Abstract
Purpose of review
The BCL6 transcriptional repressor is the most commonly involved oncogene in B-cell lymphomas. Sustained expression of BCL6 causes malignant transformation of germinal center (GC) B-cells. Understanding the mechanism of action of BCL6 is crucial for the study of how aberrant transcriptional programming leads to lymphomagenesis and development of targeted anti-lymphoma therapy.
Recent findings
Identification of BCL6 target genes indicate a critical role for BCL6 in facilitating a state of physiological genomic instability required for GC B-cells to undergo affinity maturation, and suggest its contribution to several additional cellular functions. The discovery of several layers of counter-regulatory mechanisms reveals how B-cells can control and fine-tune the potentially lymphomagenic actions of BCL6. From the biochemical standpoint, BCL6 can regulate distinct biological pathways through different cofactors. This observation explains how the biological actions of BCL6 can be physiologically controlled through separate mechanisms and affords the means for improved therapeutic targeting. The fact that patients with BCL6-dependent lymphoma can be identified based on gene signatures suggests that therapeutic trials of BCL6 inhibitors could be personalized to these individuals.
Summary
BCL6 plays a fundamental role in lymphomagenesis and is an excellent therapeutic target for development of improved anti-lymphoma therapeutic regimens.
Keywords: Transcriptional repression, BCL6, non-Hodgkin Lymphoma, transcription therapy
Introduction
BCL6 is a member of the BTB/POZ zinc finger family of transcriptional repressors and within the B-cell lineage is expressed exclusively during the GC phase of differentiation [1]. BCL6 is required for formation of GCs and its downregulation is required for B-cells to undergo further differentiation to memory cells or plasma cells [2,3]. BCL6 is often expressed constitutively in diffuse large B-cell lymphomas (DLBCLs), usually in association with translocations that fuse its coding sequence to heterologous promoters, or with point mutations in BCL6 promoter negative regulatory elements [4]. Constitutive expression of BCL6 in mice induces formation of DLBCL [5,6], and the presence of BCL6 is required for survival of human DLBCL cells [7,8]. This review will discuss recent advances in understanding the role of BCL6 in lymphomagenesis and its therapeutic targeting. Since this is a focused effort, a sampling of the most recent publications concerning BCL6 is discussed.
A double-edged sword: BCL6 facilitates a state of physiological genomic instability in GC dark zone B-cells
Germinal centers are transient structures that form within secondary lymphoid organs in response to T cell-dependent antigenic stimuli. Once B-cells enter the GC reaction they become centroblasts and acquire the ability to divide rapidly while simultaneously undergoing immunoglobulin somatic hypermutation (SHM) mediated by activation induced cytosine deaminase (AID). Coupling of mutation and proliferation allows the emergence of B-cell clones with a wide diversity of antibody mutations and increases the likelihood of some of these having high affinity for the offending antigen. Unfortunately, AID can also damage genes, especially those which are transcriptionally active in B-cells and contain somatic hypermutation hotspot sequences [9]. While AID was previously thought to mutate only a handful of genes in normal B-cells (such as BCL6) [10], Liu et. al. demonstrated that aberrant somatic hypermutation is actually widespread. 25% of transcriptionally active genes were mutated in an AID-dependent manner in normal Peyer's patch B-cells including potentially oncogenic genes such as BLK, SYK, PIM, RHOH, PAX5 and MYC. 43% of genes were mutated in B-cells from animals lacking the MSH2 and UNG components of the mismatch and base excision repair pathways, suggesting that these pathways normally oppose AID induced genomic mutagenesis [9]. These data show that GC B-cells have evolved the capability to tolerate a potentially dangerous state of physiological genomic instability. Although this may be a useful evolutionary adaptation to generate antibodies in opposition to continuous exposure to new microbes, this advantage does not come without cost, since a majority of B-cell neoplasms arise from GC derived cells. The critical role of AID and somatic hypermutation in lymphomagenesis was convincingly demonstrated by the finding that constitutive expression of BCL6 in mice failed to induce GC-type lymphomas in an AID deficient background [11].
The fact that GCs do not form in the absence of BCL6 suggested its involvement in facilitating the proliferative and genetically unstable centroblast phenotype, and along these lines, BCL6 was shown to attenuate DNA damage sensing [12]. Gain and loss of function studies in primary human B-cells and DLBCL cell lines showed that BCL6 impaired phosphorylation of histone 2AX and repair of double strand breaks after exposure to a DNA damaging agent [12]. This was due at least in part to BCL6-mediated transcriptional repression of ATR [12], which is a ubiquitously expressed gene product that normally functions as an important DNA damage sensor. ATR triggers a cascade of cellular checkpoints in response to single and double strand breaks. Through the actions of BCL6, centroblasts fail to trigger these responses. Moreover, BCL6 also represses key checkpoint genes downstream of ATR including CHEK1, TP53 and CDKN1A [8,13,14]. Sustained attenuation of DNA damage checkpoints induced by constitutive BCL6 expression is likely to play a fundamental role in lymphomagenesis. It is possible to envision a vicious circle leading to tumor initiation whereby BCL6 allows centroblasts to survive and proliferate in the presence of AID-induced genetic recombination, which can lead to constitutive expression of BCL6 due to genetic lesions; BCL6 can then in turn sustain the genomic instability and checkpoint deficient phenotype and at same time can block differentiation, allowing B-cells to undergo further AID induced mutagenesis and eventual malignant transformation (Fig. 1).
