Abstract
Agonists activating nicotinic acetylcholine receptors (nAChR) include potential therapeutic agents and also toxicants such as epibatidine and neonicotinoid insecticides with a chloropyridinyl substituent. Nicotinic agonist interactions with mollusk (Aplysia californica) acetylcholine binding protein, a soluble surrogate of the nAChR extracellular domain, are precisely defined by scanning with 17 methionine and tyrosine mutants within the binding site by photoaffinity labeling with 5-azido-6-chloropyridin-3-yl probes that have similar affinities to their nonazido counterparts. Methionine and tryrosine are the only residues found derivatized, and their reactivity exquisitely depends on the direction of the azido moiety and its apposition to the reactive amino acid side chains.
Introduction
The nicotinic acetylcholine (ACha) receptor (nAChR) is the prototypical agonist-gated ion channel responsible for rapid excitatory neurotransmission.1 The nAChR agonists include many toxicants, potential therapeutic agents acting peripherally or centrally, and the important neonicotinoid insecticides.2–5 Initial attempts to understand agonist-receptor interactions involved site-directed or chimeric mutagenesis, estimating the role of specific region(s) or amino acid(s) on ligand occupation and/or the pharmacological response. The structural biology approach of high-resolution X-ray crystallography reveals orientations of functional amino acids at atomic resolution in the ligand-bound state and conformational rearrangements of the protein upon ligand interaction. Alternatively, incorporation of an unnatural amino acid or photoaffinity labeling defines the receptor recognition properties in a physiologically-relevant, aqueous solution environment.6,7 Binding site interactions of nicotinoid and neonicotinoid agonists have been characterized by comparative structural and chemical biology approaches8–13 using mollusk ACh binding protein (AChBP), which is a suitable structural surrogate of the extracellular ligand-binding domain of the nAChR.9,14
5-Azido-6-chloropyridin-3-yl nicotinic photoaffinity probes have played an important role in studying agonist-nAChR interactions.7 In principle, these probes bind reversibly to the specific site and then the reactive nitrene intermediate, generated by photoirradiation, reacts covalently with the target protein (Chart 1). Upon photoaffinity labeling of Aplysia californica AChBP, the azidochloropyridinyl photoprobes reacted at one location which was at the interface between loop C (Y195) on the principal or (+)-face subunit and loop E (M116) on the partnering or (−)-face subunit (Figure 1),10,11 establishing a bound ligand position and conformation consistent with those observed in crystal structures.8,9,13 Interestingly, a nicotinic photoaffinity ligand of this type exclusively labeled the α4 subunit of the chick α4β2 nAChR subtype,10 in which Y225 in loop C on the α4 subunit and F137 in loop E on the β2 subunit are in homologous residue positions, spatially equivalent to Y195 and M116, respectively, of AChBP. These observations may be rationalized by the difference in the reactivity between amino acids and the photoactivated probe molecule. Therefore, the present investigation designs a methionine- and tyrosine-scanning approach on the loops C and E domains involving 17 AChBP mutants regarding photoreactivity of the 5-azido-6-chloropyridin-3-yl nicotinic photoprobes,10,15–17 ultimately mapping the specific site undergoing photoderivatization and the precise position and conformation of the bound ligand.
Chart 1.
Two Chemotypes of Nicotinic Agonists and Their 5-Azido-Substituted Photoaffinity Probes (top) and Scheme for Photoaffinity Labeling (bottom)a.
aEpibatidine, azidoepibatidine, neonicotinoid thiacloprid olefin analogue, and the azido derivative are designated by EPI, AzEPI, THO, and AzTHO, respectively. Asterisks indicate positions of tritium.
Figure 1.

AzEPI-docked structure of Aplysia AChBP, indicating the positions for the mutants studied. Relevant amino acids in yellow are from the (+)-face or principal subunit while in aquamarine are from the (−)-face or partnering subunit. This docking image is regenerated with the calculation data from our earlier report.10 Distances between the photoactive nitrene and the thiomethyl group of the methionine and the tyrosine hydroxyl group are essentially equivalent (~4 Å).
Results and Discussion
Binding Affinities of Nicotinic Agonists and the Photoaffinity Probes to the AChBP Mutants (Table 1 and Supporting Information)
Table 1.
