Abstract
The packaging of eukaryotic DNA into chromatin sterically occludes polymerases, recombinases and repair enzymes. How chromatin structure changes to allow their actions is unknown. We constructed defined fluorescently labeled trinucleosome arrays, allowing analysis of chromatin conformational dynamics via fluorescence resonance energy transfer (FRET). The arrays undergo reversible Mg2+-dependent folding like that of longer arrays studied previously. We define two intermediate conformational states in the reversible folding of the nucleosome arrays, and characterize the microscopic rate constants. Nucleosome arrays are highly dynamic even when compact, undergoing conformational fluctuations on seconds to microseconds timescales. Compact states of the arrays allow binding to DNA within the central nucleosome via site exposure. Protein binding can also drive decompaction of the arrays. Thus, our results reveal multiple modes by which spontaneous chromatin fiber dynamics allows for the invasion and action of DNA processing protein complexes.
Eukaryotic genomic DNA is wrapped into repeating chains of nucleosomes, which fold into more-compact higher order chromatin structures. DNA that is wrapped in nucleosomes is sterically occluded from large multiprotein assemblies that function on naked DNA substrates, such as RNA and DNA polymerases and repair and recombination enzyme complexes1; and both nucleosomal DNA and the linker DNA that connects nucleosomes are further occluded by chromatin folding2. Nevertheless, compact heterochromatic DNA undergoes regulated transcription3,4, repair5, and recombination6,7; gene activator proteins, RNA polymerase, and other proteins readily occupy target sites in heterochromatin8 and even in highly condensed mitotic chromosomes9; and the heterochromatin-associated protein HP1 exchanges rapidly, despite visible compaction of the heterochromatin10. How DNA becomes available for critical genetic functions, when it is wrapped in nucleosomes and further packed into compact chromatin fibers, is not known.
Nucleosomes themselves are highly dynamic, spontaneously undergoing “site exposure” conformational fluctuations that make their wrapped DNA transiently accessible to diverse DNA binding proteins11,12. Site exposure provides spontaneous access to the entire nucleosomal DNA length, with particularly rapid and efficient access to the outer-most stretches of the DNA, which spontaneously unwrap as often as once every ~250 milliseconds13. Spontaneous site exposure might play a role in photolyase-mediated repair of DNA in vivo, which occurs more quickly than can be explained by known ATP-dependent remodeling activities14; and it might contribute to genome-wide transcriptional regulation in vivo15,16, through a nucleosome-induced cooperativity17-20.
Could compact chromatin fibers, too, be intrinsically dynamic - compact in the time average, yet spontaneously undergoing transient conformational fluctuations to more-accessible states? Such intrinsic dynamics have been postulated previously21,22, but have not been demonstrated. Were such compaction/decompaction conformational dynamics to both occur and have suitable rates, they could help explain how nucleosomes in the middle of long arrays in vitro undergo spontaneous site exposure even when the arrays are highly compact on average23. More generally, such dynamics could help explain how DNA in vivo manages to be accessible to the large multiprotein assemblies that carry out the many essential genetic processes.
Here we describe studies that reveal rapid spontaneous conformational dynamics in a model reconstituted chromatin fiber, and we show how these conformational dynamics both influence and respond to the binding of a site-specific DNA binding protein. Our experiments utilize model nucleosome arrays labeled specifically with fluorescent donor and acceptor dyes, which are placed at particular locations that allow the overall compaction of the arrays, or, separately, site exposure within the central nucleosome within the array, to be monitored by FRET24. Compact nucleosome arrays prove to be highly dynamic, with significant conformational fluctuations occurring over timescales of microseconds to seconds. At least two intermediate conformational states exist in the reversible unfolding and refolding of the nucleosome arrays; we measure or place bounds on all of the corresponding microscopic rate constants. At least one compact state of the arrays allows binding to DNA inside the central nucleosome via site exposure within that nucleosome. Protein binding can also drive array decompaction.
RESULTS
FRET assay for array compaction and decompaction
We created nucleosome arrays comprising three highly positioned nucleosomes (using our “601” nucleosome positioning sequence25) separated from each other by 20 bp of linker DNA, corresponding to a nucleosome repeat length of 167 bp. Thus, our design exactly reproduces the architecture of the tetranucleosome whose structure is known from X-ray crystallography26 and the architectures of some of the longer nucleosome oligomers whose properties have been analyzed in solution and by electron microscopy26-28.
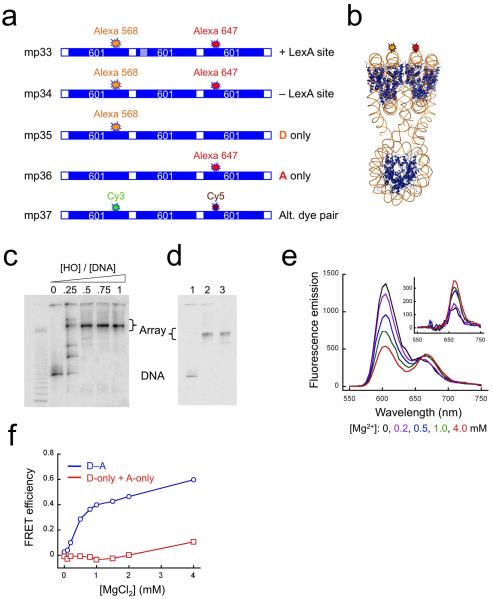
To create a FRET system for analysis of nucleosome array compaction and decompaction, we labeled our trinucleosomes at unique locations on their DNA with fluorescent Alexa568 donor and Alexa647 acceptor dyes (DNA mp34, Fig. 1a) such that if the arrays in solution adopt a compact conformation like that observed in the X-ray structure, the dyes would be close enough together in space to yield efficient FRET (Fig. 1b); and if these arrays undergo conformational changes resulting in significant changes in compactness, the FRET signal would change accordingly. Nucleosomes were reconstituted onto mp34 DNA, and the resulting nucleosome arrays purified on sucrose gradients (Fig. 1c,d).