Figure 1. BCL6 and AID contribute to lymphomagenesis by facilitating genomic instability in GC B-cells.
A: Naïve B-cells are triggered to form germinal centers by T-cell dependent immune responses. B: AID and BCL6 are upregulated as B-cells differentiate into centroblasts. AID mediates somatic hypermutation while BCL6 represses ATR and TP53 to attenuate DNA damage responses, and represses PRDM1 to block further differentiation. C: AID induces genetic lesions that lead to constitutive expression of BCL6, which in turn leads to sustained repression of ATR, TP53 and PRDM1. This forces B-cells to lock in to the GC B-cell phenotype and prevents them from undergoing terminal differentiation (D). E: The combined action of AID and BCL6 result in genomic instability and lymphomagenesis. F: Constitutive expression of BCL6 maintains DLBCL cell survival and may lead to further mutagenesis.
BCL6 mediated repression of DNA damage checkpoints is also required to maintain the survival of established lymphoma cells. Depletion of BCL6 can induce cell death and growth arrest in DLBCL cells[8,12], and this could be partially complemented by inhibiting ATR or p53 after BCL6 depletion[12,15]. Administration of a BCL6 inhibitor peptide can induce p53 and cause cell death[7,15]. The BCL6-regulated checkpoint pathway can be harnessed therapeutically since the combination of a BCL6 peptide inhibitor with pharmacologic induction of p53 activity yielded additive to synergistic increases in killing of DLBCL cells[15].
B-cells can overcome BCL6 mediated checkpoint suppression through two independent mechanisms
The germinal center compartment is highly dynamic, and centroblasts undergoing clonal expansion eventually migrate to the more heterogeneous light zone region of the GC, which also contains T-cells, macrophages and follicular dendritic cells (FDCs). In vivo imaging of germinal centers suggest that B-cell transit is bidirectional, so that B-cells can recycle back to the dark zone [16]. Among the various events occurring in the light zone, B-cells were observed to make transient contact with T-cells lasting up to 20−30 minutes [17]. T-cells are a source of CD40 ligand. CD40 signaling in B-cells was shown to disrupt the association of BCL6 with the N-CoR corepressor. This was dependent on NFkB signaling downstream of the CD40 receptor, and caused the rapid ejection of N-CoR from the BCL6 repression complex associated with the ATR promoter[12]. This resulted in induction of ATR and allowed damaged B-cells to undergo cell death[12]. The results suggest a mechanism whereby centroblasts can undergo a T-cell dependent quality control step after clonal expansion, whereby induction of ATR enhances sensing of DNA damage, which can either be repaired or could trigger apoptosis in severely damaged cells. Consistent with this scenario, in vivo imaging of GCs revealed the presence of GC T-cells attached to blebs of dead B-cells[17]. CD40 blockade of the BCL6 repression complex is reversible and BCL6 repression of target genes can be restored when CD40 signaling is timed to mimic the transient interactions of B and T-cells [18]. This recovery of BCL6 functionality suggest that B-cells might be able to continue undergoing clonal expansion even after interacting with T-cells (Fig 2A).
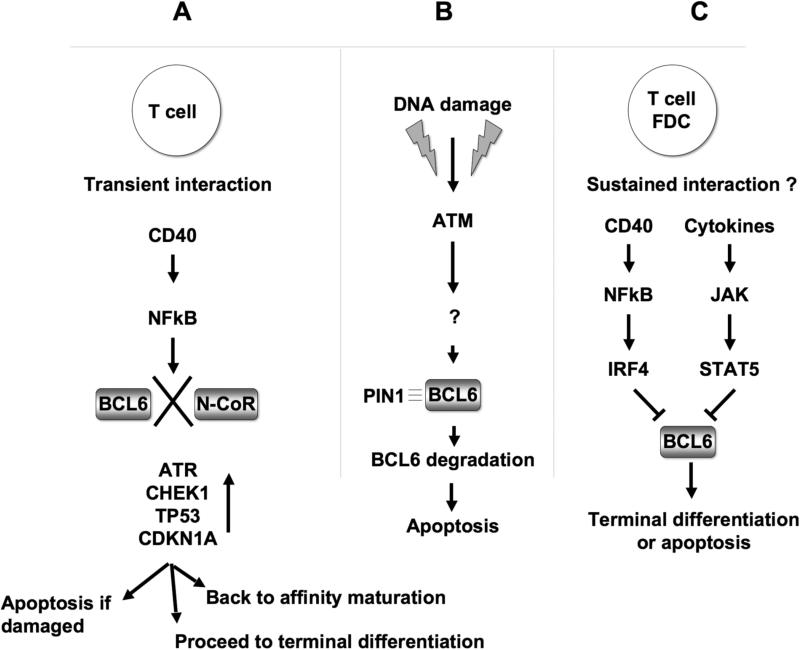
Figure 2. Inhibitory mechanisms for BCL6 function or expression.