Binding Affinities of Nicotinic Agonists and the Photoaffinity Probes and [3H]AzEPI Photoaffinity Labeling of the Aplysia AChBP Mutants
|
Kd, nMa, b |
|||||
|---|---|---|---|---|---|
| nicotinoid |
neonicotinoid |
Relative photolabeling, %a, c |
|||
| Mutant | EPI | AzEPI | THO | AzTHO | |
| WT | 8.6 | 10 | 2.8 | 1.7 | 100 |
| M116L | 9.0 | 21 | 11 | 9.6 | 121 |
| Y195F | 21 | 8.6 | 3.1 | 2.3 | 3.4 |
| M116L, Y195F | 22 | 10 | 4.7 | 3.0 | 6.9 |
| I106M, M116L | 12 | 30 | 4.3 | 3.2 | 90 |
| I106M, M116L, Y195F | 29 | 22 | 4.9 | 5.5 | 13 |
| A107M | 10 | 7.2 | 4.0 | 3.0 | 93 |
| A107M, M116L | 7.5 | 8.1 | 3.0 | 1.6 | 100 |
| A107M, M116L, Y195F | 25 | 37 | 20 | 17 | 13 |
| V108M | 7.6 | 1.5 | 9.2 | 1.1 | 33 |
| V108M, M116L | 2.9 | 0.58 | 4.8 | 0.46 | 28 |
| V108M, M116L, Y195F | 2.9 | 0.33 | 1.8 | 0.17 | 0 |
| S114M | 16 | 12 | 6.2 | 2.0 | d |
| S114M, M116L | 12 | 15 | 5.6 | 3.0 | 66 |
| S114M, M116L, Y195F | 24 | 19 | 13 | 7.2 | 13 |
| I118M | 10 | 4.5 | 239 | 70 | d |
| I118M, M116L | 5.0 | 4.8 | 299 | 68 | 55 |
| I118M, M116L, Y195F | 24 | 12 | 247 | 172 | 16 |
| M116Y | 57 | 34 | 144 | 27 | 59 |
| M116Y, Y195F | 115 | 67 | 60 | 19 | 30 |
Kd±SD (nM, n=3) and photoincorporation (pmol±SD, n=3–6) are given in Supporting Information.
Dissociation constants (Kd) determined for EPI by direct radioligand saturation and for the other three ligands by competition with [3H]EPI binding.
AChBP (38 pmol sites, 1 μg protein) was photoaffinity-labeled by [3H]AzEPI (46 pmol, 1.2-fold excess) alone (total) and protected with competitor [(±)-EPI 10000 pmol, nonspecific labeling]. Photoincorporation was quantified by scintillation counting of sliced protein bands from SDS-PAGE gels. Specific photoincorporation is defined by the difference between total and nonspecific derivatizations. Relative labeling refers to the mutant compared to the WT as 100%.
Single mutants S114M and I118M assessed for binding potency were not available for photoaffinity labeling.
Effect of Mutations on Nicotinoid and Neonicotinoid Binding
This study utilizes site-directed mutagenesis, particularly on the loop E domain in the ligand-binding pocket, to analyze interactions with the chloropyridinyl moiety of two chemotypes of nicotinic agonists: the nicotinoids [epibatidine (EPI) and azidoepibatidine (AzEPI)] and neonicotinoids [thiacloprid olefin (THO) and its azido derivative (AzTHO)]. Binding affinities (dissociation constants, Kd) were analyzed as inhibitors of [3H]EPI binding to the AChBP wild type (WT) and 19 mutants. Nicotinoids and neonicotinoids showed a rank order of binding affinity similar to Aplysia WT AChBP11 for most of the mutant AChBPs. However, neonicotinoids were clearly less active with I118M, I118M/M116L, and I118/M116L/Y195F mutants. Position 118 is not a critical point in distinguishing the two chemotype ligands (nicotinoid and neonicotinoid),11–13 thereby implying that the unexpected effect of this mutation involves a more extensive conformational alteration of the binding domain.