Figure 1. FRET analyses of nucleosome array compaction induced by Mg2+.
(a) Schematic illustrations of dye-labeled DNA constructs. “601” refers to the 147 bp-long nucleosome positioning sequence 60125,44; hatched boxes represent a specific target site for LexA protein; white boxes represent linker DNA; locations of the fluorescent donor dyes Alexa568 or Cy3, and of the acceptor dyes Alexa647 or Cy5 are indicated. (b) Expected 3-dimensional structure showing locations of the donor (orange) and acceptor (red) fluorophores. (c) Native gel analysis of the titration of 32P-labeled trinucleosome DNA with histone octamer, imaged by a Storm phosphorimager. The mass ratios of histone octamer (HO) to total DNA (including the competitor) are indicated. (d) Native gel analysis of a reconstitution reaction containing 5 μg of trinucleosome DNA, 15 μg of competitor DNA and 15 μg of HO, showing a fluorescence image of the Alexa647 dye. Lane 1, naked DNA; lane 2, after reconstitution; lane 3, after sucrose gradient purification. (e) Fluorescence spectra of the FRET labeled trinucleosomes assembled on DNA mp34 in 0.5x TE buffer plus 0-4.0 mM MgCl2. Inset: spectra after subtracting donor emission. (f) Absolute FRET efficiency from data of panel (e) (blue) and from a parallel titration of a mixture containing 50% donor-only and 50% acceptor-only labeled arrays (red), at the same total array concentration, as a control for aggregation.
In solution, long nucleosome arrays and trinucleosomes are extended (de-compacted) in the absence of Mg2+ and adopt compact conformations in the presence of increasing concentrations of Mg2+ from 0-1 mM29 or of NaCl from 5-100 mM30. Titration of our arrays with increasing concentrations of Mg2+ was accompanied by decreasing donor emission (at ~600 nm) and a concomitant increase in acceptor emission (at ~670 nm) (Fig. 1e), implying a substantial increase in FRET efficiency from ~0.0 to ~0.6 (Fig. 1f). Several experiments proved that this increase in FRET efficiency arises from a decreased distance between donor and acceptor dyes, implying a Mg2+ dependent compaction of the arrays, not from other mechanisms24. The FRET change is not due to dye-specific effects, because arrays labeled at identical locations with Cy3 and Cy5 (DNA mp37) exhibited similar Mg2+-dependent changes in FRET (data not shown; see below). Three experiments prove that the FRET increase is not due to aggregation. Titration of an equimolar mixture of arrays labeled with Alexa568 only and with Alexa647 only (DNAs mp35, mp36; Fig. 1a) exhibited no FRET increase (Fig. 1f). Titration of a different FRET system, in which the donor and acceptor dyes were placed at locations that do not neighbor in the compact structure as defined by X-ray crystallography, did not reveal significant compaction in 1 mM Mg2+ (DNA mp38; Supplementary Fig. 1), but the corresponding mixing experiment did reveal aggregation at higher Mg2+ concentration (DNAs mp39, mp310). And measurements of translational diffusion coefficients by fluorescence correlation spectroscopy (FCS) using arrays labeled with donor only (DNA mp35, Fig. 1a) showed that the diffusion coefficient of the arrays increased by ~17 ± 2% (n=3) upon addition of 1 mM Mg2+, consistent with increasing compactness, and inconsistent with aggregation (Supplementary Fig. 2). The 20bp linker DNAs in these constructs are far shorter than the DNA persistence length in 10 mM Mg2+ (~140 bp)31, so proximity of donor and acceptor dyes cannot arise from mere flexibility of the linker DNA. Finally, for both dye pairs, most of the FRET change occurred between 0-1 mM Mg2+, as expected for Mg2+-dependent compaction29. Together, these findings show that the trinucleosomes undergo a Mg2+-dependent compaction in solution, as expected. We use this system below, to monitor their spontaneous and protein-driven conformational changes.
The characteristic distance (R0) for 50% FRET efficiency with the Cy3-Cy5 pair is 6 nm32. Thus, these FRET values suggest that, in the absence of Mg2+, the arrays are extended, with the dyes on the terminal nucleosomes separated on average by greater than ~10 nm, whereas in the presence of Mg2+, the arrays are compact, with the dyes separated on average by 5-6 nm24. We do not attempt to interpret the FRET changes in terms of exact donor-acceptor distances; however, the apparent ~5-6 nm distance observed in the presence of Mg2+ exceeds the ~3-4 nm expected based on the crystallographic structure of the tetranucleosome26. As will be shown below, this difference is due in part to the compact state being in a rapid dynamic equilibrium with less-compact states, such that the time-averaged compactness is less than the greatest-possible compactness, which may be observed in the crystal.