A: Reversible inhibition of BCL6 through transient CD40 signaling: CD40 signaling triggers NFkB activity, which results in dissociation of the BCL6-N-CoR complex and release of BCL6 repression of checkpoint genes. This allows cells to undergo a DNA damage sensing quality control step leading to apoptosis if damage is severe; or once CD40 signaling ceases, to undergo additional rounds of affinity maturation, or to proceed to terminal differentiation. B: ATM induced BCL6 protein degradation. High levels of DNA damage trigger an ATM dependent cascade that results in BCL6 phosphorylation and association with PIN1, followed by proteolytic degradation of BCL6 and apoptosis. C: CD40 mediated transcriptional downregulation of BCL6. Sustained contact between FDCs and B-cells leads to NFkB induction of IRF4, which can repress BCL6, while at the same time cytokines can trigger JAK-STAT pathways whereby STAT5 can also repress BCL6. This leads to terminal differentiation of selected high affinity immunoglobulin producing cells.
BCL6 can also be regulated at the protein level in response to DNA damage. Accordingly, exposure of lymphoma cells to DNA damaging agents such as etoposide could induce phosphorylation and proteolytic degradation of BCL6. These events were dependent on activation of the ATM DNA damage sensor protein, which unlike ATR, is robustly expressed in centroblasts and plays an important role in affinity maturation. BCL6 degradation was at least partially blocked by exposure of cells to the ATM kinase inhibitor KU-55933 or to caffeine [19]. Degradation of BCL6 was dependent on the association of phosphorylated BCL6 with the PIN1 peptidyl-prolyl isomerase, and PIN1 deficient mice were observed to develop unusually large germinal centers, not unlike those observed in mice with constitutive expression of BCL6 [6,19]. The data suggest the existence of a failsafe mechanism whereby accumulation of DNA damage in centroblasts could trigger an ATM dependent pathway that can degrade BCL6 and allow damaged B-cells to undergo cell death (Fig 2B).
Failure to downregulate BCL6 upon completion of affinity maturation could contribute to lymphomagenesis
Upon completing affinity maturation B-cells expressing high affinity antibody are selected by FDCs to differentiate into memory or plasma cells. BCL6 downregulation is required for differentiation of GC-B cells into memory [20] and plasma cells [21]. Recent observations implicate CD40 and STAT5 as physiologic mediators of BCL6 transcriptional downregulation. Sustained CD40 signaling is believed to occur in B-cells only at the time of exit from the germinal center [22], most likely through contact with FDCs. Sustained CD40 stimulation was shown to induce expression of the IRF4 transcription factor via NFkB-dependent transactivation in GC cells [23]. IRF4 could in turn directly repress expression of BCL6 by binding to DNA elements within exon 1 and intron 1 of the BCL6 genomic locus. Depletion of IRF4 could at least partially antagonize CD40 mediated downregulation of BCL6. Loss of IRF4 binding sites was observed in BCL6 alleles affected by chromosomal translocations or SHM-induced point mutations, and could attenuate the repressor effect of IRF4, potentially contributing to BCL6 constitutive expression in DLBCL patients [23]. When ectopically expressed in Burkitt lymphoma cells, IRF4 was also shown to downregulate BCL6, inhibit cellular proliferation and induce plasma cell differentiation [24]. In Daudi lymphoma cells, ectopic expression of BCL6 was unable to counteract the anti-proliferative and differentiating effects of IRF4, suggesting that in addition to BCL6, other IRF4 target genes might contribute to its biological effects [24]. These data are in accordance with previous publications implicating IRF4 in plasma cell differentiation and suggest that a CD40-NFkB-IRF axis is a critical physiological contributor to BCL6 downregulation in post-GC B-cells (Fig 2C).