Effect of the Azido Substituent
To determine the accessibility of the photoactivatable azido moiety of AzEPI and AzTHO, their binding affinities were compared with their nonazido parent compounds. In general, little if any effect was observed upon introduction of the azido group as indicated by a correlation plot (Figure 2), except for three mutants V108M, V108M/M116L, and V108M/M116L/Y195F which had relatively large affinity differences between azido probes and nonazido parents (azido>nonazido). THO showed greatly diminished binding affinity with I118M, I118M/M116L, and I118M/M116L/Y195F mutants. These mutant AChBPs are the ones retaining higher affinity for AzTHO than the parent. Interestingly, M116Y and M116Y/Y195F were less sensitive to both chemotypes of parent compounds and their azido analogues also displayed an enhanced affinity. These results clearly indicate that conformational alterations of the regional binding subsite induced by these mutations influence the ligand binding. Moreover, the enhanced affinity of the azido probes compared with the nonazido parents suggests that the position of the non-azido pyridine ring in the agonist rotates to confer diminished affinity in these mutants, whereas the binding conformation of the photoprobe with an azido substituent retains the conserved direction. Therefore, the mutagenesis approach analyzed as pharmacological response in this case may have distinct limitations in precisely determining the binding position and conformation of the agonist.
Figure 2.
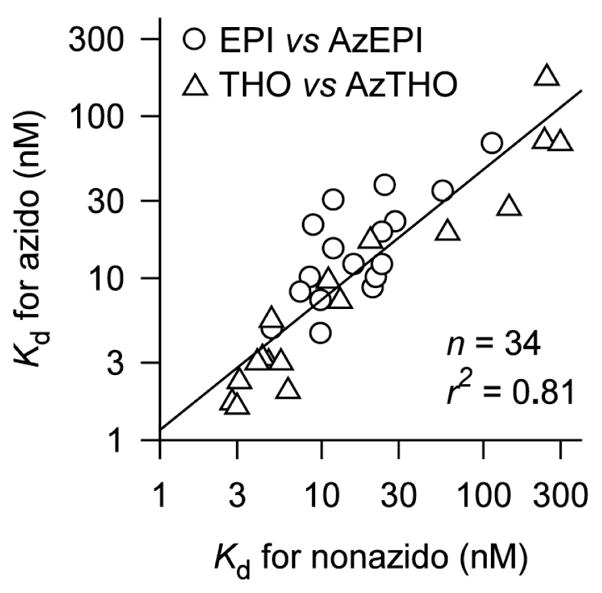
Relationship between binding affinities to the AChBP WT and mutants of nonazido and the corresponding azido nicotinoid (EPI versus AzEPI) or neonicotinoid (THO versus AzTHO). Data are from Table 1 excluding those for V108M, V108M/M116L, and V108M/M116L/Y195F mutants (see text).
Photoaffinity Labeling of AChBP Mutants
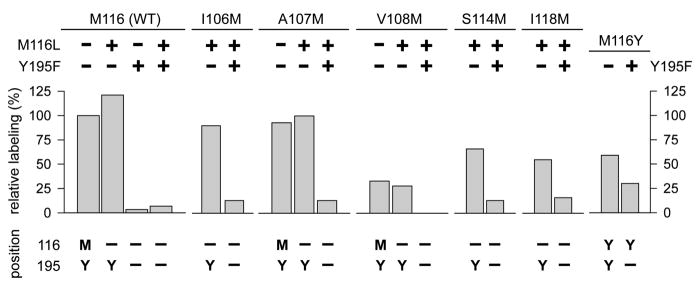
[3H]AzEPI was used to quantify or visualize photoincorporation into the AChBP WT and 17 mutants involving 7 positions. The results on methionine- or tyrosine-scanning reflect both the binding interaction of the probe and the photoreaction between the photoactivated nitrene intermediate and the amino acid with respect to spatial proximity. With the WT, [3H]AzEPI specifically photoreacted with 11.2 of 38 pmol sites (Table 1, Figure 3, and supporting information). The M116L mutation slightly enhanced the photolabeling and in contrast Y195F and double mutant M116L/Y195F almost completely abolished the photoincorporation, suggesting that the phenol side chain in Y195 is the critical site for derivatization, and M116L combined with Y195F abolishes reactivity with the photoactivated nitrene intermediate. Surprisingly M116 is not labeled in single mutant Y195F AChBP, indicating that the presence of tyrosine at 195, perhaps because of spatial positioning, is not only required for its own labeling, but also for labeling of the neighboring methionine.
Figure 3.
