frequency for CD3 with no waters is 2169.8 cm−1; addition of one H-bonding water molecule increases this by 4.5 cm−1 (to 2174.3 cm−1); a second water yields an additional increase of 3.3 cm−1 (to 2177.6 cm−1). This may result from a local increase in reaction field and/or from changes in the electron density of the sulfur d-orbitals that is communicated to the C-D σ bonds via hyperconjugation, as has been suggested with small organic molecules.50 Such H-bonds may be favored at high water to isopropanol ratios, both due to mass action and to the stabilization of any associated charge separation at the highly polarizable sulfur atom.
In all, the data suggest that observed C-D stretching frequencies are sensitive to a combination of factors that affect the polarity of their environment. In the solvent studies described above, these appear to include reaction field, H-bonding with water (including through-bond and through space effects), and/or packing with the hydrophobic t-butyl moiety of the BOC protecting group. When incorporated into a protein, this suggests the observed frequencies will be sensitive to the local electric field of the protein and/or proximal water molecules, as well as packing interactions between the labeled side chain and hydrophobic parts of the protein. Regardless of the specific physical forces involved, the data support an empirical interpretation of C-D stretching frequency in terms of overall local polarity.
To begin to explore the technique as a probe of DHFR, we next characterized the absorptions of d3Met within the enzyme. Attempts to synthesize DHFR via chemical ligation, which would allow for site-specific deuteration, proved unsuccessful (Supporting Information). However, E. coli DHFR has only five Met residues, and it is known that they may all be mutated to leucine without loss of activity.51 Thus, for the initial evaluation of C-D bonds as probes of DHFR we constructed mutants that possessed only a single Met residue (Met1) or two Met residues (combinations of Met1 and Met16, Met20, or Met42), with the remaining Met residues mutated to leucine. Each protein retained Met1, as it is required for translation. All proteins were expressed in E. coli in defined minimal medium supplemented with (methyl-d3) methionine, purified by affinity and ion exchange chromatography, and assayed by FT IR spectroscopy.
The absorption of d3Met1 in the apoenzyme was first determined using the singly labeled protein. We fit the spectra with a Gaussian function, a Lorentzian function, or a linear combination of the two (see Materials and Methods). In all cases, the spectra were best fit by a single Gaussian, with on average, only a 2% contribution from the Lorentzian function (this was also the case with the d3Met16, d3Met20, and d3Met42 absorptions). While nonlinear experiments are required to rigorously evaluate the contributions of various line broadening mechanisms to these absorptions, the essentially Gaussian lineshapes suggest that they are predominantly inhomogeneously broadened (i.e. broadened by the population of different environments, each of which results in a slightly different C-D absorption frequency, and which interconvert slowly on the experimental timescale).52 As expected for a solvent-exposed residue distant from the binding site, the absorption of d3Met1 showed virtually no changes in lineshape, frequency, or linewidth upon binding any of the complexes examined (Table 1). The absorptions of d3Met16, d3Met20, and d3Met42 were then characterized by fitting the spectra of the doubly labeled proteins and including a Gaussian representing d3Met1 (Figure 3 and Supporting Information). Spectra were collected for the apoenzyme (ligand free), the holoenzyme (bound NADPH), and complexes with folate and NADP+, MTX and NADPH, or with folate alone (Table 1). These complexes are thought to mimic the Michaelis complex, the transition state, and the product complex, respectively.10 For each spectrum, at least three independent measurements were acquired and analyzed, and the averages and standard deviations were determined. While the observed differences are generally small, those discussed below are statistically significant at the 95% confidence level.
Table 1.