The grey zone of counter-regulation between BCL6, NFkB and STAT signaling
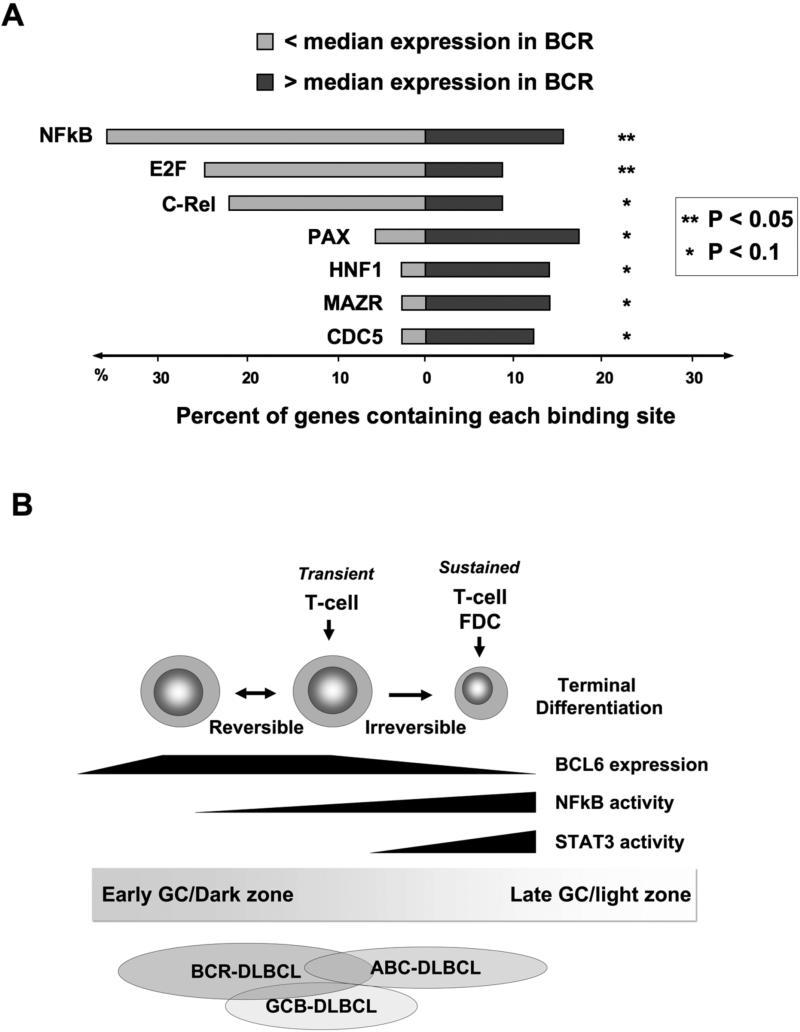
In addition to CD40, GC B-cells are also exposed to cytokines as they transit the GC light zone. These stimuli trigger the activity of NFkB and STAT signal transduction pathways, all of which are reported to be counter-regulatory to BCL6 in various ways. There appears to be somewhat of a gradient effect, whereby the expression and activity of BCL6 gradually decreases while those of NFkB and STATs increase during progression towards the GC exit. NFkB can disrupt BCL6 repression complexes [12] and downregulate its expression [23,24] as mentioned above. On the other hand, BCL6 can directly repress expression of NFkB1 (p105/p50) [25], and can physically associate and apparently inhibit the function of several NFkB subunits [26]. A TRANSFAC analysis of BCL6 target genes identified by ChIP-on-chip [27], revealed an over-representation of NFkB binding sites among those BCL6 target genes that were relatively repressed compared to median gene expression levels in DLBCL patient samples (FIG 3A); while a selection of NFkB target genes showed inverse correlation with BCL6 expression in a panel of DLBCL patients [26]. These events therefore seem to be at least in part recapitulated within DLBCL tumor cells, the origin of which is proposed to be reflective of the different stages of mature B-cell differentiation within or upon exiting GCs. One DLBCL microarray classification scheme divided tumors into germinal center B-cell type tumors (GCB) and activated B-cell tumors (ABC) [28]. The ABC DLBCLs display a gene signature consistent with NFkB activation possibly reflective of originating from late GC cells, and ABC cell lines were sensitive to killing by NFkB antagonists [29]. Nonetheless, several ABC cell lines still express BCL6 and can be killed by depleting or inhibiting BCL6 [7,15,27,30], possibly reflecting an area of the NFkB-BCL6 gradient where both pathways can co-exist and contribute to the cellular phenotype (FIG 3B). It remains to be seen how these tumor cell findings might play out in the complex milieu of the active germinal center.
Figure 3. The balance of power between BCL6, NFkB and STAT3.
A: A transcription factor binding site analysis (TRANSFAC) was performed within DNA sequences corresponding to the binding peak of BCL6 within target genes identified by ChIP-on-chip[27]. Light grey bars represent the percent of genes that are differentially expressed below the median in BCL6-dependent BCR-type (B-cell receptor signature) DLBCLs while the percent of genes above the median are represented in dark grey. The P value represents the significance of differential representation of genes above or below the median for each of the indicated transcription factor binding sites. B: BCL6 expression is upregulated when B-cells enter the germinal center reaction. In dark zone centroblasts BCL6 can block NFkB signaling through several mechanisms and directly repress STAT3 as described in the text. Transient interactions with T-cells can induce B-cell NFkB activity in a reversible fashion, which may allow B-cells to recycle back to the dark zone. Conversely, the actions of cytokines and/or more prolonged contact with FDCs or T-cells can tip the balance towards NFkB and STAT3 and downregulated BCL6 expression. GCB DLBCLs are generally proposed to reflect the biology of cells earlier in the germinal center reaction, while ABC DLBCLs are generally believed to represent later and/or exiting cells. The BCR/BCL6 dependent type of DLBCLs may partially overlap with both of these categories, but their BCL6 dependent biology suggests that they have features that overlap with GC B-cells in which BCL6 is functionally active.