[3H]AzEPI photoaffinity-labeled Aplysia AChBP mutants (shown as representative results) comparing fluorography with Coomassie blue-stained protein. The experimental condition is given in Table 1 footnote. The image consists of two separate SDS-PAGE runs with different film-exposure periods. The Y195F mutant shown as a doublet is considered to be the full length protein and the cleaved product. Similarly, in the WT protein, the cleaved product lacking the last 4 amino acids (N-terminal positions 216–219, NLFD) is sometimes observed with no influence on ligand binding and photoaffinity labeling.10,11
Methionine-scanning was then performed on the loop E domain (I106, A107, V108, S114, and I118) alone or in combination with M116L and Y195F (double and triple mutations) to examine whether the derivatization site shifts to the newly-introduced mutated-methionines (Table 1 and Figure 4). Positions I106M and A107M in the single or double mutant with M116L conferred similar or lower photoincorporation compared with that of the WT or M116L single mutant, respectively. However, the triple mutant, incorporating Y195F, greatly diminished the derivatization. V108M and double V108M/M116L mutants (i.e., with or without methionine at 116) resulted in moderately reduced incorporation, while double mutant S114M/M116L or I118M/M116L gave higher photoderivatization than those of V108 single or double mutant with M116L; combining these mutations with Y195F to give triple mutations also greatly diminished the photoincorporation. These labeling patterns are in full agreement with the crystallographic binding site structure of AChBP. A mutationally-introduced methionine at any of these positions does not enhance labeling because the reactive side chain would not face the 5-azido or 5-nitrene moiety of the bound probe in the WT. The introduced methionine at 108, although conceivably proximal to the nitrene moiety of the probe, is nevertheless spatially restrictive for derivatization. Methionine at 114 and 118 may moderately influence the binding itself or labeling due to spatial hindrance. Therefore, the direction of the nitrene group of the agonist probe is substantially conserved in the optimal binding conformation.
Figure 4.
Effects of mutagenesis on [3H]AzEPI photoaffinity labeling and positions and amino acids for photoderivatization. Relative labeling (%) was taken from Table 1.
When tyrosine was introduced at position 116, the M116Y mutant showed 46–55 % [3H]AzEPI incorporation compared with that of the WT or M116L single mutant. Interestingly, the double mutant M116Y/Y195F still retained significant photoincorporation (30 %) compared to those of other mutants with Y195F (0–16 %), providing direct evidence that tyrosine at position 116 may become a mandatory derivatization site when tyrosine at 195 is not present. Tyrosine in the M116Y mutant presumably interferes with the access of AzEPI into the binding site, leading to diminished derivatization, but the tyrosine-OH is sufficiently proximal to be derivatized by EPI nitrene in the double mutant with Y195F.
Mass Spectrometry (MS) Analysis
MS analysis of photolabeled-AChBP was used to identify the site of modification using representative single mutants M116L and M116Y and two chemotypes of probes AzEPI and AzTHO (nicotinoid and neonicotinoid, respectively) (Chart 1). With the M116L mutant, HPLC/MS of the labeling mixture for each photoprobe revealed unmodified (molecular mass 27228–27229 Da with accuracy approximately 1–2 Da for proteins in this mass range) and covalently-derivatized AChBP subunit (molecular masses 27451 and 27492 Da for EPI and THO nitrene molecules, respectively) based on the expected mass increase for the nitrene-derived product (222 and 263 Da, respectively). Similarly, the M116Y mutant gave modified proteins with the mass increase of 221 and 262 Da for EPI and THO nitrene-derived molecules, respectively, although the intensity was considerably lower than that in the M116L mutant (data not shown) or WT10,11 as observed in the [3H]AzEPI labeling patterns (Figure 4). These results on the intact modified protein indicate that the photoactivated nitrene probe is incorporated into AChBP with no more than one agonist molecule for each interfacial ligand-binding pocket. Thus, with an occupation of 30% (see supporting information) or a stoichiometry of 0.3, 30% of the subunits should have a single linked photoprobe.
MS/MS analysis for modified M116L AChBP mutant by the two nicotinic photoprobes unambiguously and exclusively pinpointed one derivatized site at Y195 on loop C of the (+)-face subunit [Table 2 and Figure 5 showing collision-induced dissociation (CID) data for the THO-nitrene-labeled peptide as an example]. With the M116Y mutant, the photoprobes modified Y195 and, in addition, the modified peptide containing tyrosine at the 116 position (TSAADIWTPDITAYSSTRPVQVLSPQIAVVTHDGSVYFIPAQR) was detected [m/z 1231.1128 (4+) − MH+ = 4921.4278 (+ 7 ppm)] for THO-nitrene-labeled peptide (+263 Da), although the derivatized site could not be identified unambiguously from the CID spectrum. These findings are fully consistent with the [3H]AzEPI labeling results.
Table 2.
