Abstract
Genes required for replication and for conjugal transfer of the Agrobacterium tumefaciens Ti plasmid are regulated by the quorum sensing transcription factor TraR, whose N-terminal domain binds to the pheromone N-3-oxooctanoyl-L-homoserine lactone (OOHL) and whose C-terminal domain binds to specific DNA sequences called tra boxes. Here, we constructed 117 mutants, altering 103 surface-exposed amino acid residues of the TraR N-terminal domain. Each mutant was tested for activation of the traI promoter, where TraR binds to a site centered 45 nucleotides upstream of the transcription start site, and of the traM promoter, where TraR binds a site centered 66 nucleotides upstream. Alteration of 18 residues blocked activity at the traI promoter. Of these, alteration at three positions impaired TraR abundance or DNA binding, leaving 15 residues that are specifically needed for positive control. Of these 15 residues, nine also blocked or reduced activity at the traM promoter, while six had no effect. Amino acid residues required for activation of both promoters probably contact the carboxy terminal domain of the RNA polymerase ∀ subunit, while residues required only for traI promoter activation may contact another RNA polymerase component.
Introduction
Many species of bacteria use diffusible pheromones to coordinate a wide range of physiology, including pathogenesis, sporulation, the formation of biofilms, and horizontal transfer of DNA (Whitehead et al., 2001; Winans and Bassler, 2002). One such regulatory system is composed of the LuxI and LuxR proteins of Vibrio fischeri, where LuxI synthesizes 3-oxo-hexanoylhomoserine lactone (OHHL) (Eberhard et al., 1981), while LuxR is an OHHL receptor and OHHL-dependent transcription factor of the organism’s bioluminescence operon. In the past 15 years, a wide variety of related systems have been discovered, and a small number of them have been intensively studied. In most cases, a LuxI homolog is functionally paired with a LuxR homolog, in that the former synthesizes an acylhomoserine lactone (AHL) while the latter is a transcription factor that binds the cognate AHL. In most cases, DNA binding activity requires the AHL (Pappas et al., 2004), while in a few cases, the AHL has the opposite effect, blocking DNA binding (Castang et al., 2006; Cui et al., 2006; Fineran et al., 2005; Sjoblom et al., 2006). Biochemical, genetic, and structural studies of a number of LuxR homologues have revealed that they are two-domain proteins, whose N-terminal domain (NTD) binds AHLs and whose C-terminal domain (CTD) binds DNA (Pappas et al., 2004). Both domains contribute to protein dimerization (Luo et al., 2003; Vannini et al., 2002; Zhang et al., 2002).
The Ti plasmid of the plant pathogen Agrobacterium tumefaciens encodes such a regulatory system, consisting of the AHL synthase TraI and the transcription factor TraR. TraI synthesizes primarily 3-oxo-octanoylhomoserine lactone (OOHL) while TraR is an OOHL-dependent activator of three closely spaced promoters of the repABC operon, which directs plasmid replication and partitioning, and of three promoters of the tra and trb operons, which are required for conjugative transfer of the Ti plasmid (Fuqua and Winans, 1994; Li and Farrand, 2000; More et al., 1996; Pappas and Winans, 2003; Piper et al., 1993; Zhang et al., 1993). TraR-OOHL complexes bind to 18-nucleotide dyad-symmetrical sequences called tra boxes to activate transcription of target promoters (Winans et al., 1999).
Genetic, biochemical and structural studies of TraR and its interactions with OOHL and DNA have made this protein an intensively studied representative of the LuxR family. TraR monomers bind to OOHL in a 1: 1 mole ratio and form homodimers that bind to tra boxes with high affinity and specificity (Luo and Farrand, 1999; Zhu and Winans, 1999). The N-terminal pheromone-binding domain of TraR is sufficient for OOHL binding and dimerization, as TraR fragments containing just this domain are able to form inactive heterodimers with full-length protein (Chai et al., 2001; Luo et al., 2003; Qin et al., 2000; Zhu and Winans, 1998). Binding of OOHL to TraR is virtually irreversible, as this pheromone is buried deeply within the protein and makes no contact with bulk solvent. Activity of TraR, LuxR, and a few other members of this family have been reconstituted in vitro, and require only promoter DNA, the activator complexed with its AHL, and RNA polymerase (RNAP) (Urbanowski et al., 2004; Zhu and Winans, 1999).
The quaternary structure of TraR, OOHL, and tra box DNA has been solved by X-ray crystallography (Vannini et al., 2002; Zhang et al., 2002). This structure confirmed the domain structure of this dimeric protein. Protein dimerization is achieved largely by a rather long alpha helix on each of the two the N terminal domains, which together create a coiled coil, and by two shorter helices in the C terminal domains (CTD), which create a second coiled coil. The CTD is a four-helix bundle with a helix-turn-helix DNA binding motif (HTH) common to many prokaryotic regulators (Nelson, 1995). This four-helix domain structure is common to all members of the LuxR-NarL-FixJ superfamily of prokaryotic transcriptional regulators (Fuqua and Greenberg, 2002). Several amino acid residues found on the recognition helix make specific contact with tra box DNA sequences (Vannini et al., 2002; White and Winans, 2007; Zhang et al., 2002).
There are seven known TraR-dependent promoters on the octopine-type Ti plasmid, all of which contain tra boxes. At five of these promoters (PtraA, PtraC, PtraI, PrepA1, and PrepA3) the TraR-binding site is centered approximately 45 nucleotides upstream of the transcription start site, adjacent to the -35 elements of these promoters. At the remaining two promoters (PrepA2 and PtraM), the tra box is located approximately 66 nucleotides upstream.
The first class of promoters are reminiscent of class II-type promoters first described for CAP (Busby and Ebright, 1999) while the second class of promoters resemble class I-type promoters. Activators of class II-type promoters can interact with several different subunits of RNAP, while activators of class I-type promoters are generally thought to interact with the C-terminal domain of the alpha subunit of RNAP (∀-CTD), which is connected to ∀-NTD by a flexible linker (Busby and Ebright, 1999). A number of other activators have been shown to contact either the Φ or ∀ subunits via their DNA binding domains (Busby and Ebright, 1999; Bushman et al., 1989; Crater and Moran, 2002; Danot et al., 1996; Stibitz, 1994).
In a previous study (White and Winans, 2005), we used TraR structural data to alter all surface exposed amino acids of the C-terminal domain, and tested each for defects in activating a class I promoter and a class II promoter (White and Winans, 2005). Alteration of six amino acid residues abolished activation of both promoters, without significantly affecting protein accumulation or DNA binding. As these mutants were defective at both classes of promoter, we concluded that the residues of the wild type TraR most likely interacted with the ∀CTD.