NFkB can induce the expression of IL6 and IL10 in ABC cell lines, leading to autocrine activation of JAK/STAT3 pathways [31]. Higher expression of STAT3 was associated with a subset of ABC tumors with particularly high NFkB signaling. This is of interest from the therapeutic standpoint, since ABC tumors are more chemoresistant, and a pan-JAK kinase inhibitor could reduce STAT3 phosphorylation and induce cell death in ABC cell lines[31]. The importance of the NFkB-JAK/STAT axis was underlined by the fact that an IKKβ kinase inhibitor (MLN120B) could synergistically kill lymphoma cells in combination with JAK inhibition[31]. Interestingly, STAT3 is a BCL6 target gene and is expressed in GC B-cells at the edges of the germinal center as BCL6 becomes downregulated [32]. STAT3 expression was inversely correlated with BCL6 in DLBCLs [32]. BCL6 can also antagonize the actions of STAT3 and STAT6 by binding to overlapping BCL6/STAT binding elements as well as through physical interactions (e.g.[2,33]). Activation of STAT5 can occur in response to cytokine signaling for example through IL2 and IL5 in the germinal center light zone. STAT5 can bind directly to sites within BCL6 exon1 and intron1 and repress its expression[34]. The fact that these STAT5 binding sites can be ablated by point mutations in primary DLBCLs suggests that loss of STAT5 repressor activity can contribute to lymphomagenesis[34]. MAD1, another transcription factor induced by IL5 signaling can also bind to and repress the BCL6 promoter[35]. Taken together, the data suggest that BCL6 can hold NFkB and STAT signaling in check during the dark zone clonal expansion phase of the GC reaction. It is likely that many BCL6 positive DLBCLs arise from dark zone centroblasts and some of the lymphomas currently classified as GCB-type DLBCL presumably arise from these cells. However, events taking place in the light zone such as direct cellular contacts with FDCs and cytokine signaling can tip the balance away from BCL6 by transducing powerful activation signals that induce transcriptional downregulation of BCL6 and terminal differentiation. Thus, even though BCL6 is not usually thought of as associated with ABC type lymphoma, it probably still plays a critical role in their initiation since BCL6 translocations are more common in this DLBCL subtype.
BCL6 controls different biological functions through distinct biochemical mechanisms (Fig 4A)
Figure 4. BCL6 regulates different biological functions through distinct corepressors.
A: The graphical representation depicts two molecules of BCL6 forming a homodimer through the BTB domain. The BCL6 BTB domain can recruit SMRT, N-CoR and BCoR through the lateral groove. Repression of ATR, CHEK1, TP53 and CDKN1A is dependent on these corepressors and mediates survival and proliferation during AID induced somatic hypermutation. A region of BCL6 proximal to the BTB domain recruits CtBP, which can mediate negative autoregulation of BCL6. The central second repression domain (RD2) of BCL6 recruits an MTA3/NuRD complex, which is required for repression of PRDM1 and thus mediates differentiation blockade. The zinc finger (ZF) domain can bind directly to ETO [52], although a specific biological function for this corepressor is not yet known. B: A gene ontology analysis of BCL6 target genes identified by ChIP-on-chip shows them to be associated with specific biological functions as indicated in the pie chart.
As a transcription factor, BCL6 must recruit a series of corepressor proteins in order to repress its target genes. The BCL6 BTB domain forms an obligate dimer with autonomous repressor activity, and can bind directly and in a mutually exclusive manner to the SMRT, N-CoR and BCoR corepressors [36,37]. N-CoR and SMRT are highly conserved scaffold/adaptor proteins both of which can individually form a stable complex with HDAC3, TBL1, TBLR1 and GPS2 [38]. Both N-CoR and SMRT bind to BCL6 via a conserved 17-residue BCL6 binding domain (BBD) [37]. A BCL6 BTB lateral groove motif makes extensive contact with the SMRT BBD via amino acid side chains that emerge from the BTB dimer interface. These residues are not conserved in other BTB domain transcription factors, which therefore cannot bind to the SMRT/N-CoR BBD [37]. Remarkably, a 17-mer peptide corresponding to residues 498 to 514 of BCoR can bind to the same BCL6 BTB lateral groove position, even though it has is practically no similarity to the SMRT/N-CoR BBD [39]. The BCL6 BTB lateral groove adopts a different conformation when binding BCoR, thus simultaneously demonstrating the properties of high specificity and remarkable flexibility for ligand binding [39]. From the biochemical standpoint BCoR forms a completely different type of complex than N-CoR and SMRT. Specifically, BCoR associates with a polycomb/ubiquitin ligase complex containing RING1, RYBP, NSPC1, RNF2, FBXL10 and Skp1[40,41]. Among other activities, RNF2 can mono-ubiquitylate histone2A, FBXL10 can demethylate histone 3, lysine 36 and Skp1 is an SCF ubiquitin ligase component. Binding of BCoR could thus introduce completely different chromatin modifications than N-CoR and SMRT to BCL6 target genes. It is not clear whether the association of BCL6 with N-CoR/SMRT vs. BCoR is somehow regulated in order to bring in each complex for a specific function, or whether recruitment is stochastic. However, cell penetrating peptides containing either the SMRT BBD or the BCoR BBD had similar biological effects in inducing BCL6 target genes and killing DLBCL cells[7,39].