Tryptic Peptides of AChBP M116L and M116Y Mutants Photoaffinity Labeled by Two Nitrene Probes Identified From CID Data and Accurate Masses Determined by FTICR Mass Spectrometry
| Molecular mass, Da |
|||
|---|---|---|---|
| Nitrene probe | Precursor m/z (z) | Measured | Theoretical |
| M116L, Q184-K203 with Y195 modified | |||
| EPI | 1335.1294 (2+) | 2668.2432 (+4 ppm)a | 2668.2328 |
| THO | 1356.0891(2+) | 2710.1626 (<1 ppm)a | 2710.1640 |
| M116Y, Q184-K203 with Y195 modified | |||
| EPI | 890.4220 (3+) | 2668.2426 (+4 ppm)a | 2668.2328 |
| THO | 1356.0944 (2+) | 2710.1732 (+3 ppm)a | 2710.1640 |
The error of the mass measurement is shown in parenthesis.
Figure 5.
Low energy CID spectrum of the Q184-K203 peptide of M116L AChBP mutant which is THO-nitrene-modified at Y195. Data were acquired in a LC/MS/MS experiment on a QSTAR Elite mass spectrometer, precursor ion at m/z 904.38(3+). The “M” subscript indicates fragments featuring the modification. From this spectrum the modification site can be identified unambiguously: C-terminal fragment y8 was detected unmodified while the next member of this fragment series already displays the mass shift corresponding to the modification. C* stands for carbamidomethyl cysteines.
When taken together, our studies show labeling at two positions, exemplified by tyrosine 195 and methionine 116 in the wild type,10,11 with labeling at M116 requiring the presence of Y195. Interestingly, other methionines on the complementary face will not substitute as a labeling acceptor for 116. The phenolic hydroxyl on tyrosine and the side chain of methionine are suitable photoreactive acceptors owing to the polarizability of the aromatic ring and the thiomethyl moiety, respectively.
Docking Simulations (Figure 6)
Figure 6.
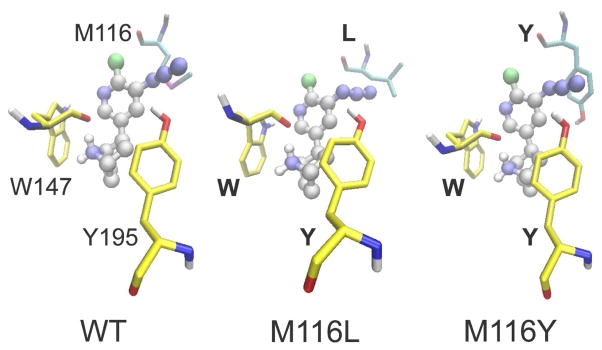
Docking simulations of AzEPI into the AChBP agonist binding site of WT (left) and M116L (middle) and M116Y (right) mutants emphasizing methionine, leucine, and tyrosine at 116 (loop E on partnering subunit in aquamarine) and Y195 (loop C on principal subunit in yellow) positions. Loop B W147 (in yellow) on the principal subunit, important for interactions with the AzEPI +NH2 (bridged nitrogen) moiety via H-bonding and cation-π contact, is also displayed. The nitrene nitrogen from azido photolysis is approximately equal distant (4 Å) from the methionine thiomethyl moiety and the tyrosine hydroxyl oxygen.
AzEPI is calculated to dock into the WT AChBP (2BYQ, EPI-bound crystal structure) and in silico mutants M116L and M116Y with energies of −9.69, −9.75, and −10.10 kcal/mol, respectively. In the WT, the azido group of AzEPI is suitably positioned at the interface between Y195 and M116, allowing derivatization by photoactivated EPI-nitrene. The nitrene nitrogen arising from azido photolysis is approximately equal distant (4 Å) from the methionine thiomethyl and the tyrosine hydroxyl oxygen. The AzEPI binding location and conformation is fully superimposable onto the EPI captured in the AChBP crystal structure.9 The leucine side chain of M116L faces the outside [similar to that of L112 in Lymnaea AChBP liganded with nicotine (1UW6)8] compared with the methionine of the WT, presumably conferring easy access for binding of the azido moiety. The azido group can rotate at the C-N bond during binding and thus accommodate differing side chain steric requirements in the mutants relative to the WT. The Y195 is appropriately positioned for this exclusive modification. The tyrosine of M116Y was minimized to give the energetically-optimal geometry which may spatially impair the access of the azido moiety but is sufficiently proximal (about 4 Å) to be derivatized by the nitrene intermediate as observed in binding and photolabeling experiments. Although the tyrosine side chain at the 116 position could possibly twist out of the binding pocket, the calculated lowest energy geometry has this tyrosine side chain superimposed with that of the methionine in the WT (image not shown). In all three cases, the AzEPI cationic bridged nitrogen (+NH2) H-bonds with the backbone carbonyl oxygen of W147 (<2 Å) and is positioned for a cation-π interaction with the indole side chain (3–4 Å). These simulations fully account for the binding and photoaffinity labeling results.