Spectroscopic dataa.
| Residue | Complex | ν,cm−1 | FWHMb, cm−1 |
||
|---|---|---|---|---|---|
| Met1 | Apoenzyme | 2131.4 | (0.4) | 10.9 | (0.5) |
| NADPH | 2132.2 | (0.3) | 10.3 | (0.3) | |
| Folate/NADP+ | 2132.3 | (0.2) | 10.5 | (0.3) | |
| MTX/NADPH | 2132.4 | (0.2) | 10.5 | (0.3) | |
| Folate | 2132.3 | (0.2) | 10.4 | (0.4) | |
| Met16 | Apoenzyme | 2129.7 | (0.4) | 12.3 | (0.2) |
| NADPH | 2132.6 | (0.4) | 9.1 | (0.5) | |
| Folate/NADP+ | 2132.5 | (0.2) | 9.3 | (1.1) | |
| MTX/NADPH | 2133.2 | (0.6) | 9.6 | (1.1) | |
| Folate | 2132.9 | (0.4) | 10.3 | (0.2) | |
| Met20 | Apoenzyme | 2128.6 | (0.4) | 9.8 | (1.0) |
| NADPH | 2132.8 | (0.4) | 9.4 | (1.0) | |
| Folate/NADP+ | 2132.2 | (0.5) | 10.6 | (0.7) | |
| MTX/NADPH | 2130.3 | (1.0) | 12.2 | (0.7) | |
| Folate | 2129.8 | (0.6) | 10.8 | (1.1) | |
| Met42 | Apoenzyme | 2126.2 | (0.2) | 7.3 | (0.3) |
| NADPH | 2125.9 | (0.1) | 7.5 | (0.5) | |
| Folate/NADP+ | 2126.2 | (0.2) | 7.0 | (0.3) | |
| MTX/NADPH | 2125.7 | (0.1) | 7.1 | (0.4) | |
| Folate | 2125.4 | (0.1) | 7.1 | (0.3) | |
Standard deviations are in parentheses.
Full-width at half maximum.
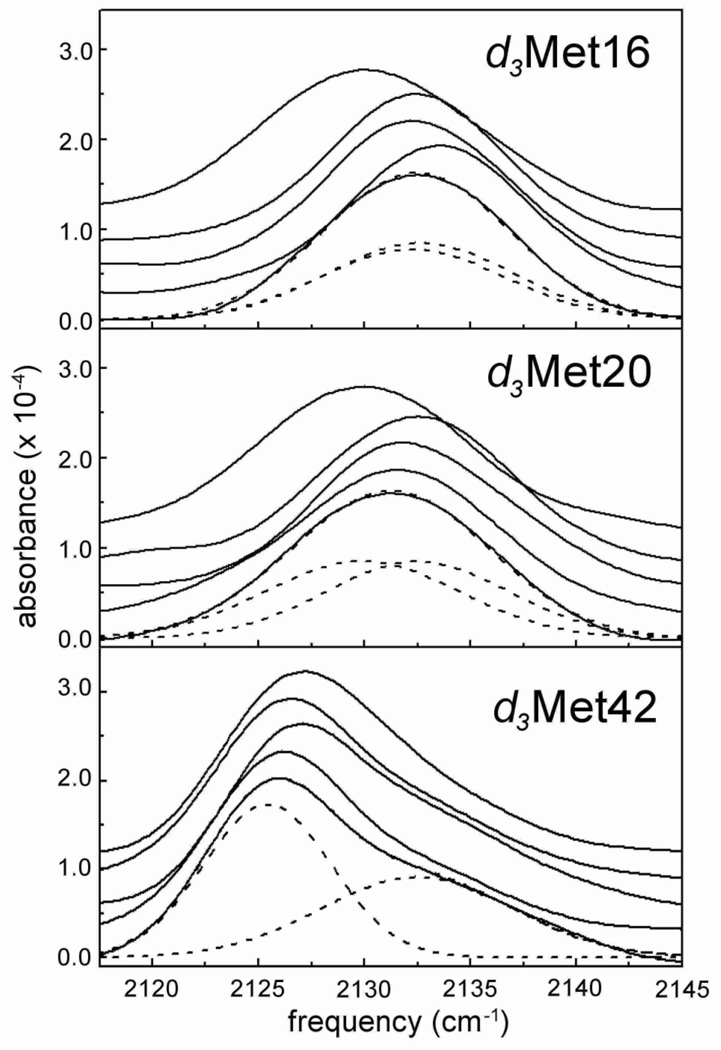
Figure 3.
Spectra of deuterated proteins. Shown from top to bottom of each panel are the spectra for the apoenzyme, the NADPH complex, the folate/NADP+ complex, the MTX/NADPH complex, and the folate complex. For the folate complex, the components and total fit of absorption band are shown with dashed lines. Fits of all other spectra are included in Supporting Information.