Interestingly, blockade of the lateral groove with a SMRT-based cell penetrating BCL6 peptide inhibitor (BPI) could induce expression of genes such as ATR, TP53 and CDKN1A involved in DNA damage and cell cycle checkpoints, but did affect expression of BCL6 target genes involved in differentiation such as PRDM1 [7,12,15,27,42]. Accordingly, when administered to a panel of DLBCL cell lines, BPI induced cell death and cell cycle arrest but not differentiation, suggesting that BCL6 regulates differentiation related genes through a different biochemical mechanism. Along these lines, BCL6 can also interact with the MTA3/NuRD corepressor complex, which was shown to be involved in blocking plasmacytic differentiation in lymphoma cells[43]. MTA3 associated with BCL6 through its unstructured middle region rather than the BTB domain[43]. MTA3 expression was limited to B-cells within germinal centers, and was tightly correlated with BCL6 in B-cell lymphomas, regardless of whether they were GCB or ABC type[42,44]. MTA3 could form a complex with BCL6 on the PRDM1 locus at a site located within intron 4, corresponding to a conserved region which might also regulate expression of the PRDM1 short isoform[42]. MTA3 depletion from DLBCL cells induced PRDM1 expression but not ATR, TP53 or CDKN1A. Unlike BPI, MTA3 depletion induced plasmacytic differentiation of DLBCL cells but did not cause cell death[42]. Collectively, these data show that even within the same cells, BCL6 uses distinct corepressor complexes to repress genes involved in survival and proliferation vs. those involved in differentiation (Fig 4A). When considered in the context of germinal center physiology, the biochemical dissociation of BCL6 functions suggests a model whereby transient CD40 signaling through disruption of the BCL6-N-CoR/SMRT complex can induce checkpoint genes but leave the BCL6 differentiation blockade function intact; thus making it possible for centroblasts to maintain their phenotype and return for more rounds of affinity maturation after undergoing DNA damage quality control.
In another example of the functional dissociated of BCL6 transcriptional complexes, BCL6 was shown to mediate its own negative autoregulation independently of whether it was able to bind to SMRT, N-CoR, BCoR or MTA3 [45]. Instead, BCL6 was shown to also bind directly to the CtBP corepressor through a region adjacent and possibly overlapping with the BTB domain. CtBP was recruited to the BCL6 locus in a BCL6 dependent manner and its depletion induced BCL6 expression[45]. A recent study performed BCL6 ChIP-on-chip in the Ramos B-cell lymphoma cell line and identified ∼500 BCL6 targets [27]. The chief cellular functions associated with these genes included protein ubiquitylation, cell cycle and growth control, DNA repair, mitochondrial energy metabolism, transcriptional regulation, signal transduction and immune system regulation (Fig 4B). While some of these functions were expected, others particularly in protein ubiquitylation and metabolism were not previously known, and raise the possibility that additional corepressor specific BCL6 pathways might be discovered in the future.
BCL6 as a therapeutic target in DLBCL
Given the role of BCL6 in DLBCL pathogenesis it seems clear that BCL6 is an excellent therapeutic target. However, BCL6 is a fairly ubiquitous protein, and its functions in other organs must also be considered. Targeting the BTB domain of BCL6 provides a route to specifically disrupt the survival functions of BCL6 without affecting some of its other functions. The lateral groove of BCL6 can be effectively inhibited using cell-penetrating BPI based peptidomimetics designed to mimic the SMRT BBD. These peptides kill DLBCLs in vitro and in vivo without inducing toxicity in animals or generating anti-peptide antibodies [27,30]. In contrast, BCL6 null mice develop a fatal inflammatory disease due to loss of BCL6 functions in macrophages and T-cells[46]. The lack of inflammatory findings in BPI treated mice suggest that either these actions of BCL6 may not be fully dependent on the BTB lateral groove, or that more profound and sustained loss of function of BCL6 is required. A peptide aptamer screening approach yielded a BTB domain inhibitor that did not disrupt SMRT and N-CoR interaction with BCL6 but still could inhibit its actions[47]. The mechanism of action of the aptamer inhibitor remains to be determined. More recently, a computer aided drug design strategy was used to generate lateral groove blocking small molecules with antilymphoma activity in vitro and in vivo[48]. While lateral groove inhibitors kill DLBCL cells mostly due to checkpoint protein reactivation, targeting MTA3 might serve as a form of differentiation therapy for lymphomas, which could also have utility as a therapeutic modality as has been shown to be useful in leukemia therapy.
A more comprehensive review of approaches for therapeutic targeting of BCL6 was recently published [49]. However it is worth mentioning that a major obstacle to translating BCL6-targeted therapy into the clinical setting involves the ability to select patients most likely to respond. Given the molecular heterogeneity of DLBCL this could be quite challenging. Although the GCB type of DLBCL is generally associated with BCL6, both GCB and ABC cell lines respond to BPI. BCL6 translocations occur more frequently in ABC than GCB patients but point mutations were more frequent in GCB patients, and altogether these lesions did not correlate well with BCL6 expression level [50]. A second effort at classifying DLBCL patients according to gene expression profiles used comprehensive consensus clustering in a cohort of 175 patients, and found that patients segregated into three subtypes. One of these featured expression of genes associated with B-cell receptor signaling and proliferation (BCR-DLBCL), another featured expression of genes involved in oxidative phosphorylation (OxPhos), while the third was determined by infiltrating host immune response cells (HR) [51]. These signatures are unrelated to and do not overlap with the GCB and ABC subtypes. Based on the reasoning that BCL6 target genes would be coordinately regulated in lymphomas that were biologically dependent on BCL6, a gene set enrichment analysis was performed using the BCL6 target genes identified by ChIP-on-chip[27]. The data showed that BCL6 target genes were coordinately regulated in BCR vs. OxPhos lymphomas, but not in GCB vs. ABC tumors[27]. In a blinded test of cell response to BPI, only BCR cell lines were sensitive to BPI, while OxPhos cells (regardless of whether they expressed BCL6) were resistant[27]. BCR patients would thus be the best candidates for clinical trials with BCL6 inhibitors.