An overlay of two neonicotinoid and one nicotinoid crystal structures (Figure 7) reveals a linkage of entrapped water molecules extending from the first which is hydrogen bonded to the pyridine nitrogen at the subunit interface. The extension of the three other water molecules reaches into the channel vestibule. The connection plus a similar one for the bound nicotine agonist in the Lymnaea AChBP8 suggests a network that could be disrupted through mutagenesis or the attachment of a bulky substituent. All of this points to the necessity of the pyridine and the azido- or chlorine-substituted pyridines maintaining a fixed position in the bound state. Mutations in this region may alter the conduit or linkage of these water molecules.
Figure 7.
Overlay of nicotinoid EPI (purple) and neonicotinoids thiacloprid (silver) and imidacloprid (orange) bound to Aplysia AChBP WT.9,13 The pyridine nitrogens in the AChBP bound nicotinoids and neonicotinoids are associated with one water molecule (red ball) that forms a series of polar contacts [W147-εN, I106-O, I118-O, and I118-εN] plus three other water molecules extending to the vestibule surface (S103-OH, S103-O, P104-O, and P119-N). The amino acids in aquamarine are from the complementary or (−)-face subunit and the one in yellow is from the principal or (+)-face subunit. The distances indicated by dashed-lines are less than 4 Å. The neonicotinoids imidacloprid and thiacloprid are analogues of THO and EPI (see Chart 1).
Conclusion
In site-directed or chimeric mutagenesis, attenuated or enhanced pharmacological responses could be directly attributable to modified interacting determinant side chains in the binding site or indirectly arise from altered conformational states of the receptor. Crystallization may not reveal the exact binding orientations observed in physiological medium, since crystal packing and the position of the symmetry related molecule may influence the selection of conformational states. On the other hand, photoaffinity labeling may preferentially involve the more reactive residues and stable derivatization, such as with tyrosine and methionine, leading to a possible interpretative bias, since other proximal side chains may be far less reactive. Accordingly, comparisons of solution- (mutagenesis and chemical approaches) and crystal-based structures offer a more comprehensive and precise picture of multiple binding site interactions. The present investigation defines the binding position and conformation of nicotinic agonists at the AChBP binding pocket based on methionine- or tyrosine-scanning with respect to azidochloropyridinyl nicotinoid and neonicotinoid photoaffinity labeling, i.e. the influence of the neighboring amino acids and the efficiency of photoderivatization. The agonist photoprobe is nestled in the fully-conserved binding position and conformation and the direction of the azido or nitrene moiety of the probe is independent of that for the potentially-reacting amino acid side chains. These findings, combined with the structural biology results9,13 may prompt structure-guided design of nicotinic therapeutic agents and insecticides with enhanced effectiveness and safety.
Experimental Section
Chemicals
AzEPI or [3H]AzEPI, THO and AzTHO were available from our earlier studies.10,15,17 (±)-EPI HCl and [3H]EPI were purchased from TOCRIS (Ellisville, MO) and Amersham Biosciences (Piscataway, NJ), respectively.
Expression, Mutagenesis, and Purification of AChBPs
WT AChBP from Aplysia californica was expressed from a cDNA synthesized from oligonucleotides selected for mammalian codon usage in HEK cells, as previously described.18,19 The AChBP gene was inserted into a pFLAG-CMV-3 expression vector (Sigma, St. Louis, Mo) containing a preprotrypsin leader peptide followed by a NH2-terminal FLAG epitope. Mutant AChBPs were generated by polymerase chain reaction-mediated standard mutagenesis procedures, and cassettes containing the mutation were subcloned into the WT vector and verified by double-stranded sequencing. WT and mutant AChBP-transfected HEK-293S cells lacking the N-acetylglucosaminyltransferase I gene20 were selected with G418 to generate stably expressing cell lines. Cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 3% fetal bovine serum and media was harvested at 3-day intervals for up to 4 weeks. Adsorption onto a FLAG antibody column followed by elution with the FLAG peptide yielded purified protein in quantities between 0.5 and 2 mg/L. Purity and assembly of subunits as a pentamer were assessed by SDS-PAGE and fast protein liquid chromatography.