FRET assay for site exposure in a nucleosome array
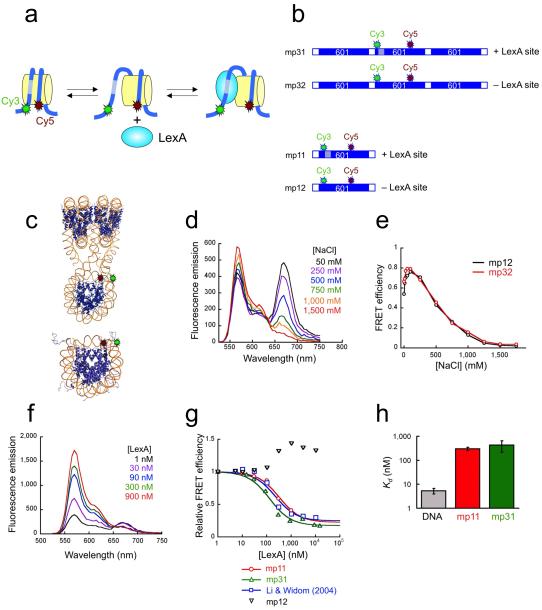
To study how a nucleosome within an array accommodates the binding of an exogenous site-specific DNA binding protein (we chose for convenience the Escherichia coli repressor protein LexA), we designed a second FRET-labeled array system (Fig. 2a,b), in which both dyes are placed on the central nucleosome, at locations such that the resulting FRET signal is sensitive to site exposure conformational changes in this nucleosome (shown schematically in Fig. 2a for the case of a mononucleosome; see also refs.12,13). When the DNA of this labeled nucleosome is in the fully wrapped state, a high FRET signal will be observed. Site exposure increases the distance between donor and acceptor, decreasing the FRET and freeing up the LexA binding site. Binding of LexA protein to the exposed target site traps the nucleosome in this site-exposed low-FRET state.
Figure 2. FRET analyses of site exposure in mononucleosomes and in nucleosome arrays, in the absence of Mg2+.
(a) Schematic illustration of FRET system for detecting site exposure. A mononucleosome system is shown. The approximate locations of Cy3 donor and Cy5 acceptor dyes are indicated. Site exposure occurs via unwrapping from a DNA end12, increasing the distance between donor and acceptor and thus decreasing the FRET, while freeing up the LexA target site (hatched). Binding of LexA traps the nucleosome in this open state. (b) Schematic illustrations of dye-labeled DNA constructs. (c) Expected 3-dimensional structure showing locations of the donor (green) and acceptor (red) fluorophores. (d) Fluorescence spectra of mp32 trinucleosome arrays titrated with NaCl from 0-1.5M. (e) Absolute FRET efficiencies from the data of panel (d) and from a parallel titration on mononucleosomes (black). (f) Fluorescence spectra of mp31 trinucleosome arrays titrated with LexA protein from 1-900 nM. (g) Quantitative FRET analysis of LexA titrations from (e) together with parallel titrations on mononucleosome systems with (mp11) or without (mp12) LexA target sites, showing also results obtained on a related LexA site-containing mononucleosome system having the acceptor dye on the histone core (residue 35 of histone H3)12 instead of on the DNA. Curves are fits to simple binding isotherms12. (h) Apparent binding affinities (dissociation constants) of LexA from measurements such as those in (f) obtained by fitting the FRET data to a simple binding isotherm. Error bars, standard deviations, n = 2.
We created variants of this FRET-labeled nucleosome array with and without a binding site for LexA protein inside the labeled nucleosome (DNAs mp31, mp32; Fig. 2b) at the identical location within this nucleosome as analyzed previously12,13. FRET changes that we might observe could be due to changes in nucleosome packing within the array rather than to conformational changes within the central nucleosome itself. To control for this possibility, we created FRET-labeled mononucleosomes identical to the labeled central nucleosome of the array (with or without a LexA site) using DNAs mp11 and mp12 (Fig. 2b,c). Nucleosomes were reconstituted onto FRET-labeled array or mononucleosome DNA and further purified purified (Supplementary Fig. 3).
Two titration experiments confirmed that these FRET systems properly monitor site exposure conformational changes within the central nucleosome of the array: first, using NaCl, which destabilizes the wrapping of the nucleosomal DNA (and, at sufficiently high concentrations, drives the histones off altogether)12,13; and second, using LexA protein, which binds to its nucleosomal DNA target site only when that nucleosome undergoes a site exposure conformational change12,13.
Titration of these arrays with NaCl (Fig. 2d) leads to increasing Cy3 emission and decreasing Cy5 emission, implying a substantial decrease in FRET (Fig. 2e), as expected for a system monitoring the unwrapping of DNA in the central nucleosome. The decreasing FRET arises from the progressively increasing fraction of time that the DNA in the labeled nucleosome spends partially (or, at very high NaCl concentrations, completely) unwrapped off the histone surface12,13,33. Similar titrations on mononucleosomes gave identical results (Fig. 2e). These results confirm that the FRET changes observed with the arrays are not due to changes in packing of nucleosomes within the array, but reflect conformational changes within the labeled central nucleosome.
Titration of these arrays with LexA protein (here done in the absence of Mg2+, so that the arrays are extended) also led to a decrease in FRET (Fig. 2f,g). Thus, as expected, LexA binds to the central nucleosome within the array, and binding is coupled to partial unwrapping (site exposure) of the DNA of this nucleosome. The DNA unwrapping is only partial, since the FRET does not decrease to zero, even at near-saturating LexA concentrations. This FRET system is sensitive only to site-specific binding of LexA, not to nonspecific binding, since titration of a FRET-labeled nucleosome lacking a specific LexA target site did not decrease the FRET (Fig. 2g). We obtained quantitatively identical results for specific binding by LexA with the mononucleosome and array systems (Fig. 2g,h). Thus, site exposure in the central nucleosome of extended arrays is negligibly influenced by the existence of neighboring nucleosomes.