Conclusions
Over the past few years the function of BCL6 in lymphomagenesis has become much clearer, especially in regards to the discoveries that BCL6 suppresses DNA damage checkpoints and of physiological mechanisms that regulate and modulate the actions of BCL6 as B-cells exit the germinal center. The biochemical mechanism of action of BCL6 and intersecting pathways such as NFkB and STATs during germinal center differentiation is reflected in the different molecular phenotypes of DLBCL, and is leading to the identification of biological pathways amenable to therapeutic targeting in specific patient cohorts. Finally, BCL6 is emerging as a major therapeutic target in DLBCL, and increasingly sophisticated inhibitors of BCL6 should soon make the transition to clinical trials.
Acknowledgements
AM is supported by NCI R01 CA104348, the Chemotherapy Foundation, the Sam Waxman Cancer Research Foundation, the G&P Foundation and is a Leukemia and Lymphoma Society Scholar. •
References
- 1.Allman D, Jain A, Dent A, et al. BCL-6 expression during B-cell activation. Blood. 1996;87:5257–5268. [PubMed] [Google Scholar]
- 2.Dent AL, Shaffer AL, Yu X, et al. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 3.Ye BH, Cattoretti G, Shen Q, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 4.Ye BH. BCL-6 in the pathogenesis of non-Hodgkin's lymphoma. Cancer Invest. 2000;18:356–365. doi: 10.3109/07357900009012179. [DOI] [PubMed] [Google Scholar]
- 5.Baron BW, Anastasi J, Montag A, et al. The human BCL6 transgene promotes the development of lymphomas in the mouse. Proc Natl Acad Sci U S A. 2004;101:14198–14203. doi: 10.1073/pnas.0406138101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cattoretti G, Pasqualucci L, Ballon G, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 7.Polo JM, Dell'Oso T, Ranuncolo SM, et al. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat Med. 2004;10:1329–1335. doi: 10.1038/nm1134. [DOI] [PubMed] [Google Scholar]
- 8.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 9.Liu M, Duke JL, Richter DJ, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 10.Pasqualucci L, Neumeister P, Goossens T, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 11.Pasqualucci L, Bhagat G, Jankovic M, et al. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- 12.Ranuncolo SM, Polo JM, Dierov J, et al. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol. 2007;8:705–714. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- 13.Ranuncolo SM, Polo JM, Melnick A. BCL6 represses CHEK1 and suppresses DNA damage pathways in normal and malignant B-cells. Blood Cells Mol Dis. 2008 doi: 10.1016/j.bcmd.2008.02.003. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phan RT, Saito M, Basso K, et al. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol. 2005;6:1054–1060. doi: 10.1038/ni1245. [DOI] [PubMed] [Google Scholar]
- 15.Cerchietti L, Polo J, DaSilva G, et al. Sequential transcription factor targeting for diffuse large B-cell lymphomas. Cancer Res. 2008 doi: 10.1158/0008-5472.CAN-07-5817. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwickert TA, Lindquist RL, Shakhar G, et al. In vivo imaging of germinal centres reveals a dynamic open structure. Nature. 2007;446:83–87. doi: 10.1038/nature05573. [DOI] [PubMed] [Google Scholar]
- 17.Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007;315:528–531. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- 18.Ci W, Polo JM, Dell'Oso T, Melnick A. A “Survival of the fittest” mechanism for weeding out potentially lymphomagenic B-cells during germinal center B-cell differentiation. Blood. 2007:110. [Google Scholar]
- 19.Phan RT, Saito M, Kitagawa Y, et al. Genotoxic stress regulates expression of the proto-oncogene Bcl6 in germinal center B cells. Nat Immunol. 2007;8:1132–1139. doi: 10.1038/ni1508. [DOI] [PubMed] [Google Scholar]
- 20.Kuo TC, Shaffer AL, Haddad J, Jr., et al. Repression of BCL-6 is required for the formation of human memory B cells in vitro. J Exp Med. 2007;204:819–830. doi: 10.1084/jem.20062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tunyaplin C, Shaffer AL, Angelin-Duclos CD, et al. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173:1158–1165. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- 22.Basso K, Klein U, Niu H, et al. Tracking CD40 signaling during germinal center development. Blood. 2004;104:4088–4096. doi: 10.1182/blood-2003-12-4291. [DOI] [PubMed] [Google Scholar]
- 23.Saito M, Gao J, Basso K, et al. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 2007;12:280–292. doi: 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 24.Teng Y, Takahashi Y, Yamada M, et al. IRF4 negatively regulates proliferation of germinal center B cell-derived Burkitt's lymphoma cell lines and induces differentiation toward plasma cells. Eur J Cell Biol. 2007;86:581–589. doi: 10.1016/j.ejcb.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Li Z, Wang X, Yu RY, et al. BCL-6 negatively regulates expression of the NF-kappaB1 p105/p50 subunit. J Immunol. 2005;174:205–214. doi: 10.4049/jimmunol.174.1.205. [DOI] [PubMed] [Google Scholar]
- 26.Perez-Rosado A, Artiga M, Vargiu P, et al. BCL6 represses NFkappaB activity in diffuse large B-cell lymphomas. J Pathol. 2008;214:498–507. doi: 10.1002/path.2279. [DOI] [PubMed] [Google Scholar]
- 27.Polo JM, Juszczynski P, Monti S, et al. Transcriptional signature with differential expression of BCL6 target genes accurately identifies BCL6-dependent diffuse large B cell lymphomas. Proc Natl Acad Sci U S A. 2007;104:3207–3212. doi: 10.1073/pnas.0611399104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 29.Lam LT, Davis RE, Pierce J, et al. Small molecule inhibitors of IkappaB kinase are selectively toxic for subgroups of diffuse large B-cell lymphoma defined by gene expression profiling. Clin Cancer Res. 2005;11:28–40. [PubMed] [Google Scholar]
- 30.Cerchietti L, Melnick A. 2008. Unpublished data.