Two residues in the N-terminal domain of the pTiC58 TraR protein are critical for activation of a class II type promoter (Luo and Farrand, 1999). Mutations of these residues (D10 and G123) disrupted contacts with purified ∀-CTD (Qin et al., 2004). It seemed plausible that other residues in the N-terminal domain of TraR might also make specific contacts with RNAP. In order to study the role of the TraR N-terminal domain in transcription activation, we performed saturating site-directed mutagenesis of all surface-exposed TraR residues. Each mutant was tested for in vivo activity at a class II promoter, for its accumulation in vivo, and for ability to bind DNA in vitro. We identified 15 mutations that accumulate and bind to DNA but that fail to activate the class II promoter. Of these, 9 mutants also abolish activation of the class I promoter, while the remaining 6 mutants affected only the class II promoter. From these data, and from the positions of these mutations on the surface of TraR, we speculate that the residues needed for both class I and class II promoters are likely to interact with ∀CTD of RNAP, while the residues needed only for the class II promoter are likely to interact with another RNAP subunit.
Results
Mutagenesis of residues in the N-terminal domain of TraR
As described above, we sought to determine whether the N-terminal domain of TraR contains regions likely to make direct contact with RNA polymerase. We used the X-ray crystal structure of TraR (Vannini et al., 2002; Zhang et al., 2002) to identify all solvent-exposed residues on this part of the protein, and used site-directed mutagenesis to alter each residue (Vannini et al., 2002; Zhang et al., 2002). Seven residues of the linker domain were also included in this study. We have previously found that many alterations of surface residues of TraR can cause a significant decrease in protein accumulation, indicating that the mutants were sensitive to proteolysis (White and Winans, 2005). In an effort to minimize this problem, we made substitutions wherever possible that preserved interactions with adjacent amino acid residues, but that altered the surface exposed to solvent. In all, 117 point mutants were constructed at 103 positions, saturating the surface of the TraR NTD. These mutants are listed in Table S1 and also indicated in Fig. 1.
Figure 1.
Saturation mutagenesis of the surface of the NTD of TraR. Residues in white were altered by site-directed mutagenesis.
Activity of TraR mutants in vivo
Each TraR mutant was tested for its ability to activate transcription of a TraR-dependent promoter. We used A. tumefaciens strain NTL4 (pCEW260), which lacks a Ti plasmid and thus does not have the native traR or traI genes (Luo et al., 2001). Plasmid pCW260 contains a PtraI-lacZ fusion, and has a wild type traI promoter except for a one-nucleotide mutation in its tra box. This mutation creates a consensus tra box with perfect dyad symmetry (White and Winans, 2005). The wild type TraR protein expressed this fusion at high levels (approximately 3000 Miller units) in the presence of saturating levels of OOHL (100 nM), while its activity in the absence of either TraR or OOHL was barely detectable (less than 5 Miller units). OOHL was added at four different concentrations (0.1 nM, 1 nM, 10 nM and 100 nM) to each culture and assayed for ∃-galactosidase activity 8 hours later. The resulting expression levels for all 117 mutants are shown in Table S1. Twenty mutants, together altering 18 residues, showed less than 50% of wild type activity in the presence of 100 nM OOHL (Table 2).
Table 2.
Mutations in the TraR N Terminal Domain that are defective in transcriptiona
| Mutation | traI Promoter Activity at the Indicated OOHL Concentrations | |||
|---|---|---|---|---|
| 0.1 nM | 1 nM | 10 nM | 100 nM | |
| Wild type | (100)b | (100)b | (100)b | (100)b |
| Vector Control | <1 | <1 | <1 | <1 |
| D6E | 20 | 15 | 28 | 42 |
| D6G | <1 | <1 | <1 | <1 |
| K7R | 9 | 31 | 28 | 44 |
| D10N | <1 | <1 | <1 | <1 |
| A13L | <1 | 1 | 20 | 7 |
| E15K | 6 | 18 | 31 | 46 |
| D17E | 12 | 11 | 34 | 21 |
| I20W | 5 | 12 | 38 | 14 |
| H44K | 5 | 18 | 34 | 42 |
| P71H | <1 | <1 | <1 | 8 |
| K74E | <1 | 6 | 16 | 8 |
| R75E | 8 | 32 | 41 | 48 |
| R77E | <1 | <1 | 2 | 1 |
| S78E | 12 | 26 | 22 | 28 |
| R79E | 32 | 48 | 36 | 36 |
| K80E | 3 | 19 | 24 | 23 |
| N122A | <1 | 3 | 22 | 29 |
| N122D | <1 | 1 | 10 | 16 |
| G123R | <1 | <1 | <1 | <1 |
| D144R | <1 | <1 | 5 | 23 |
All mutations included in this list functioned at levels 50% or lower than wild type TraR in the presence of 100 nM OOHL. Data are presented as a percentage of wild type.
Wild type TraR expressed the promoter, a traI-lacZ fusion, at 700, 1500, 2600 and 3000 units of ∃-galactosidase activity at 0.1 nM, 1 nM, 10 nM and 100 nM of OOHL, respectively.
In vivo accumulation of mutant proteins
In previous studies, we found that some TraR point mutants defective in activation also failed to accumulate in vivo (White and Winans, 2005). We therefore used semi-quantitative Western immunoblotting to test all mutants for their ability to accumulate in vivo. These assays were done using the same A. tumefaciens strain as used for activity assays described above. Equivalent amounts of crude cell extracts were loaded into each lane. A representative Western is shown in Fig. 2, and the data expressed in per cent accumulation compared with wild type for all mutants are summarized in Table S1. Results for the 20 mutants described above are shown in Table 3.
Figure 2.
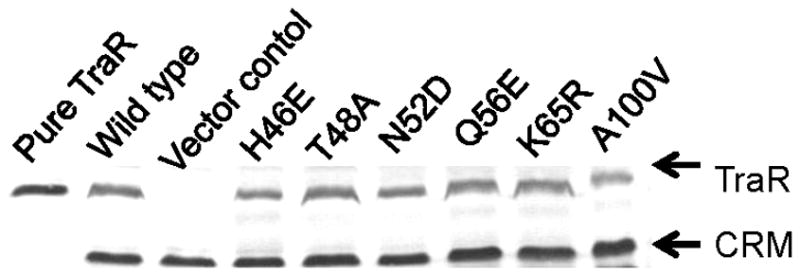
Western immunoblot data of TraR point mutants in A. tumefaciens strain NTL4 (pCEW260). Strains containing pYC335 or pPZP201served as positive and negative controls, respectively. Purified TraR was also used as a positive control for all westerns (left lane). The cross-reacting material (CRM) was used to normalize the intensity of each TraR band.
Table 3.