One structural feature of DHFR whose dynamics are thought to be critical for catalysis is the Met20 loop (comprised of residues 9–24).10,14 Structural8,10 and NMR53–55 studies show that the loop is disordered in the absence of ligands and suggest that this disorder results from the population of at least two equilibrating conformations.10 To characterize this loop, we first examined the absorptions of d3Met16 and d3Met20 in the apoenzyme. Single absorptions were observed that are significantly red-shifted compared to their absorptions in the various complexes (see below), with absorption frequencies of 2129.7 cm−1 (d3Met16) and 2128.6 cm−1 (d3Met20). In comparison to the same absorptions in the different complexes and based on the solvent studies described above, these absorption frequencies indicate that the Met16 and Met20 side chains experience a relatively apolar environment in the apoenzyme. The absorptions d3Met16 and d3Met20 are also relatively broad and for d3Met16, they are broader in the apoenzyme than in any of the ligand-bound states (see below). As discussed above, the Gaussian shape of these absorptions suggests that they are predominantly inhomogeneously broadened, which in turn suggests that the greater linewidth observed for d3Met16 reflects increased static inhomogeneity resulting either from increased coupling of the C-D transition dipole moment to its environment or from the presence of multiple conformations of the Met20 loop. However, because the absorptions of d3Met16 in the occluded and closed states have the same frequency and linewidth (see below), which are different from that observed in the apoenzyme, the Met20 loop in the apoenzyme cannot simply be equilibrating between these two conformations.
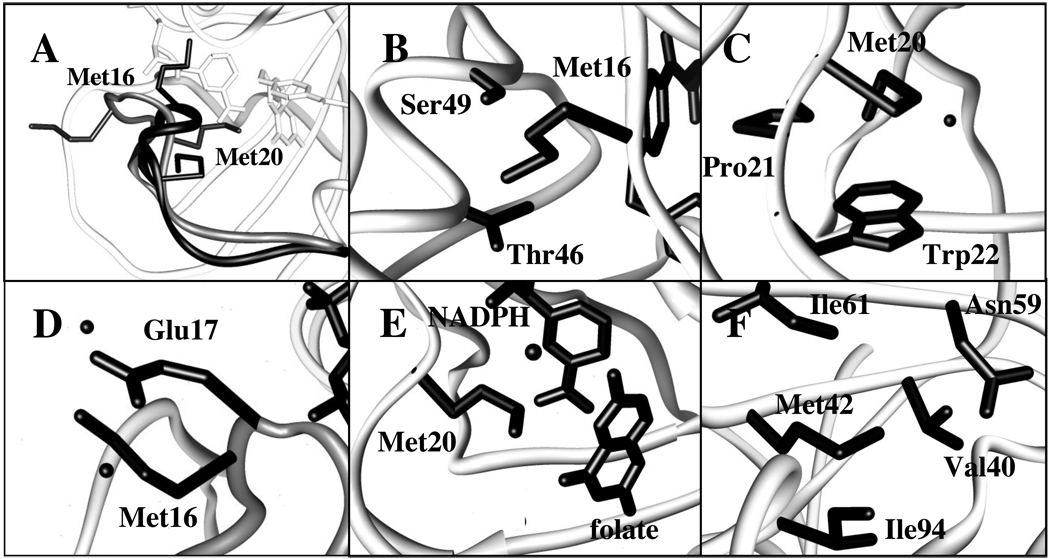
In order to characterize the effects of ligand binding, we next examined the absorptions of d3Met16 and d3Met20 in complexes that are thought to mimic the different states of the catalytic cycle. In complexes where only substrate (or a substrate analog such as folate) is bound, the Met20 loop assumes the ‘occluded’ conformation (Figure 4A), occupying part of the active site, and this conformation is thought to be a mimic of the product complex.10,56 The single symmetric stretching absorption observed for both d3Met16 and d3Met20 when bound to folate reveals that in the occluded conformation each side chain experiences a well-defined microenvironment. No significant frequency shift or change in linewidth is observed with d3Met20 upon folate binding, indicating that the dynamics and electrostatic environment of the residue are similar in the apo and occluded state. In contrast, folate binding induces a significant narrowing and blue-shift of the d3Met16 absorption. The blue-shift reveals that the environment of this residue is more polar in the occluded state than it is in the ligand-free state, and more polar than is the environment of Met20 in either state. This data is consistent with available structural data10 which show in the folate-induced occluded state that the Met16 side chain is in van der Waals contact with the polar backbone oxygen atom of His45 and the side chains of Thr46 and Ser49 (Figure 4B), while the Met20 side chain packs with the hydrophobic side chains of Trp22 and Pro21 (Figure 4C). While line narrowing in the heterogeneous limit may result from reduced coupling between the C-D transition dipole and its environment and/or from reduced sampling of different environments, the apparently anisotropic and polar environment of the Met16 side chain suggests that in this case it results from the latter. Thus, the differences between the apo and occluded loop conformations are position specific, with little difference apparent at Met20, while the Met16 side chain appears to be positioned in a more polar and more restricted environment in the folate-induced occluded state.