Radioligand Binding and Photoaffinity Labeling
Affinities of test chemicals (reciprocals of the Kd values) for the AChBP mutants were evaluated by an adaptation of a scintillation proximity assay with [3H]EPI.19 Photoaffinity labeling experiments and MS measurements of derivatized intact AChBP subunit and MS/MS analyses of tryptic fragments pinpointing the sites of modification were performed according to our previous methodologies.10–12
Calculations
Mutants M116L and M116Y of 2BYQ (EPI-bound AChBP crystal structure) were made in silico using Maestro 8.5 (Schrodinger, LLC, Portland, OR). In each case, several side chain rotamers were generated as starting structures for minimizations and conformational searches based on their representation. The EMBRACE algorithm in Macromodel 9.6, including the OPLS 2005 force field and a GB/SA water solvent model,21–23 was used to explore the conformational space of the mutant side chain along with all residues within 8 Å of the bound EPI.9 One hundred conformational search steps were interleaved with 500 step minimizations on each of the starting mutant rotamers in order to generate reasonable structural models for the receptor mutants. The best mutant receptor models for M116L and M116Y generated in this fashion were subsequently used for docking with AzEPI. Autodock424,25 with 100 steps of the Lamarkian genetic algorithm in a 40 point grid centered on the active site was used for each ligand-receptor pair. Top hits were selected within 1 kcal/mole of the best calculated binding energy.
Supplementary Material
Acknowledgments
T.T.T. and P.T. were funded by NIH grants R37-GM18360 and UO1-NS05846; M.T. and J.E.C. by NIEHS grant R01 ES08424; and D.M., K.F.M., and A.L.B. by NIH NCRR grants RR015084, RR001614, and RR019934. J.E.C. was supported by the William Muriece Hoskins Chair in Chemical and Molecular Entomology and K.A.D. by NSF grant CHE-0233882. J.M.C.-B. acknowledges a 2007-2008 UC MEXUS-CONACYT Faculty Fellowship.
Footnotes
Abbreviations: ACh, acetylcholine; AChBP, ACh binding protein; AzEPI or [3H]AzEPI, azidoepibatidine photoprobe or its radioligand; AzTHO, azidoneonicotinoid photoprobe; CID, collision-induced dissociation; EPI or [3H]EPI, epibatidine or its radioligand; Kd, dissociation constant; nAChR, nicotinic ACh receptor; THO, neonicotinoid thiacloprid olefin; WT, wild type.
Supporting Information Available. One table giving binding affinities (Kd±SD, n=3) of nicotinic agonists and photoaffinity probes and a second table giving photoincorporation (pmol±SD, n=3–6) for total, nonspecific and specific labeling of the Aplysia AChBP mutants. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Changeux J-P, Edelstein SJ. Nicotinic Acetylcholine Receptors: From Molecular Biology to Cognition. Vol. 284. Odile Jacob; New York: 2005. [Google Scholar]
- 2.Cassels BK, Bermúdez I, Dajas F, Abin-Carriquiry JA, Wonnacott S. From ligand design to therapeutic efficacy: The challenge for nicotinic receptor research. Drug Discov Today. 2005;10:1657–1665. doi: 10.1016/S1359-6446(05)03665-2. [DOI] [PubMed] [Google Scholar]
- 3.Daly JW. Nicotinic agonists, anatagonists, and modulators from natural sources. Cell Mol Neurobiol. 2005;25:513–552. doi: 10.1007/s10571-005-3968-4. [DOI] [PubMed] [Google Scholar]
- 4.Tomizawa M, Casida JE. Neonicotinoid insecticide toxicology: Mechanisms of selective action. Annu Rev Pharmacol Toxicol. 2005;45:247–268. doi: 10.1146/annurev.pharmtox.45.120403.095930. [DOI] [PubMed] [Google Scholar]
- 5.Taylor P. Agents acting at the neuromuscular junction and autonomic ganglia. In: Brunton L, Lazo JS, Parker KL, editors. Goodman and Gilman’s Pharmacological Basis of Therapeutics. 11. McGraw-Hill; New York: 2006. pp. 217–236. [Google Scholar]
- 6.Dougherty DA. Cys-loop neuroreceptors: structure to the rescue? . Chem Rev. 2008;108:1642–1653. doi: 10.1021/cr078207z. [DOI] [PubMed] [Google Scholar]