- 31.Lam LT, Wright G, Davis RE, et al. Cooperative signaling through the STAT3 and NF-{kappa}B pathways in subtypes of diffuse large B cell lymphoma. Blood. 2008 doi: 10.1182/blood-2007-09-111948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding BB, Yu JJ, Yu RY, et al. Constitutively activated STAT3 promotes cell proliferation and survival in the activated B-cell subtype of diffuse large B-cell lymphomas. Blood. 2008;111:1515–1523. doi: 10.1182/blood-2007-04-087734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reljic R, Wagner SD, Peakman LJ, Fearon DT. Suppression of signal transducer and activator of transcription 3-dependent B lymphocyte terminal differentiation by BCL-6. J Exp Med. 2000;192:1841–1848. doi: 10.1084/jem.192.12.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker SR, Nelson EA, Frank DA. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 2007;26:224–233. doi: 10.1038/sj.onc.1209775. [DOI] [PubMed] [Google Scholar]
- 35.Lee SC, Bottaro A, Chen L, Insel RA. Mad1 is a transcriptional repressor of Bcl-6. Mol Immunol. 2006;43:1965–1971. doi: 10.1016/j.molimm.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 36.Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000;14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmad KF, Melnick A, Lax S, et al. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol Cell. 2003;12:1551–1564. doi: 10.1016/s1097-2765(03)00454-4. [DOI] [PubMed] [Google Scholar]
- 38.Karagianni P, Wong J. HDAC3: taking the SMRT-N-CoRrect road to repression. Oncogene. 2007;26:5439–5449. doi: 10.1038/sj.onc.1210612. [DOI] [PubMed] [Google Scholar]
- 39.Ghetu AF, Corcoran CM, Cerchietti L, et al. Structure of a BCOR Corepressor Peptide in Complex with the BCL6 BTB Domain Dimer. Mol Cell. 2008;29:384–391. doi: 10.1016/j.molcel.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol Cell Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanchez C, Sanchez I, Demmers JA, et al. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Parekh S, Polo JM, Shaknovich R, et al. BCL6 programs lymphoma cells for survival and differentiation through distinct biochemical mechanisms. Blood. 2007;110:2067–2074. doi: 10.1182/blood-2007-01-069575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujita N, Jaye DL, Geigerman C, et al. MTA3 and Mi-2/NuRD Complex Regulate Cell Fate During B-Lymphocyte Differentiation. Cell. 2004;119:75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 44.Jaye DL, Iqbal J, Fujita N, et al. The BCL6-associated transcriptional co-repressor, MTA3, is selectively expressed by germinal centre B cells and lymphomas of putative germinal centre derivation. J Pathol. 2007;213:106–115. doi: 10.1002/path.2199. [DOI] [PubMed] [Google Scholar]
- 45.Mendez LM, Polo JM, Yu JJ, et al. CtBP is an essential corepressor for BCL6 autoregulation. Mol Cell Biol. 2008 doi: 10.1128/MCB.01400-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toney LM, Cattoretti G, Graf JA, et al. BCL-6 regulates chemokine gene transcription in macrophages. Nat Immunol. 2000;1:214–220. doi: 10.1038/79749. [DOI] [PubMed] [Google Scholar]
- 47.Chattopadhyay A, Tate SA, Beswick RW, et al. A peptide aptamer to antagonize BCL-6 function. Oncogene. 2006;25:2223–2233. doi: 10.1038/sj.onc.1209252. [DOI] [PubMed] [Google Scholar]
- 48.Da Silva G, Ghetu AF, Cerchietti L, et al. Design and Development of Small Molecules for Specific Targeted Therapy of Diffuse Large B-Cell Lymphoma. Blood. 2007:110. [Google Scholar]
- 49.Parekh S, Prive GG, Melnick A. Therapeutic targeting of the BCL6 oncogene for diffuse large B-cell lymphomas. Leuk Lymphoma. 2008:1–9. doi: 10.1080/10428190801895345. iFirst. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iqbal J, Greiner TC, Patel K, et al. Distinctive patterns of BCL6 molecular alterations and their functional consequences in different subgroups of diffuse large B-cell lymphoma. Leukemia. 2007;21:2332–2343. doi: 10.1038/sj.leu.2404856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monti S, Savage KJ, Kutok JL, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005;105:1851–1861. doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- 52.Chevallier N, Corcoran CM, Lennon C, et al. ETO protein of t(8;21) AML is a corepressor for Bcl-6 B-cell lymphoma oncoprotein. Blood. 2004;103:1454–1463. doi: 10.1182/blood-2003-06-2081. [DOI] [PubMed] [Google Scholar]