Figure 4.
Environment of Met residues in crystal structures of DHFR bound to folate (PDB 1rx7) or to and folate and NADP+ (PDB 1rx2). (A) Superposition of Met20 loop in the occluded (black) and closed (grey) states with side chains of Met16 and Met20 shown; (B) Met16 in occluded state (His45 backbone carbonyl not shown for clarity); (C) Met20 in occluded state; (D) Met16 in closed state; (E) Met20 in closed state (the folate (p-aminobenzoyl)glutamate group has been omitted for clarity); (F) Met42 (same environment in occluded and closed states). Panels (B) – (F) include crystallographically observable water molecules that are within 4 Å of the labeled ε-methyl group.
When DHFR binds cofactor (or a cofactor analog), the Met20 loop assumes the ‘closed’ conformation, in which Met16 is flipped out of the active site and Met20 packs on the bound cofactor and substrate (Figure 4A).10,14 When folate or MTX are also bound, the complexes are thought to mimic the Michaelis complex and transition state, respectively.10,14 For d3Met16, the FT IR data reveal a significant blue-shift upon formation of any of these complexes, revealing that in the closed state, as in the occluded state, the side chain experiences a more polar environment. For d3Met20, binding NADPH or folate and NADP+ also induces a blue-shift, while binding MTX and NADPH does not. These changes in environment can again be rationalized structurally.10 In the closed conformation, the Met16 methyl group appears to be in a polar environment, regardless of the ligands bound (Figure 4D). Specifically, it is proximal to two water molecules as well as to the carboxylate side chain of Glu17. In contrast, the Met20 methyl group in the closed conformation is oriented into the active site (Figure 4E), and the structures predict that its environment should be more sensitive to the nature of the ligands bound. Specifically, in the NADPH complex, several crystallographically observable water molecules are proximal to the Met20 methyl group. In the complex with cofactor and folate, the methyl group is proximal to one water molecule and the folate carbonyl group. Finally in the MTX and NADPH complex, the structure predicts that the Met20 methyl group is in a relatively less polar environment due to the absence of a corresponding carbonyl group in MTX.
There are also differences in how complex formation affects the linewidths of d3Met16 and d3Met20. The absorption linewidth of d3Met20 increases upon binding MTX and NADPH, but not upon binding any of the other ligands (Table 1). In contrast, for d3Met16, binding any of the ligands induces a small, but significant and consistent decrease in linewidth. Thus, when considering both the observed frequencies and linewidths, we conclude that as with folate, cofactor binding produces a more polar environment and a general reduction in the variability of the environments sampled at Met16, but that the changes at Met20 are smaller and/or ligand-dependent.
In addition to residues within the Met20 loop, we examined Met42, which is positioned in the hydrophobic core of DHFR (Figure 4F). In the apoenzyme, the spectrum reveals a d3Met42 absorption with a center frequency of 2126.4 cm−1 and a linewidth of 7.8 cm−1. The strong red-shift and narrowing of the absorption, relative to the other absorptions, indicates an environment that is less polar and more homogeneous. Little change is observed in either absorption frequency or linewidth upon ligand binding. This data suggests that the core of DHFR, at least that experienced by the side chain of Met42 is apolar and relatively rigid, and that unlike the Met20 loop, no significant structural or dynamic changes are induced by ligand binding. These conclusions are consistent with the apparently well-packed and hydrophobic environment of the Met42 side chain observed in all DHFR crystal structures.