Protein binding via spontaneous site exposure in a compact nucleosome array
The above data show that we have established two different kinds of FRET system: one that monitors overall compaction of the array (Fig. 1); and one that monitors site exposure conformational changes within the central nucleosome (Fig. 2). We used these systems to ask whether LexA binding couples to changes in the conformation of nucleosome arrays when the arrays start out in their compact state. Our previous work23 allowed us to ask how array compaction influenced protein binding, but did not allow analysis of what happens to the array when the proteins bind. Our new FRET systems provide both kinds of information directly.
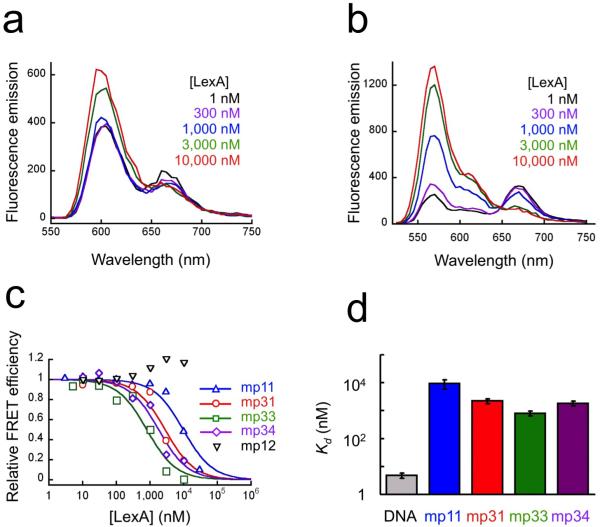
We titrated FRET-labeled arrays that are sensitive to array compactness (mp33) or to site exposure in the central nucleosome (mp31) with increasing concentrations of LexA protein in solutions containing 1mM Mg2+, such that, prior to addition of LexA, the arrays are compact. As expected, in zero or very low LexA concentrations, the compactness-sensitive arrays exhibit high FRET (Fig. 3a), showing that they are indeed compact. Correspondingly, in the same conditions, the site exposure-sensitive arrays exhibit high FRET (Fig. 3b), showing that the central nucleosome’s DNA is fully wrapped. Increasing concentrations of LexA lead to decreasing FRET in both systems (Fig. 3a,b).
Figure 3. FRET analyses of site exposure and decompaction for nucleosome arrays in 1 mM Mg2+.
(a) LexA titration of compaction-sensitive nucleosome arrays (DNA mp33) in 1 mM Mg2+, monitored by FRET. (b) As in (a) except using site exposure-sensitive arrays (DNA mp31). (c) Quantitative analysis of titrations from (a,b) and additional systems: compaction-sensitive arrays lacking a LexA site (mp34), and site exposure-sensitive mononucleosomes having (mp11) or lacking (mp12) a LexA site. (d) Apparent binding affinities (dissociation constants) of LexA from measurements such as those in (c). Error bars, standard deviations, n = 2-3.
Consistent with our analysis of restriction enzyme accessibility in nucleosome 17-mer arrays23, quantitative analysis of the data from the site exposure-sensitive arrays (Fig. 3c,d) shows that LexA binds to its target site in the middle of the compact array with no reduction in affinity (if anything, a slight increase in affinity) compared to binding to the same site in a mononucleosome (compare DNA mp31 with mp11). Binding is accompanied by loss of the intra-nucleosomal FRET signal, but only if the nucleosome contains a specific LexA target site (compare DNA mp11 with mp12). Thus, these data establish that proteins can bind site-specifically to target sites inside a nucleosome in the middle of a compact nucleosome array, and show for the first time that this binding occurs via site exposure within the central nucleosome.
Array decompaction driven by nonspecific LexA binding
To assess whether LexA binding influenced the overall compaction of the arrays we carried out parallel LexA titrations to those described above except now using the compaction-sensitive FRET system. Binding of LexA is accompanied by de-compaction of the arrays (Fig. 3c,d). Importantly, however, equivalent results obtain regardless of whether the arrays contain a specific LexA binding site within the central nucleosome, or not (compare DNA mp33 with mp34). Thus, whereas the site-exposed state is driven to high occupancy only by site-specific LexA binding, decompaction of the arrays is driven by nonspecific LexA binding. We presume that the relevant nonspecific binding is to the linker DNA (see Discussion); indeed, as we shall discuss there, this outcome is predicted from our analyses of restriction enzyme accessibility within nucleosome 17-mer arrays23.
Dynamics of spontaneous nucleosome array compaction and decompaction
How is it that DNA binding proteins are able to bind via site exposure to a nucleosome inside a compact array? The simplest possibility is that perhaps the compact state of the arrays is not a frozen, inert conformational state, as imaged by X-ray crystallography26, but instead, it might be dynamic, fluctuating between more- and less-compact conformations in some manner that makes site exposure possible. This idea is analogous to how we presently understand the binding of proteins to target sites inside isolated nucleosomes: binding occurs via intrinsic nucleosome conformational dynamics (site exposure), which happens spontaneously and with high frequency, preceding and allowing the subsequent binding of a protein to a now freely-accessible nucleosomal DNA target site13.