- 7.Tomizawa M, Casida JE. Molecular recognition of neonicotinoid insecticides: The determinants of life or death. Accounts Chem Res. 2009;42:260–269. doi: 10.1021/ar800131p. [DOI] [PubMed] [Google Scholar]
- 8.Celie PHN, van Rossum-Fikkert SE, van Dijk WJ, Brejc K, Smit AB, Sixma TK. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–914. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 9.Hansen SB, Sulzenbacher G, Huxford T, Marchot P, Taylor P, Bourne Y. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 2005;24:3635–3646. doi: 10.1038/sj.emboj.7600828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomizawa M, Maltby D, Medzihradszky KF, Zhang N, Durkin KA, Presley J, Talley TT, Taylor P, Burlingame AL, Casida JE. Defining nicotinic agonist binding surfaces through photoaffinity labeling. Biochemistry. 2007;46:8798–8806. doi: 10.1021/bi700667v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomizawa M, Talley TT, Maltby D, Durkin KA, Medzihradszky KF, Burlingame AL, Taylor P, Casida JE. Mapping the elusive neonicotinoid binding site. Proc Natl Acad Sci USA. 2007;104:9075–9080. doi: 10.1073/pnas.0703309104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomizawa M, Maltby D, Talley TT, Durkin KA, Medzihradszky KF, Burlingame AL, Taylor P, Casida JE. Atypical nicotinic agonist bound conformations conferring subtype selectivity. Proc Natl Acad Sci USA. 2008;105:1728–1732. doi: 10.1073/pnas.0711724105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talley TT, Harel M, Hibbs RH, Radić Z, Tomizawa M, Casida JE, Taylor P. Atomic interactions of neonicotinoid agonists with AChBP: Molecular recognition of the distinctive electronegative pharmacophore. Proc Natl Acad Sci USA. 2008;105:7606–7611. doi: 10.1073/pnas.0802197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 15.Tomizawa M, Wen Z, Chin HL, Morimoto H, Kayser H, Casida JE. Photoaffinity labeling of insect nicotinic acetylcholine receptors with a novel [3H]azidoneonicotinoid. J Neurochem. 2001;78:1359–1366. doi: 10.1046/j.1471-4159.2001.00518.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang N, Tomizawa M, Casida JE. Structural features of azidopyridinyl neonicotinoid probes conferring high affinity and selectivity for mammalian α4β2 and Drosophila nicotinic receptors. J Med Chem. 2002;45:2832–2840. doi: 10.1021/jm010508s. [DOI] [PubMed] [Google Scholar]
- 17.Zhang N, Tomizawa M, Casida JE. 5-Azidoepibatidine: An exceptionally potent photoaffinity ligand for neuronal α4β2 and α7 nicotinic acetylcholine receptors. Bioorg Med Chem Lett. 2003;13:525–527. doi: 10.1016/s0960-894x(02)00936-8. [DOI] [PubMed] [Google Scholar]
- 18.Hibbs RE, Talley TT, Taylor P. Acrylodan-conjugated cysteine side chains reveal conformational state and ligand site locations of the acetylcholine-binding protein. J Biol Chem. 2004;279:28483–28491. doi: 10.1074/jbc.M403713200. [DOI] [PubMed] [Google Scholar]
- 19.Talley TT, Yalda S, Ho KY, Tor Y, Soti FS, Kem WR, Taylor P. Spectroscopic analysis of benzylidene anabaseine complexes with acetylcholine binding proteins as models for ligand-nicotinic receptor interactions. Biochemistry. 2006;45:8894–8902. doi: 10.1021/bi060534y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reeves PJ, Callewaeer N, Contreras R, Khorana HG. Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohamadi F, Richard NGJ, Guida WC, Liskamp R, Lipton M, Caufield C, Chang G, Hendrickson T, Still WC. Macromodel, an integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J Comput Chem. 1990;11:440–467. [Google Scholar]
- 22.Still WC, Tempczyk A, Hawley RC, Hendrickson T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J Am Chem Soc. 1990;112:6127–6129. [Google Scholar]
- 23.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 24.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- 25.Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J Comput Chem. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.