Accumulation in A. tumefaciens and DNA-binding affinity in vitro of all transcription-defective mutants.
| Mutation | Accumulation (percent of wild type) | DNA binding (percent of wild type) |
|---|---|---|
| Wild type | (100) | (100) |
| Vector Control | <1 | <1 |
| D6E | 120 | 89 |
| D6G | 102 | 70 |
| K7R | 78 | 80 |
| D10N | 101 | 109 |
| A13L | 100 | 65 |
| E15K | 45 | 74 |
| D17E | 70 | 30 |
| I20W | 49 | 57 |
| H44K | 27 | 20 |
| P71H | <10 | NTa |
| K74E | 76 | 74 |
| R75E | 88 | 41 |
| R77E | 77 | 72 |
| S78E | 100 | 103 |
| R79E | 68 | 63 |
| K80E | 92 | 65 |
| N122A | 61 | 100 |
| N122D | 33 | 35 |
| G123R | 72 | 110 |
| D144R | 82 | 74 |
NT, not determined.
Of the 20 mutants described in Table 2, three mutants accumulated at levels less than 45% of wild type (Table 3). Conversely, 17 mutations, altering 16 different residues, did not cause a decrease in abundance commensurate with their decrease in activity. These 16 mutations are expected to cause defects in some other aspect of transcription activation, possibly either DNA binding or interactions with RNAP.
Ability of TraR mutants to bind DNA fragments containing tra box sequences
We conducted electrophoretic mobility shift assays (EMSA) using a radiolabeled DNA fragment and cleared cell lysates containing TraR point mutants. The strains were cultured in the presence of OOHL and Western blots were performed with each cleared cell lysate (data not shown). The volumes of each lysate added to the gel were adjusted using the Western immunoblot data described above. The DNA fragment used in these experiments contained the traA-traC intergenic region, which contains a consensus binding site for TraR. Mutants that had severe defects in accumulation in A. tumefaciens (see above) also did not accumulate well in the protease-deficient strain KY2347, and were therefore not included in the gel mobility shift assays. Representative EMSA data are shown in Fig. 3, while the data for all mutants are summarized in Table S1. Complexes were not detected using an extract lacking TraR (Fig. 3, lanes labeled vector control). Of the 20 transcription-defective mutants described in Table 2, three bound the tra box DNA fragment at levels less than 40% of wild type (Table 3). For these mutants, D17E, H44K, and N122D, the defect in DNA binding could explain the defect in transcription. However, two of these mutants were also defective in accumulation. In all, a total of 16 mutants, altering 15 residues, showed defects in transcription that could not be accounted for by a lack of accumulation or an inability to bind tra box DNA. These mutants are therefore candidates for having defects in interactions with RNA polymerase.
Figure 3.

Electrophoretic mobility shift assays with TraR in crude cell extracts. The amount of full-length, soluble TraR in each extract was normalized using western immunoblots.
Activity of site-directed mutants at a class I promoter
The activity assays described above were all done with a class II promoter, where TraR could interact with several different subunits of RNAP. We also tested these mutants at the traM promoter, which is a class I promoter. TraR activating such a promoter should in principle interact only with ∀CTD. The promoter we used was identical to the wild type PtraM promoter except that its tra box contained the consensus TraR binding sequence (White and Winans, 2005).
This promoter has a TraR-independent basal expression of approximately 120 Miller units, and is activated only approximately seven-fold by TraR in the presence of saturating OOHL concentrations. Since this induction ratio is rather low, we tested the mutants using saturating levels of OOHL (100 nM) and deducted the TraR-independent activity of this promoter. Many of the TraR mutants that were impaired at traI were also impaired at PtraM (Table 4). The clearest examples of this include D6G, D10N, K74E, R77E, and G123R, while other possible examples include R75E, S78E, R79E, and D144R. In a previous study, additional positive control mutations were isolated in the TraR CTD, all of which blocked activity at both types of promoters (White and Winans, 2005).
Table 4.
Activity of TraR mutants at the TraM promoter.
| Mutation | Activity at PtraI 100 nM | Activity at PtraM 100 nM | TraR-dependent PtraM activity |
|---|---|---|---|
| Wild Type | (100) | (100) | 86 |
| Vector | <1 | 14 | 0 |
| D6E | 42 | 78 | 64 |
| D6G | <1 | 16 | 2 |
| K7R | 44 | 75 | 61 |
| D10N | <1 | 15 | 1 |
| A13L | 7 | 104 | 90 |
| E15K | 46 | 98 | 84 |
| D17E | 21 | 70 | 56 |
| I20W | 14 | 100 | 86 |
| H44K | 42 | 27 | 13 |
| K74E | 8 | 36 | 22 |
| R75E | 48 | 40 | 26 |
| R77E | 1 | 15 | 1 |
| S78E | 28 | 37 | 23 |
| R79E | 36 | 46 | 32 |
| K80E | 23 | 102 | 88 |
| N122D | 16 | 33 | 19 |
| N122A | 29 | 62 | 48 |
| G123R | <1 | 35 | 21 |
| D144R | 23 | 47 | 33 |
A few of the mutants that were defective at the traI promoter were unimpaired at the PtraM promoter (Table 4). The clearest examples of this are A13L, I20W, and K80E, while other possible examples include K7R, E15K, and N122A. These phenotypes suggest that the wild type residues at these positions contact RNAP at the traI promoter but make no critical contacts with RNAP at the traM promoter.
Intragenic complementation of TraR mutants
Dimeric transcription factors ought in principle to have two activating regions (ARs) per dimer. If an AR is composed of amino acids from both subunits, it may be possible to restore protein function by co-expressing two defective proteins (see Fig. 4). The data described above, taken in conjunction with structural information, suggested that the AR that contacts ∀-CTD might be composed of amino acid residues of both TraR subunits. To test this, we co-expressed various TraR PC mutants and assayed for TraI promoter activity. One of these mutations, W184H, lies in the TraR-CTD and has a strong positive control defect (White and Winans, 2005). When mutant D10N was co-expressed with W184H, no complementation was observed (Table 5). This suggests that one mutation disrupted one of the two ARs in the TraR dimer, while the other mutation disrupted the second AR (Fig. 4). Residues W184 and D10 of each AR must therefore be contributed by the same subunit. In contrast, when mutants G123R and W184H were co-expressed, activity was almost fully restored (Table 5). This suggests that W184 and G123 of each AR are contributed by different subunits, and that in the merodiploid strain, each heterodimer has one functional site and one doubly non-functional one (Fig. 4).
Figure 4.