If this reasoning is correct, it follows that nucleosome arrays should exhibit rapid transient conformational fluctuations between decompacted and re-compacted states, even in the absence of any other exogenous DNA binding proteins, and even in solution conditions (such as 1 mM Mg2+), in which the arrays are compact in the time average. If compaction/decompaction fluctuations were limited by diffusion, they might occur very rapidly, since nucleosomes diffuse a distance comparable to their diameter in just microseconds34. However, if a large energy barrier separates the compact and de-compacted states, the actual timescale could be far slower.
Since there is at present no information even on the existence of such potential nucleosome array conformational dynamics, let alone on their timescales, we took two complementary approaches, which together span the entire relevant range of timescales. FRET-FCS allows the analysis of conformational fluctuations on timescales of microseconds to tens of milliseconds, while stopped-flow FRET allows analyses for timescales of milliseconds or longer. Both experiments are sensitive only to conformational changes that are large enough to yield significant changes in FRET.
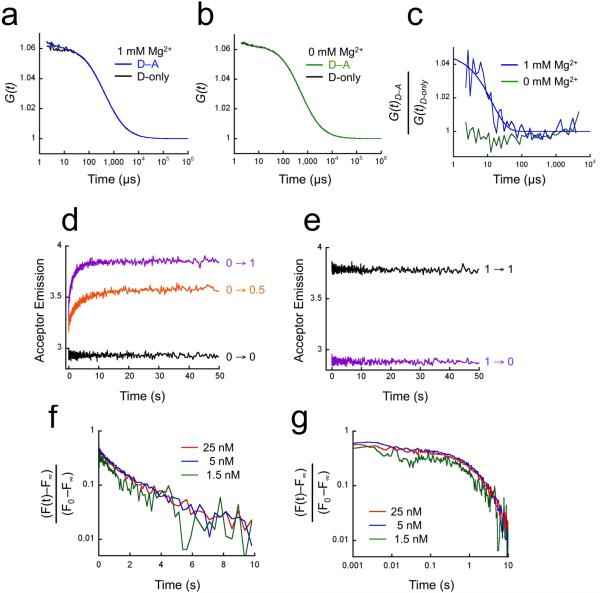
The FRET-FCS experiment measures the ratio of fluorescence donor intensity autocorrelation functions for nucleosome arrays labeled with both donor and acceptor (DNA mp34), and, separately, for arrays labeled with donor only (DNA mp35), thereby eliminating contributions from diffusion. If nucleosome arrays undergo reversible interconversion between two conformational states, one more-compact (high FRET), and one less-compact (low FRET), and if these fluctuations occur within microseconds to tens of milliseconds, this will be manifested as an exponentially decaying ratio function with a characteristic decay time equal to the sum of the decompaction plus re-compaction rate constants13.
Indeed, FRET-FCS analysis (Fig. 4) revealed an exponentially decaying ratio autocorrelation function with a decay time of ~10-5 seconds, implying that nucleosome arrays that, on average, are compacted (in 1 mM Mg2+), undergo spontaneous rapid but transient fluctuations to less-compact conformations and back, remarkably quickly, ~105 times per second. The small amplitude of the ratio function (Fig. 4c) implies either that the two states are roughly equally populated but that the more-compact state is only slightly more-compact (slightly greater FRET) or alternatively that the more-compact state is significantly more compact (significantly greater FRET) but is populated only with low probability. As expected, no decay is observed for arrays in the absence of Mg2+, where compact states have negligible probability.
Figure 4. Conformational dynamics of nucleosome arrays.
(a) Fluorescence donor (Alexa568) autocorrelation decay curves for compact nucleosome arrays in 1 mM Mg2+. Blue: arrays labeled with both FRET donor and acceptor dyes (mp34); black, arrays labeled with donor only (mp35). (b) Fluorescence donor autocorrelation decay curves for extended nucleosome arrays (0 mM Mg2+). Green: arrays labeled with both FRET donor and acceptor dyes (mp34); black, arrays labeled with donor only (mp35). (c) Ratios of donor autocorrelation decay in presence of acceptor to decay with donor-only, from the data of (a; blue) and (b; green). (d) Stopped-flow FRET analysis of kinetics of nucleosome array compaction. Equal volumes of compaction-sensitive nucleosome arrays (DNA mp37) that were in their extended state (0 mM Mg2+) were rapidly mixed with MgCl2, and Cy5 emission was monitored. The final concentration of the arrays was 5 nM; final concentrations of MgCl2 are indicated. (e) Stopped-flow FRET analysis of array decompaction. As in (d) except the arrays were initially compact in 1 mM Mg2+, and then they were rapidly mixed with an excess of EDTA (final concentration 2 mM), so that the effective final Mg2+ concentration is zero. (f) Quantitative analysis of array compaction kinetics, for 1 mM Mg2+ (final) and final concentrations of arrays of 1.5, 5, and 25 nM (indicated). F(t), measured Cy5 fluorescence at time (t); F0, intensity at time 0; F∞, fluorescence at long time. (g) As in (f) except plotted with time on a log scale.
Our stopped-flow FRET experiments start with extended nucleosome arrays (in 0 mM Mg2+); we then rapidly add 1 mM Mg2+, and measure the rate with which the arrays compact, as monitored by the corresponding increase in FRET. The resulting data reveal a large instantaneous increase in FRET, together with a slower (but still rapid), further FRET increase of comparable magnitude (Fig. 4d). The instantaneous FRET increase implies a corresponding increase in array compactness that occurs faster than the mixing deadtime of the stopped-flow instrument, which is ~10-3 seconds, while the subsequent FRET increase, having a time constant of ~2 seconds, implies a subsequent further increase in array compactness. Both the instantaneous and slower phases of these kinetics reflect intramolecular conformational changes, not aggregation/disaggregation, since both rate constants are independent of array concentration over a 17-fold range (Fig. 4f,g).