Conclusions
Electrostatics and dynamics contribute to all protein functions, including protein folding and structure, molecular recognition, and catalysis. Unfortunately, despite increased interest in understanding their contributions to catalysis, both have been challenging to characterize experimentally. For these purposes, C-D bonds have several advantages. Within the Born-Oppenheimer approximation, they do not perturb the protein but provide a spectrally resolved absorption that is largely free of influence from other protein vibrations. This renders C-D absorptions excellent reporters of their specific environment. Towards the development of C-D probes dynamics and electrostatics in enzymes, we have shown that the C-D bonds of (methyl-d3) methionine are approximately linearly dependent on solvent dielectric. While the origins of this behavior remain to be fully understood, the observation facilitates at least an empirical interpretation of the data. Our preliminary analysis of DHFR suggests that the environment of Met20 is sensitive to ligand binding only when it is in the closed state and oriented directly into the binding site. In contrast, the environment experienced by Met16 appears to be much more sensitive to ligand binding, in all cases becoming apparently more polar and rigid. This is consistent with the previous speculation that Met20 loop fluctuations contribute to catalysis through active site compression.57 In contrast, the core of the enzyme, at least as sampled by the side chain of Met42, appears to remain well packed, rigid, and apolar under all conditions examined, perhaps providing a stable core upon which the loops are free to fluctuate as required for ligand binding and catalysis. These preliminary studies suggest that the method should find important applications in the study of DHFR. For example, much interest has been focused on the contribution of correlated motions within DHFR to its catalytic activities, and correlated changes in linewidth would support this hypothesis, and help define the parts of the protein involved. These studies, as well as a more quantitative understanding of the data will facilitated by both a more rigorous analysis of the relationship between C-D absorption frequency, electric field, and solvation as well as modeling and time resolved studies to help understand the observed linewidths. Such efforts are currently underway.
Supplementary Material
Acknowledgement
This work was supported by an NSF graduate fellowship (M.C.T) and Grant MCB-0346967 (F.E.R) and NIH Grant RR12255 (D.A.C.).
Footnotes
Supporting Information Available Experimental and computational details. This information available free of charge from http://pubs.acs.org.
References
- 1.Honig B, Nicholla A. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 2.Perutz MF. Science. 1978;201:1187–1191. doi: 10.1126/science.694508. [DOI] [PubMed] [Google Scholar]
- 3.Villa J, Warshel A. J. Phys. Chem. B. 2001;105:7887–7907. [Google Scholar]
- 4.Warshel A, Papzyan A. Curr. Opin. Struct. Biol. 1998;8:211–217. doi: 10.1016/s0959-440x(98)80041-9. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Viloca M, Truhlar DG, Gao J. Biochemistry. 2003;42:13558–13575. doi: 10.1021/bi034824f. [DOI] [PubMed] [Google Scholar]