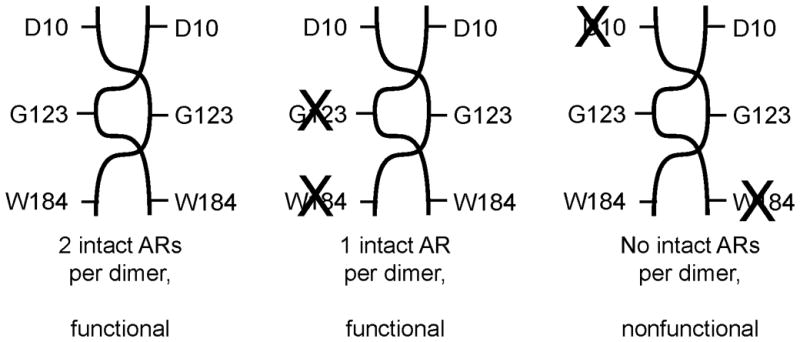
Intragenic complementation of mutations of D10, G123, and W184. Successful complementation was taken as an indication that the amino acid residues are contributed by opposite subunits.
Table 5.
Intragenic complementation of positive control mutations of TraR.
| pPZP201 derivative | pBBR1MCS5 derivative | ∃-galactosidase Activity |
|---|---|---|
| wild type | wild type | 2343 |
| vector | vector | 2 |
| D10N | G123R | 425 |
| G123R | D10N | 334 |
| D10N | W184H | 45 |
| W184H | D10N | 34 |
| G123R | W184H | 1556 |
| W184H | G123R | 1408 |
In a third experiment, mutants D10N and G123R were co-expressed. Activity was restored, but not to wild type levels (Table 5). The restoration of activity suggests that D10 and G123 of each AR are contributed by different subunits. The fact that restoration was incomplete suggests that the two mutations on one AR somehow weakened the activity of the opposite AR, possibly via a perturbation of the quaternary structure of the TraR dimer.
Discussion
In a previous study, we saturated the surface of the TraR CTD with point mutations, and identified a contiguous patch of residues that are essential for activation of Class I and Class II promoters (White and Winans, 2005). In the present study, we sought to identify one or more similar patches on the TraR NTD. Two positive control mutations in the TraR NTD have previously been described by another group (Qin et al., 2004). Saturation mutagenesis of the surface of the TraR NTD led to the discovery of a significant number of residues that are required for positive control. Each such residue may make direct contacts with RNAP.
TraR is probably a highly flexible molecule, with two NTDs tethered to two CTDs by a flexible linker. If so, then the spatial position of the two NTDs of the dimer with respect to the two CTDs could be highly variable at different promoters. We believe that the CTDs of the TraR dimer, once bound to tra box DNA, can then recruit RNAP to the promoter. Different surfaces of the TraR NTDs could then “explore” the surface of any proximal RNAP surface until a patch of TraR binds a patch of RNAP.
Assuming that residues required for positive control do contact RNAP, we would like to begin to map these interactions, identifying which subunit of RNAP is contacted by each residue. Presumably, residues required for activation of both class I and class II promoters most likely contact the ∀-CTD, while residues required only for class II promoters are more likely to contact some other RNAP subunit. We have divided all positive control mutants into two groups: (i) those that affect the Class I and Class II promoters, and (ii) those that affect only the Class II promoter. Residues of the former class are likely to contact ∀-CTD and are shown in white in Figures 5 and 6, while the latter class are likely to contact another RNAP subunit and are shown in black.
Figure 5.
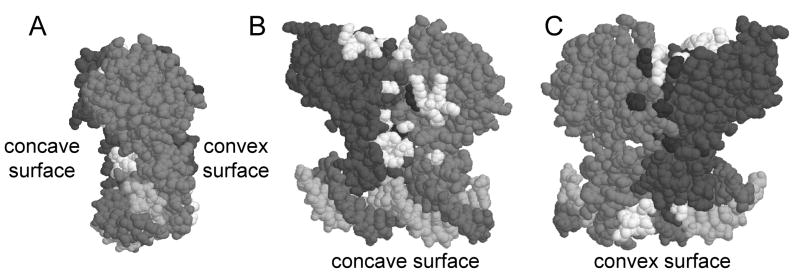
Positive control mutants isolated in this study (in the TraR NTD) and in a previous study (TraR-CTD) (White and Winans, 2005). Residues in white affect both PtraI and PtraM, while residues in black affected only PtraI. (A) View of TraR along the DNA axis, showing the concave and convex faces of the crystal structure. (B and C) TraR rotated to show the concave surface and convex surfaces, respectively.
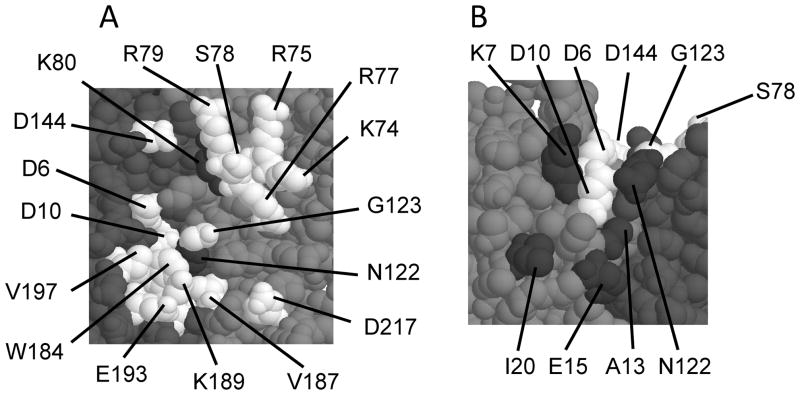
Figure 6.
Closeup views of TraR residues required for positive control. Residues in white are needed for both promoters, while residues in black are needed only for PtraI. (A) A patch on the concave surface, composed of residues from both subunits and both domains of each subunit. (B) A patch on the convex surface, composed of residues from the NTDs of both subunits.
The asymmetry of crystallized TraR creates what we will refer to as a “concave” surface and a “convex” surface (Fig. 5). In many cases, residues that are quite closely packed on the concave surface are widely dispersed on the convex surface. For example, W184 of one subunit and G123 of the other subunit lie less than 5 angstroms apart on the concave surface, but lie 55 angstroms apart on the convex surface. All the amino acid residues that are thought to contact ∀CTD are closely spaced on the concave surface, while they are widely separated on the convex surface. As a working model, we propose that the asymmetry of cyrstalized TraR resembles to some degree the active form in vivo, and that the concave surface of TraR resembles the surface that contacts ∀CTD. If so, all four domains of the TraR dimer contribute residues to this putative patch. Three are contributed by the NTD of the A subunit (D6, D10 and D144), six are contributed by the NTD of the B subunit (K74, R75, R77, S78, R79, and G123), five are contributed by the CTD of the A subunit (W184, V187, K189, E193 and V197), and one is contributed by the CTD of the B subunit (D217). Some of these residues are somewhat sheltered from solvent, especially D6, D10, but would be far more exposed to solvent if the NTD and CTD were to separate slightly. Intragenic complementation analysis supports this model, as it demonstrated that residues G123 and D10 are contributed by opposite subunits, as are residues G123 and W184, while W184 and D10 are contributed by the same subunit.