Stopped-flow FRET also allows us to investigate the kinetics of decompaction. We start with compact arrays (in 1 mM Mg2+), then rapidly add excess EDTA, chelating the Mg2+, such that the stable conformation of the arrays is now the extended state. Decompaction of the arrays is monitored by the corresponding loss of FRET. (Note that this experiment measures decompaction in 0 mM Mg2+.) The decompaction reaches completion within the mixing deadtime (~10-3 seconds; Fig. 4e).
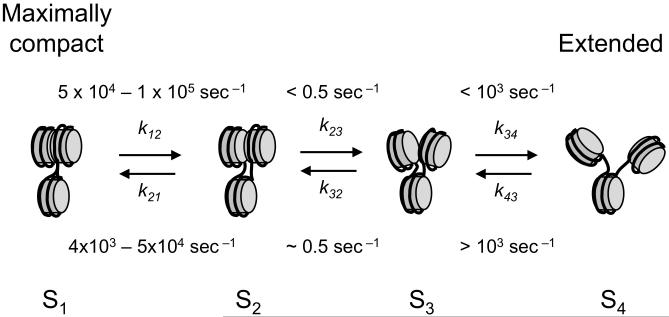
The simplest kinetic mechanism that integrates the equilibrium and kinetic data for 1 mM Mg2+ (Figs. 1 and 4, respectively) requires a minimum of four conformational states (i.e., states with differing FRET efficiencies) (Fig. 5, and Supplemental text), with at least two conformational intermediates between the most-compact and most-extended states of the arrays.
Figure 5. Minimal kinetic scheme for nucleosome array dynamics in 1 mM Mg2+.
The stopped-flow FRET experiments (Fig. 4d,f,g) exhibit biphasic compaction kinetics, requiring a minimum of three conformational states, with the second forward step (S3 → S2, rate k32) and reverse step (S2 → S3, rate k23) being slower than the first forward step (S4 → S3, rate k43). The equilibrium titrations (Fig. 1) demonstrate that the arrays are compact (in the time average) in 1 mM Mg2+; yet the FRET-FCS experiment (Fig. 4a,c) demonstrates rapid interconversion between two states, with a relaxation time of ~10-5 sec, much faster than the rates for S3 ←→ S2, therefore requiring at least one additional compact state (S1) connected reversibly to S2. The corresponding rates (or the bounds on them) imply that only states S1 and S2 are significantly populated in 1 mM Mg2+; yet the central nucleosomes of compact arrays undergo nucleosomal site exposure and bind LexA protein, just as for mononucleosomes. Thus states S1 and/or S2 are competent for nucleosomal site exposure. See Supplementary text for further details of the kinetic analysis. The structures shown are intended only to represent that compactness increases progressively (decreasing distance from nucleosome 1 to nucleosome 3) as the arrays evolve from state S4 → S1.
DISCUSSION
A model system for analysis of conformational dynamics of nucleosome arrays
Our array system exactly reproduces the architecture of three consecutive nucleosomes that adopt a compact structure in the tetranucleosomes investigated by X-ray crystallography26, as well as of 12-mer nucleosome arrays analyzed in solution and by electron microscopy26-29. Consistent with expectation from those studies and from another study of trinucleosomes30, our arrays exhibit a reversible Mg2+ concentration-dependent compaction (Fig. 1). Moreover, the ability of proteins to bind site-specifically to target sites inside the central nucleosome of our trinucleosomes (Fig. 2) agrees quantitatively with our results on binding inside the central nucleosome in a closely similar 17-mer nucleosome array23. Thus, while highly simplified, our trinucleosomes capture essential aspects of the behavior of the tetramer and longer nucleosome arrays, which are widely used as model systems for studies of chromatin structure and function23,26,27,29,35,36. Like many of these other studies, our system lacks the “linker histone” H1, which influences chromatin structure and accessibility30,37. Omission of H1 is justified by the findings that H1 is not essential for viability in some eukaryotes38 and is not a stoichiometric structural component of chromatin39,40.
Spontaneous conformational dynamics and site exposure in compact nucleosome arrays
In our earlier studies on mononucleosomes we proposed11 and later proved12,13,41 that site specific DNA binding proteins can bind to target sites that (in the time average) are buried inside nucleosomes, and that the mechanism of this binding is via spontaneous partial unwrapping (site exposure) of the nucleosomal DNA. The rate of unwrapping sufficient to expose a LexA site at the same location as in this present study is 4 sec-1 (DNA remains fully-wrapped for only ~250 milliseconds before spontaneously unwrapping); and the re-wrapping rate is 20-90 sec-1 (once unwrapped, DNA re-wraps after just 10-50 milliseconds). (Note that the actual rate of binding depends on a protein’s concentration. At low concentrations, e.g., restriction enzyme assays, site exposure occurs in a rapid pre-equilibrium regime, with re-wrapping competing kinetically with protein binding11,41-43. However, at high concentrations, easily reached in vitro, the binding rate exceeds the re-wrapping rate, and unwrapping becomes rate-limiting13.) Our subsequent studies showed that buried nucleosomal DNA target sites remain similarly accessible to restriction enzymes even when those nucleosomes are present in long arrays, and even when these arrays are (in the time-average) highly compact23. Thus it was natural to imagine that binding of proteins to target sites inside nucleosomes in long nucleosome arrays might also be made possible by spontaneous site exposure; but whether and how site exposure could actually occur inside highly compact arrays was not known. Appropriate conformational dynamics within the arrays could certainly help; but whether such dynamics actually occur was not known either.