- 6.Wong KF, B WJ, Hammes-Schiffer S. J. Phys. Chem. B. 2004;108:12231–12241. [Google Scholar]
- 7.Li ea. Biochemistry. 1992;31:7826–7833. doi: 10.1021/bi00149a012. [DOI] [PubMed] [Google Scholar]
- 8.Bystroff C, Kraut J. Biochemistry. 1991;26:2227–2239. doi: 10.1021/bi00222a028. [DOI] [PubMed] [Google Scholar]
- 9.Bystroff C, Oatley SJ, Kraut J. Biochemistry. 1990;29:3263–3277. doi: 10.1021/bi00465a018. [DOI] [PubMed] [Google Scholar]
- 10.Sawaya MR, Kraut J. Biochemistry. 1997;36:586–603. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- 11.Daniel RM, Dunn RV, Finney JL, Smith JC. Annu. Rev. Biophys. Biomol. Struct. 2003;32:69–92. doi: 10.1146/annurev.biophys.32.110601.142445. [DOI] [PubMed] [Google Scholar]
- 12.Eisenmesser EZ, Bosco DA, Akke M, Kern D. Science. 2002;295:1520–1523. doi: 10.1126/science.1066176. [DOI] [PubMed] [Google Scholar]
- 13.Maglia G, Allemann RK. J. Am. Chem. Soc. 2003;125:13372–13373. doi: 10.1021/ja035692g. [DOI] [PubMed] [Google Scholar]
- 14.Schnell JR, Dyson HJ, Wright PE. Annu. Rev. Biophys. Biomol. Struct. 2004;33:119–140. doi: 10.1146/annurev.biophys.33.110502.133613. [DOI] [PubMed] [Google Scholar]
- 15.Sikorski RS, Wang L, Markham KA, Rajagopalan PT, Benkovic SJ, Kohen A. J. Am. Chem. Soc. 2004;126:4778–4779. doi: 10.1021/ja031683w. [DOI] [PubMed] [Google Scholar]
- 16.Swanwick RS, Maglia G, Tey LH, Allemann RK. Biochem J. 2006;394:259–265. doi: 10.1042/BJ20051464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim HS, Damo SM, Lee SY, Wemmer D, Klinman JP. Biochemistry. 2005;44:11428–11439. doi: 10.1021/bi050630j. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Goodey NM, Benkovic SJ, Kohen A. Proc. Natl. Acad. Sci. USA. 2006;103:15753–15758. doi: 10.1073/pnas.0606976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cleland WW. Acc. Chem. Res. 1975;8:145–151. [Google Scholar]
- 20.Dahlberg ME, Benkovic SJ. Biochemistry. 1991;30:4835–4843. doi: 10.1021/bi00234a002. [DOI] [PubMed] [Google Scholar]
- 21.Miller GP, Wahnon DC, Benkovic SJ. Biochemistry. 2001;40:867–875. doi: 10.1021/bi001608n. [DOI] [PubMed] [Google Scholar]
- 22.Thorpe IF, Brooks CL., III J. Am. Chem. Soc. 2005;127:12997–13006. doi: 10.1021/ja053558l. [DOI] [PubMed] [Google Scholar]
- 23.McElheny D, Schnell JR, Lansing JC, Dyson HJ, Wright PE. Proc. Natl. Acad. Sci. USA. 2005;102:5032. doi: 10.1073/pnas.0500699102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quack M. In: Mode Selective Chemistry. Jortner J, Levine RD, Pullman B, editors. Boston: Kluwer Academic Press; 1991. [Google Scholar]
- 25.Sibert EL, III, Hynes JT, Reinhardt WP. J. Chem. Phys. 1984;81:1135–1144. [Google Scholar]
- 26.Sibert EL, III, Reinhardt WP, Hynes JT. J. Chem. Phys. 1984;81:1115–1134. [Google Scholar]
- 27.Cohen BE, McAnaney TB, Park ES, Jan YN, Boxer SG, Jan LY. Science. 2002;296:1700–1703. doi: 10.1126/science.1069346. [DOI] [PubMed] [Google Scholar]
- 28.Suydam IT, Boxer SG. Biochemistry. 2003;42:12050–12055. doi: 10.1021/bi0352926. [DOI] [PubMed] [Google Scholar]
- 29.Lockhart DJ, Kim Peter S. Science. 1992;257:947–951. doi: 10.1126/science.1502559. [DOI] [PubMed] [Google Scholar]