Of the fifteen residues of this patch, five are basic and five are acidic. As described above, these residues most likely interact with ∀-CTD of RNAP. Given the flexibility of the TraR linker, the NTD and CTD of this protein should be able to separate, so that the residues contributed by the two CTDs could bind one face of ∀-CTD of RNAP, while the residues contributed by the two NTDs could bind another face. The ∀-CTD of RNAP of A. tumefaciens has a pronounced acidic patch that could conceivably bind the basic residues K74, R75, R77, and R79 (data not shown).
Four residues (W184, V187, K189, and V197) are located at or near the turn between ∀-helix 10 and ∀-helix 11 (the scaffold helix of the helix-turn-helix motif). Very similar activating regions have been identified in a number of other regulators with HTH motifs, including CAP, FNR, SoxS, and 8C1 (Bell and Busby, 1994; Busby and Ebright, 1999; Bushman et al., 1989; Griffith and Wolf, 2002). In all of these cases, the activating region includes a surface-exposed loop similar to that of TraR. Similar ARs have been identified in the DNA-binding domains of the NarL homologues GerE and BvgA (Crater and Moran, 2002; Stibitz, 1994). The activating region of FNR that contacts ∀CTD is also extensive and spans across three surface-exposed loops, while the FNR homologue CAP contacts the ∀CTD with just one loop (Bell and Busby, 1994; Busby and Ebright, 1999; Li et al., 1998; Williams et al., 1997).
Although we predict that the two activating regions described above contact the ∀-CTD, the positions of these patches are intriguing. In the CAP-∀CTD-DNA ternary structure, the ∀CTD binds in the minor groove directly downstream of CAP in class I promoters, and directly upstream of CAP in class II promoters. Contacts are therefore made using the “downstream surface” or “upstream surface” of CAP, respectively. In contrast, the activating regions of TraR lie neither on the upstream nor downstream surfaces of the protein, but rather on a side surface, suggesting that ∀-CTD could bind DNA to one side of TraR (Fig. 5B). This was reported to be the case for the BvgA protein of Bordetella pertussis (Boucher et al., 2003).
Some of the positive control mutants isolated in this study affected only the Class II promoter (indicated in black in Figures 5 and 6). The simplest interpretation is that all or at least most of these mutations block interactions between TraR and a portion of RNAP other than ∀-CTD. One problem with this interpretation is that some of these mutants overlap regions that were interpreted as binding ∀-CTD. For example, the mutation K80E blocked PtraI expression, but had no effect on PtraM (Table 4). However, residue K80 is adjacent to residues K74-R79, which were needed for both promoters. Similarly, residues K7, A13, E15, I20 of one subunit, and N122 from the other subunit, are needed only for PtraI, yet they roughly encircle residues D10 and D6, which are needed for both promoters. Nevertheless, it is tempting to speculate that K7, A13, E15, I20, and N122 form a contiguous patch on the convex surface of the TraR dimer, and that this patch contacts RNAP. If so, alteration of D6 or of D10 might conceivably cause a more general loss of function of the protein, blocking activity at both promoters.
The mutations that affect both promoters may be somewhat analogous to the AR1 region of the CAP protein of E. coli (Busby and Ebright, 1999; Lawson et al., 2004; Niu et al., 1996). AR1 (residues 156-164 and R209) of CAP interacts with the “287 determinant” of ∀ (residues 283-288 and 314-317, all lying within the ∀CTD). In contrast, TraR residues that contact ∀-CTD are thought to lie on both the TraR CTD and the TraR NTD. AR2 of CAP (residues 19, 21, 96, and 101, which form a contiguous patch) interacts with residues 162-165 of ∀ (within the ∀NTD). AR3 of CAP (residues 52-58) interacts with residues 593-603 of Φ (lying within region 4, which also decodes the -35 promoter element). At Class I promoters, only AR1 contacts RNAP, while at Class II promoters, AR1, AR2 and AR3 can all make productive contacts. AR1 of CAP acts by recruiting RNA polymerase to promoters, while AR2 and AR3 are thought to function by a combination of recruitment and postrecruitment mechanisms such as promoter melting.
The LuxR CTD has been extensively mutagenized in a search of positive control mutants (Egland and Greenberg, 2001; Trott and Stevens, 2001). Three residues essential for activation but not for DNA binding were described, K198, W201, and I206, which align with residues K189, E192, and V197 of TraR, respectively. K189, E193, and V197 of TraR are essential for positive control, indicating that these positive control regions overlap. The LuxR-NTD has not so far been studied at this level. The fact that LuxR functions in E. coli has been exploited by using libraries of alanine scanning mutants of the RNAP ∀ and Φ subunits. Mutations of Φ residues 591, 595, 597, 602, and 603 strongly inhibited LuxR-dependent gene expression (Johnson et al., 2003). Many of these residues were also critical in interactions between Φ and AR3 of CAP (Lawson et al., 2004). In a separate study, alteration of ∀-CTD residues 262, 265, 290, 295, 296 and 314 inhibited LuxR activity. These residues overlap the 287 determinant, which interacts with AR1 of CAP (Lawson et al., 2004). These studies using screening of alanine-scanning mutants of LuxR or RNAP subunits thus provided evidence that LuxR binds both the ∀CTD and Φ subunits of the RNAP (Finney et al., 2002; Johnson et al., 2003; Stevens et al., 1999).
Experimental Procedures
Bacterial strains and plasmids
Bacterial strains and plasmids used in this study are listed in Table 1. A. tumefaciens strains were cultured in AT minimal medium at 28°C (Tempe et al., 1977). Escherichia coli strains were cultured in LB medium. Synthetic OOHL was provided by A. Eberhard (Cornell University). Antibiotics were added to maintain plasmids at the following concentrations: 100 μg/mL spectinomycin, and 50 μg/mL kanamycin for E. coli; and 200 μg/mL spectinomycin and 200 μg/mL kanamycin for A. tumefaciens.
Table 1.