Our new results answer these questions. Nucleosome arrays are compact in 1 mM Mg2+ (Figs. 1, 4, Supplementary Figs. 1, 2), driven near-completely into a pair of compact states, S1 and S2 (Fig. 5). Moreover, these compact states are highly dynamic, interconverting on an exceptionally fast timescale, ~10 microseconds. Significant FRET fluctuations imply significant changes in distance. (Note that the existence of a populated but less than maximally compact state in 1 mM Mg2+ (S2) could also cause the steady state FRET (Fig. 1) to be lower than expected based on the X-ray structure.) And not only are the compact nucleosome arrays highly dynamic, they are dynamic in a way that allows for binding of LexA protein via site exposure within the central nucleosome of the compact arrays (Fig. 3). DNA unwrapping (the hallmark of site exposure) is demonstrated by the accompanying LexA-dependent decrease in intra-nucleosomal FRET, which implies that the end segment of nucleosomal DNA (where the donor is attached) moves away from the middle stretch of that same nucleosome’s DNA (where the acceptor is attached) coupled to LexA binding. Finally, for both compact and extended arrays, the quantitative extents of binding via site exposure were similar for the arrays compared to mononucleosomes (Figs. 2, 3), as we found previously with longer arrays 23.
Protein binding can drive decompaction of nucleosome arrays
Binding of LexA protein to its target site within the central nucleosome is accompanied by decompaction of the array (Fig. 3c). However this decompaction is not attributable to the site exposure conformational change, because, when nucleosomes lack a LexA target site, addition of LexA protein does not drive formation of the site exposed state (Fig. 2g; and refs.12,13), yet the arrays nevertheless decompact (Fig. 3c). It follows that array decompaction is driven by nonspecific LexA binding. Moreover the concentrations of LexA required to induce array decompaction exceed the dissociation constant for nonspecific binding of LexA to naked DNA, (~300 nM; ref.12), so nonspecific binding is expected.
This nonspecific LexA binding is most likely to linker DNA, because linker DNA more closely resembles ordinary B-form DNA than does nucleosomal DNA26 (and thus is more-recognizable by LexA), and because linker DNA is much-more accessible than is wrapped nucleosomal DNA23. Moreover, we expect on thermodynamic grounds that binding of proteins to linker DNA should cause array decompaction: our studies on 17-mer nucleosome arrays23 showed that array compaction strongly reduced binding to linker DNA compared to binding to naked DNA. Since chromatin compaction destabilizes protein binding to linker DNA, it follows from thermodynamic linkage that the opposite should equally be true: binding to linker DNA should destabilize the compact state of the nucleosome array, driving array decompaction.
Indeed, both of these effects are simultaneously manifested in the present study. We see a large reduction in LexA binding affinity for nonspecific binding to the compact array (~1800 nM for half-maximal effect for the arrays lacking a specific LexA site, versus ~300 nM for nonspecific binding to naked DNA); and at the same time, when binding is driven by sufficiently high LexA concentrations, it is accompanied by decompaction of the array.
In summary, our results demonstrate that protein binding to nucleosome arrays, most likely to the linker DNA, can cause complete decompaction of the arrays. This decompaction could in principle be regulated and have significant consequences for the binding of other proteins to chromatin and ultimately for biological function.
Supplementary Material
ACKNOWLEDGMENTS
We thank Prof. Elliot Elson (Washington University, St. Louis) and Dr. Edmund Matayoshi (Abbott Laboratories) for access to Zeiss Confocor 2 instruments, on which preliminary FCS experiments were carried out. We thank Drs. Elson and Matayoshi, and members of the Widom lab, for discussions, Dr. Karen Swinger (Northwestern University) for help with the trinucleosome structure figures, and the Keck Biophysics and Biological Imaging Facilities at Northwestern University for the use of instruments. M.G.P. acknowledges support from US National Institutes of Health postdoctoral fellowship F32 GM072306 and a Career Award in the Biomedical Sciences from the Burroughs-Wellcome Fund. J.W. acknowledges research support from US National Institutes of Health grants R01 GM54692 and R01 GM58617.
Appendix
METHODS
Labeling DNA oligos
We incorporated amine-reactive Alexa568 and 647 (Invitrogen), or Cy3 and Cy5 (Amersham) into amino-dT-containing oligonucleotides by PCR with labeled DNA oligos. Oligonucleotides were ethanol precipitated, washed, and resuspended in NaH2BO3, pH8.5. The reactive dye was dissolved in DMSO, added to the oligonucleotide in 5-fold molar excess, and incubated at room temperature overnight. The labeled oligos were ethanol precipitated twice, then purified by reverse-phase HPLC on a C18 column with an acetonitrille gradient. The gradient resolved free dye, unlabeled oligonucleotide, and the desired dye-labeled oligonucleotide.