- 30.Macgregor RB, Weber Gregorio. Nature. 1986;319:70–73. doi: 10.1038/319070a0. [DOI] [PubMed] [Google Scholar]
- 31.Park ES, Andrews Steven S, Hu Robert B, Boxer Steven G. J. Phys. Chem. B. 1999;103:9813–9817. [Google Scholar]
- 32.Getahun Z, Huang CY, Wang T, De Leon B, DeGrado WF, Gai F. J. Am. Chem. Soc. 2003;125:405–411. doi: 10.1021/ja0285262. [DOI] [PubMed] [Google Scholar]
- 33.Suydam IT, Snow CD, Pande VS, Boxer SG. Science. 2006;313:200–204. doi: 10.1126/science.1127159. [DOI] [PubMed] [Google Scholar]
- 34.Fawcett WR, et al. J. Phys. Chem. 1993;97:9293–9298. [Google Scholar]
- 35.Barth A, Zscherp C. Q. Rev. Biophys. 2002;35:369–430. doi: 10.1017/s0033583502003815. [DOI] [PubMed] [Google Scholar]
- 36.Chin JK, Jimenez R, Romesberg FE. J. Am. Chem. Soc. 2001;123:2426–2427. doi: 10.1021/ja0033741. [DOI] [PubMed] [Google Scholar]
- 37.Cremeens ME, Fujisaki H, Zhang Y, Zimmermann J, Sagle LB, Matsuda S, Dawson PE, Straub JE, Romesberg FE. J. Am. Chem. Soc. 2006;128:6028–6029. doi: 10.1021/ja061328g. [DOI] [PubMed] [Google Scholar]
- 38.Griffiths PR, de Haseth James. Fourier Transform Infrared Spectroscopy. New York: John Wiley & Sons, Inc.; 1986. [Google Scholar]
- 39.Smith BC. Fundamentals of Fourier Transform Infrared Spectroscopy. New York: CRC Press; 1996. [Google Scholar]
- 40.Akerlof G. J. Am. Chem. Soc. 1932;54:4125–4139. [Google Scholar]
- 41.Fraser RDB, Suzuki E. Anal. Chem. 1969;41:37–39. [Google Scholar]
- 42.Rao CNR, Singh S, Senthilnathan VP. Chem. Soc. Rev. 1976;5:297–316. [Google Scholar]
- 43.Reimers JR, Hall LE. J. Am. Chem. Soc. 1999;121:3730–3744. [Google Scholar]
- 44.Gutmann V. Coord. Chem. Rev. 1976;18:225–255. [Google Scholar]
- 45.Bluman AG. Elementary Statistics. New York: McGraw Hill; 2004. [Google Scholar]
- 46.Onsager L. J. Am. Chem. Soc. 1936;58:1486–1493. [Google Scholar]
- 47.Allen FH, Bird CM, Rowland RS, Raithby PR. Acta Cryst. 1997;B53:696–701. [Google Scholar]
- 48.Klaholz BP, Mitschler A, Moras D. J. Mol. Biol. 2000;302:155–170. doi: 10.1006/jmbi.2000.4032. [DOI] [PubMed] [Google Scholar]
- 49.Wennmohs F, Schindler M. J. Comput. Chem. 2005;26:283–293. doi: 10.1002/jcc.20163. [DOI] [PubMed] [Google Scholar]
- 50.Nakai H, Kawai M. J. Chem. Phys. 2000;113:2168–2174. [Google Scholar]
- 51.Shaw D, Odom JD, Dunlap RB. Biochim. Biophys. Acta. 1999;1429:401–410. doi: 10.1016/s0167-4838(98)00250-7. [DOI] [PubMed] [Google Scholar]
- 52.Schmidt JR, Sundlass N, Skinner JL. Chem. Phys. Lett. 2003;378:559–566. [Google Scholar]
- 53.Falzone CJ, Wright PE, Benkovic SJ. Biochemistry. 1991;30:2184–2191. doi: 10.1021/bi00222a023. [DOI] [PubMed] [Google Scholar]
- 54.Li L, Falzone CJ, Wright PE, Benkovic SJ. Biochemistry. 1992;31:7826–7833. doi: 10.1021/bi00149a012. [DOI] [PubMed] [Google Scholar]
- 55.Falzone CJ, Wright PE, Benkovic SJ. Biochemistry. 1994;33:439–442. doi: 10.1021/bi00168a007. [DOI] [PubMed] [Google Scholar]
- 56.Schnell JR, Dyson HJ, Wright PE. Biochemistry. 2004;43:374–383. doi: 10.1021/bi035464z. [DOI] [PubMed] [Google Scholar]
- 57.Cannon WR, Garrison BJ, Benkovic SJ. J. Am. Chem. Soc. 1997;119:2386–2395. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.