Strains and plasmids used in this study.
| Strains and plasmids | Relevant features | References |
|---|---|---|
| Strains | ||
| DH5∀ | E. coli, ∀-complementation | Stratagene |
| KY2347 | E. coli strain MG1655, (clpPX-lon) 1196::cat | (Herman et al., 1998) |
| NTL4 | A. tumefaciens, Ti-plasmidless derivative of strain C58 | (Luo et al., 2001) |
| WCF47 | A. tumefaciens containing a non polar internal deletion of traI | (Zhu et al., 1998) |
| Plasmids | ||
| pYC335 | traR cloned into EcoRI and BamHI sites of pPZP201 | (Chai and Winans, 2004) |
| pPZP201 | broad-host range cloning vector, SpR | (Hajdukiewicz et al., 1994) |
| pCEW105 | Consensus PtraM-lacZ fusion, KmR | (White and Winans, 2005) |
| pCEW250 | Consensus tra box at PtraI | (White and Winans, 2007) |
| pBBR1MCS5 | Broad host range vector, GmR | (Kovach et al., 1995) |
| pCEW260 | Consensus PtraI-lacZ fusion, KmR | (White and Winans, 2007) |
DNA manipulations and strain constructions
Recombinant DNA techniques were performed using standard procedures (Sambrook and Russell, 2001). Plasmid DNA was isolated from E. coli with QIAprep spin miniprep kits (Qiagen) for DNA sequence analysis. DNA sequences of constructs that were obtained by PCR were verified using automated DNA sequencing (Cornell Biotechnology Resource Center) and analyzed using the LaserGene program (DNASTAR). Plasmids were introduced into E. coli by transformation (Sambrook and Russell, 2001) and into A. tumefaciens by electroporation (Cangelosi et al., 1991). E. coli strain DH5∀ was used for all plasmid constructions.
Site-directed mutagenesis
Site-directed mutagenesis of TraR was performed using synthetic overlap extension PCR (Sambrook and Russell, 2001). A 978 bp fragment of plasmid pYC335 was amplified using Taq polymerase High Fidelity (Invitrogen). The restriction sites for EcoRI and SacII were used to introduce mutated DNA fragments into the wild type gene. For mutations T167S, A168V and E169Q, we used EcoRI and MfeI. All oligonucleotides used in this study are listed in Table S2 and were obtained from Integrated DNA Technologies (Coralville, Iowa). Restriction enzymes were obtained from New England Biolabs.
In vivo assays for TraR activity
Bioassays were conducted with derivatives of A. tumefaciens strain NTL4 harboring plasmid pCEW260, which carries PtraI-lacZ reporter or pCEW105, which carries a PtraM-lacZ reporter. Each strain also contained plasmid pYC335, which expresses Plac-traR, or a derivative of pYC335 expressing a traR point mutant. Strains were cultured in At minimal medium to an OD600 of 0.3 to 0.4. Each was then diluted 20 fold into fresh AT medium containing the indicated concentrations of OOHL, and incubated with vigorous aeration for 8 hours. Assays for ∃-galactosidase activity were performed as previously described (Miller, 1972). All experiments were conducted in triplicate and repeated at least three times.
Immunodetection of TraR
The abundance of each TraR protein was determined in parallel with the activity assays described above. A portion of each culture was centrifuged and the cell pellets were resuspended in 5% of their original volume in 1x cracking buffer (125 mM Tris pH 6.8, 4% SDS, 20% glycerol, 200 mM DTT, 0.02% bromphenol blue). Cells were disrupted by boiling for 5 min, cooling to −80° C, and boiling for another 5 min. A fraction of each sample was size-fractionated using 12% SDS polyacrylamide gels, and electroblotted onto nitrocellulose membranes (BIORAD). The membranes were blocked using TBS (20 mM Tris pH 7.9, 500 mM NaCl, 0.05% Tween20) with 5% skim milk, and immunodetected in TBS with pre-adsorbed polyclonal anti-TraR rabbit antiserum (Chai and Winans, 2004). Goat anti-rabbit IgG conjugated with alkaline phosphatase (BIORAD) was used as the secondary antibody, and the membranes were stained with BCIP (5-bromo-4-chloro-3-indoyl phosphate p-toluidine salt) and NBT (p-nitro blue tetrazolium chloride) (BIORAD). Westerns were performed with fresh cell lysates for each strain at least three times. Data were analyzed using ImageJ (http://rsb.info.nih.gov/ij/) (Rasband, 2004), and normalized against cross-reacting material in each lane.
Gel mobility shift assays
Clarified cell extracts were used for all gel mobility shift assays. Extracts were prepared from the strain KY2347 (a clp, lon mutant) carrying pYC335 or derivatives of it carrying each of the traR mutants. Strains were cultured at 28° C in LB broth supplemented with 100 ug/mL spectinomycin to an OD600 of 0.2, treated with 500 uM IPTG and 200 nM OOHL, and incubated for an additional 6 hours at 28°C. Cells were then harvested, resuspended in SEDG buffer (Pappas and Winans, 2003), disrupted using a French pressure cell (20,000 psi), and clarified by ultracentrifugation. TraR abundance in each extract was estimated using Western immunoblots, which were analyzed using ImageJ (Rasband, 2004). Equivalent amounts of soluble full-length TraR were added to each binding reaction.
A 247 nucleotide fragment containing the consensus tra box sequence was constructed by PCR amplification, using pCEW250 as template and oligonucleotides Ptra-box For and Ptra-box Rev as primers (Table S2). A negative control fragment of 211 bp in length was PCR amplified from pCEW250 using the primers Pcontrol gel shift For and Pcontrol gel shift Rev (Table S2). Both fragments were end-labeled with [(-32P]dATP (Pelkin Elmer) using T4 polynucleotide kinase (New England Biolabs) and incubated with protein extracts as previously described (Zhu and Winans, 1999). Reactions were size-fractionated in 8% polyacrylamide gels at 4°C as previously described (Pappas and Winans, 2003). Results were analyzed using a Storm B840 PhosphorImager (Molecular Dynamics). Gel shifts were performed with independent clarified lysates at least twice for each strain.
Intragenic complementation
Bioassays were conducted with derivatives of A. tumefaciens strain WCF47 (Zhu et al., 1998) carrying a PtraI-lacZ fusion in the Ti plasmid. The TraR mutants, W184H, D10N and G123R were cloned in both pPZP201 and pBBR1MCS5 using the restriction sites for EcoRI and BamHI. Every possible mutant/plasmid combination was tested. Assays for ∃-galactosidase activity using 100 nM of OOHL were performed as previously described (Miller, 1972).
Structural analyses
SWISSPROT PDB Viewer (Guex and Peitsch, 1997) (http://www.expasy.org/spdbv/) and Protein Explorer (Martz, 2002) (http://proteinexplorer.org) were used to identify all surface residues of the TraR NTD, and to map the point mutants onto the structure of TraR. There are two crystal structures of TraR-OOHL-DNA complexes available from RCSB (http://www.rcsb.org) (accession codes are 1L3L and 1HOM).
Acknowledgments
The authors gratefully acknowledge Dr. Anatol Eberhard (Cornell University) for the synthesis and purification of OOHL used in this study, and Tom Winans for help with mutagenesis. We also thank all members of our laboratory for helpful discussions and critical review of this manuscript. This work was supported by grant GM42893. EDC acknowledges the financial support of the Brazilian government through a fellowship grant from the “Coordenacao de Aperfeicoamento de Pessoal de Nivel Superior (Capes)”.
References
- Bell A, Busby S. Location and orientation of an activating region in the Escherichia coli transcription factor, FNR. Mol Microbiol. 1994;11:383–390. doi: 10.1111/j.1365-2958.1994.tb00318.x. [DOI] [PubMed] [Google Scholar]
- Boucher PE, Maris AE, Yang MS, Stibitz S. The response regulator BvgA and RNA polymerase alpha subunit C-terminal domain bind simultaneously to different faces of the same segment of promoter DNA. Mol Cell. 2003;11:163–173. doi: 10.1016/s1097-2765(03)00007-8. [DOI] [PubMed] [Google Scholar]
- Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- Bushman FD, Shang C, Ptashne M. A single glutamic acid residue plays a key role in the transcriptional activation function of lambda repressor. Cell. 1989;58:1163–1171. doi: 10.1016/0092-8674(89)90514-x. [DOI] [PubMed] [Google Scholar]
- Cangelosi GA, Best EA, Martinetti G, Nester EW. Genetic analysis of Agrobacterium. Methods Enzymol. 1991;204:384–397. doi: 10.1016/0076-6879(91)04020-o. [DOI] [PubMed] [Google Scholar]
- Castang S, Reverchon S, Gouet P, Nasser W. Direct evidence for the modulation of the activity of the Erwinia chrysanthemi quorum-sensing regulator ExpR by acylhomoserine lactone pheromone. J Biol Chem. 2006;281:29972–29987. doi: 10.1074/jbc.M601666200. [DOI] [PubMed] [Google Scholar]
- Chai Y, Zhu J, Winans SC. TrlR, a defective TraR-like protein of Agrobacterium tumefaciens, blocks TraR function in vitro by forming inactive TrlR: TraR dimers. Mol Microbiol. 2001;40:414–421. doi: 10.1046/j.1365-2958.2001.02385.x. [DOI] [PubMed] [Google Scholar]
- Chai Y, Winans SC. Site-directed mutagenesis of a LuxR-type quorum-sensing transcription factor: alteration of autoinducer specificity. Mol Microbiol. 2004;51:765–776. doi: 10.1046/j.1365-2958.2003.03857.x. [DOI] [PubMed] [Google Scholar]
- Crater DL, Moran CP., Jr Two regions of GerE required for promoter activation in Bacillus subtilis. J Bacteriol. 2002;184:241–249. doi: 10.1128/JB.184.1.241-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Chatterjee A, Hasegawa H, Chatterjee AK. Erwinia carotovora subspecies produce duplicate variants of ExpR, LuxR homologs that activate rsmA transcription but differ in their interactions with N-acylhomoserine lactone signals. J Bacteriol. 2006;188:4715–4726. doi: 10.1128/JB.00351-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danot O, Vidal-Ingigliardi D, Raibaud O. Two amino acid residues from the DNA-binding domain of MalT play a crucial role in transcriptional activation. J Mol Biol. 1996;262:1–11. doi: 10.1006/jmbi.1996.0493. [DOI] [PubMed] [Google Scholar]
- Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- Egland KA, Greenberg EP. Quorum sensing in Vibrio fischeri: analysis of the LuxR DNA binding region by alanine-scanning mutagenesis. J Bacteriol. 2001;183:382–386. doi: 10.1128/JB.183.1.382-386.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineran PC, Slater H, Everson L, Hughes K, Salmond GP. Biosynthesis of tripyrrole and beta-lactam secondary metabolites in Serratia: integration of quorum sensing with multiple new regulatory components in the control of prodigiosin and carbapenem antibiotic production. Mol Microbiol. 2005;56:1495–1517. doi: 10.1111/j.1365-2958.2005.04660.x. [DOI] [PubMed] [Google Scholar]
- Finney AH, Blick RJ, Murakami K, Ishihama A, Stevens AM. Role of the C-terminal domain of the alpha subunit of RNA polymerase in LuxR-dependent transcriptional activation of the lux operon during quorum sensing. J Bacteriol. 2002;184:4520–4528. doi: 10.1128/JB.184.16.4520-4528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C, Greenberg EP. Listening in on bacteria: acyl-homoserine lactone signalling. Nat Rev Mol Cell Biol. 2002;3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- Fuqua WC, Winans SC. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith KL, Wolf RE., Jr A comprehensive alanine scanning mutagenesis of the Escherichia coli transcriptional activator SoxS: identifying amino acids important for DNA binding and transcription activation. J Mol Biol. 2002;322:237–257. doi: 10.1016/s0022-2836(02)00782-9. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Herman C, Thevenet D, Bouloc P, Walker GC, D’Ari R. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH) Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DC, Ishihama A, Stevens AM. Involvement of region 4 of the sigma70 subunit of RNA polymerase in transcriptional activation of the lux operon during quorum sensing. FEMS Microbiol Lett. 2003;228:193–201. doi: 10.1016/S0378-1097(03)00750-X. [DOI] [PubMed] [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, 2nd, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Lawson CL, Swigon D, Murakami KS, Darst SA, Berman HM, Ebright RH. Catabolite activator protein: DNA binding and transcription activation. Curr Opin Struct Biol. 2004;14:10–20. doi: 10.1016/j.sbi.2004.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Wing H, Lee D, Wu HC, Busby S. Transcription activation by Escherichia coli FNR protein: similarities to, and differences from, the CRP paradigm. Nucleic Acids Res. 1998;26:2075–2081. doi: 10.1093/nar/26.9.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PL, Farrand SK. The replicator of the nopaline-type Ti plasmid pTiC58 is a member of the repABC family and is influenced by the TraR-dependent quorum-sensing regulatory system. J Bacteriol. 2000;182:179–188. doi: 10.1128/jb.182.1.179-188.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZQ, Farrand SK. Signal-dependent DNA binding and functional domains of the quorum-sensing activator TraR as identified by repressor activity. Proc Natl Acad Sci U S A. 1999;96:9009–9014. doi: 10.1073/pnas.96.16.9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZQ, Clemente TE, Farrand SK. Construction of a derivative of Agrobacterium tumefaciens C58 that does not mutate to tetracycline resistance. Mol Plant Microbe Interact. 2001;14:98–103. doi: 10.1094/MPMI.2001.14.1.98. [DOI] [PubMed] [Google Scholar]
- Luo ZQ, Smyth AJ, Gao P, Qin Y, Farrand SK. Mutational analysis of TraR. Correlating function with molecular structure of a quorum-sensing transcriptional activator. J Biol Chem. 2003;278:13173–13182. doi: 10.1074/jbc.M210035200. [DOI] [PubMed] [Google Scholar]
- Martz E. Protein Explorer: easy yet powerful macromolecular visualization. Trends Biochem Sci. 2002;27:107–109. doi: 10.1016/s0968-0004(01)02008-4. [DOI] [PubMed] [Google Scholar]
- Miller J. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- More MI, Finger LD, Stryker JL, Fuqua C, Eberhard A, Winans SC. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science. 1996;272:1655–1658. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- Nelson HC. Structure and function of DNA-binding proteins. Curr Opin Genet Dev. 1995;5:180–189. doi: 10.1016/0959-437x(95)80006-9. [DOI] [PubMed] [Google Scholar]
- Niu W, Kim Y, Tau G, Heyduk T, Ebright RH. Transcription activation at class II CAP-dependent promoters: two interactions between CAP and RNA polymerase. Cell. 1996;87:1123–1134. doi: 10.1016/s0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas KM, Winans SC. A LuxR-type regulator from Agrobacterium tumefaciens elevates Ti plasmid copy number by activating transcription of plasmid replication genes. Mol Microbiol. 2003;48:1059–1073. doi: 10.1046/j.1365-2958.2003.03488.x. [DOI] [PubMed] [Google Scholar]
- Pappas KM, Weingart CL, Winans SC. Chemical communication in proteobacteria: biochemical and structural studies of signal synthases and receptors required for intercellular signalling. Mol Microbiol. 2004;53:755–769. doi: 10.1111/j.1365-2958.2004.04212.x. [DOI] [PubMed] [Google Scholar]
- Piper KR, Beck von Bodman S, Farrand SK. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature. 1993;362:448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- Qin Y, Luo ZQ, Smyth AJ, Gao P, Beck von Bodman S, Farrand SK. Quorum-sensing signal binding results in dimerization of TraR and its release from membranes into the cytoplasm. Embo J. 2000;19:5212–5221. doi: 10.1093/emboj/19.19.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Luo ZQ, Farrand SK. Domains formed within the N-terminal region of the quorum-sensing activator TraR are required for transcriptional activation and direct interaction with RpoA from agrobacterium. J Biol Chem. 2004 doi: 10.1074/jbc.M405299200. [DOI] [PubMed] [Google Scholar]
- Rasband WS. Image J. Bethesda, Maryland, USA: National Institutes of Health; 2004. [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- Sjoblom S, Brader G, Koch G, Palva ET. Cooperation of two distinct ExpR regulators controls quorum sensing specificity and virulence in the plant pathogen Erwinia carotovora. Mol Microbiol. 2006;60:1474–1489. doi: 10.1111/j.1365-2958.2006.05210.x. [DOI] [PubMed] [Google Scholar]
- Stevens AM, Fujita N, Ishihama A, Greenberg EP. Involvement of the RNA polymerase alpha-subunit C-terminal domain in LuxR-dependent activation of the Vibrio fischeri luminescence genes. J Bacteriol. 1999;181:4704–4707. doi: 10.1128/jb.181.15.4704-4707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stibitz S. Mutations in the bvgA gene of Bordetella pertussis that differentially affect regulation of virulence determinants. J Bacteriol. 1994;176:5615–5621. doi: 10.1128/jb.176.18.5615-5621.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempe J, Petit A, Holsters M, Van Montagu M, Schell J. Thermosensitive step associated with transfer of the Ti plasmid during conjugation: possible relation to transformation in crown gall. Proc Natl Acad Sci U S A. 1977;74:2848–2849. doi: 10.1073/pnas.74.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott AE, Stevens AM. Amino acid residues in LuxR critical for its mechanism of transcriptional activation during quorum sensing in Vibrio fischeri. J Bacteriol. 2001;183:387–392. doi: 10.1128/JB.183.1.387-392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski ML, Lostroh CP, Greenberg EP. Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein. J Bacteriol. 2004;186:631–637. doi: 10.1128/JB.186.3.631-637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini A, Volpari C, Gargioli C, Muraglia E, Cortese R, De Francesco R, Neddermann P, Marco SD. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. Embo J. 2002;21:4393–4401. doi: 10.1093/emboj/cdf459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CE, Winans SC. Identification of amino acid residues of the Agrobacterium tumefaciens quorum-sensing regulator TraR that are critical for positive control of transcription. Mol Microbiol. 2005;55:1473–1486. doi: 10.1111/j.1365-2958.2004.04482.x. [DOI] [PubMed] [Google Scholar]
- White CE, Winans SC. The quorum-sensing transcription factor TraR decodes its DNA binding site by direct contacts with DNA bases and by detection of DNA flexibility. Mol Microbiol. 2007;64:245–256. doi: 10.1111/j.1365-2958.2007.05647.x. [DOI] [PubMed] [Google Scholar]
- Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev. 2001;25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- Williams SM, Savery NJ, Busby SJ, Wing HJ. Transcription activation at class I FNR-dependent promoters: identification of the activating surface of FNR and the corresponding contact site in the C-terminal domain of the RNA polymerase alpha subunit. Nucleic Acids Res. 1997;25:4028–4034. doi: 10.1093/nar/25.20.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans SC, Zhu J, More MI. Cell density-dependent gene expression by Agrobacterium tumefaciens during colonization of crown gall tumors. In: Dunny GM, Winans SC, editors. Cell-Cell Signaling in Bacteria. Washington, D. C: ASM Press; 1999. [Google Scholar]
- Winans SC, Bassler BL. Mob psychology. J Bacteriol. 2002;184:873–883. doi: 10.1128/jb.184.4.873-883.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Murphy PJ, Kerr A, Tate ME. Agrobacterium conjugation and gene regulation by N-acyl-L-homoserine lactones. Nature. 1993;362:446–448. doi: 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]
- Zhang RG, Pappas T, Brace JL, Miller PC, Oulmassov T, Molyneaux JM, Anderson JC, Bashkin JK, Winans SC, Joachimiak A. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature. 2002;417:971–974. doi: 10.1038/nature00833. [DOI] [PubMed] [Google Scholar]
- Zhu J, Beaber JW, More MI, Fuqua C, Eberhard A, Winans SC. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J Bacteriol. 1998;180:5398–5405. doi: 10.1128/jb.180.20.5398-5405.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Winans SC. Activity of the quorum-sensing regulator TraR of Agrobacterium tumefaciens is inhibited by a truncated, dominant defective TraR-like protein. Mol Microbiol. 1998;27:289–297. doi: 10.1046/j.1365-2958.1998.00672.x. [DOI] [PubMed] [Google Scholar]
- Zhu J, Winans SC. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc Natl Acad Sci U S A. 1999;96:4832–4837. doi: 10.1073/pnas.96.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]