Preparation of Labeled dsDNA
See Supplemental Text for details. We used PCR to prepare labeled mononucleosome DNA and each nucleosome DNA of the trinucleosome templates. Fluorophore-labeled DNA oligos were used to incorporate Cy3 at basepair 3 and Cy5 at the basepair 83 in the 147 bp-long nucleosome positioning sequence (NPS) of DNAs mp11 and mp12, and within the central NPS of DNAs mp31 and mp32. Some sequences contained the 20 bp binding site of LexA between basepairs 8-27 of the NPS. Fluorophore labeled DNA oligos were also used to incorporate Cy3 or Alexa568 at basepair 112th base pair of the left-hand NPS and/or Cy5 or Alexa647 at basepair 35 of the right-hand NPS (DNAs mp31-37). Two different asymmetric TspRI sites were incorporated outside each NPS sequence. Each NPS sequence was digested by TspRI, and phenol-chloroform extracted. The three NPSs were combined at equal molar ratios, ligated, phenol-chloroform extracted, and PAGE purified.
Proteins
We purified histone octamer from chicken erythrocytes as described45. We expressed LexA from plasmid pJWL228 (gift from J. Little) and purified it as described46.
Nucleosome Array Reconstitutions
We reconstituted mononucleosomes by double dialysis from 0.5xTE, 2M NaC, 1mM BZA, 5μg of 601 DNA, 15μg of long salmon sperm DNA (10-50 kb), 10μg of histone octamer, and trace amounts of 32P-labeled 601 DNA47 in 50 μl, then purified away from salmon sperm DNA and any aggregates on sucrose gradients25 and assayed by native 5% PAGE, visualized by phosphorimager (32P) or fluorimager (Cy5, Alexa647). We reconstituted trinucleosomes as above except using 5μg of 601 trimer DNA, 15μg core particle DNA, 15μg histone octamer and trace amounts of 32P-labeled trimer DNA, purified on sucrose gradients, and assayed by native 4% PAGE
LexA binding
We estimated the binding affinity of LexA to the DNA constructs by EMSA. Radiolabeled DNA at 0.1nM was incubated with LexA for 5 minutes in 0.5xTE or 5mM Tris + 1mM MgCl2. Ficoll was added to 3% and the samples analyzed by native 5% PAGE run in 1/3xTBE or 1/3xTB + 1mM MgCl2, then analyzed by phosphorimager.
Fluorescence measurements
We used an ISS PC1 photon-counting spectrometer to acquire fluorescence spectra, with additional 550nm or 570nm cut-on filters for Cy3/Cy5 or Alexa568/Alexa647 arrays, respectively, in the emission channel. Cy3 was excited at 515nm; Cy5 was directly excited at 610 nm. Alexa568 was excited at 545nm; Alexa647 was directly excited at 625nm. All MgCl2 and LexA titrations were carried out with 2nM trinucleosomes or 6nM mononucleosomes.
Fret efficiency measurements
We used the (ratio)A method24 to determine the FRET efficiency, E:
| (1) |
FA(λ) is the fluorescence emission of the acceptor (Cy5, Alexa647), when excited by the wavelength, λ. εD(λ) and εA(λ) are extinction coefficients of the donor (Cy3, Alexa568) and acceptor (Cy5, Alexa647), at λ; d+ the fraction of molecules labeled with acceptor; λ’ the wavelength for donor excitation; λ” the wavelength for direct acceptor excitation. E does not depend on the percent of acceptor labeled molecules. We set d+=1 since we used purified labeled oligos. We noticed a loss of acceptor emission during trinucleosome preparation, likely from photobleaching. This explains the larger than expected Cy3 donor emission12.
Stopped flow measurements
We used an Applied Photophysics instrument for the stopped flow experiments, allowing analyses on timescales of 1msec and longer. Cy3 was excited using an Argon arc lamp with a 500-530nm bandpass filter, and monitored Cy5 emission through a 650nm cut-on filter. Trinucleosomes and MgCl2 were separately diluted into 5mM Tris, pH8.0, and mixed at 1:1 (v/v) ratio. Final concentrations were 25nM, 10nM and 1.5nM trinucleosomes and 0, 0.5mM and 1mM MgCl2. We acquired Cy5 fluorescence emission intensity data on a log(time) scale from 1msec to 100sec. For experiments with 0.5 or 1mM MgCl2 concentrations, the fluorescence intensity at time=0, F0, is obtained from parallel reactions in which the nucleosomes are mixed with 0 mM MgCl2 instead.
FCS measurements
We used a Zeiss Confocor 3 instrument for free solution FCS experiments. We acquired donor (Alexa568) correlation functions for A568/A647 double-labeled (DNA mp34) and A568 donor only-labeled (DNA mp35) trinucleosome arrays. Fluctuations in donor fluorescence emission arise from both diffusion and from conformational fluctuations causing changes in FRET, and are described by the correlation function 13,48-51:
| (2) |
Here, t is the lag time of the donor autocorrelation, τ the diffusion time, α the form factor of the confocal volume, N the average number of molecules in the confocal volume, kbq and k qb the forward and backward rates between the dark and bright fluorescent states, and λ = (kbq + kqb). Diffusion is removed by taking the ratio of donor autocorrelation functions: G(t)donor&acceptor / G(t)donor-only. This analysis requires that the translational diffusion coefficients of singly- and doubly-labeled arrays be identical. While not strictly true, this is a good approximation, since translational diffusion coefficients are weak functions of molecular volume, and the additional fluorophore itself represents only a small increase in molecular volume. Alexa568 was excited using a 514.5nm argon laser at 50 microwatts, and the emission isolated with a 530-590nm bandpass filter. Samples contained 5mM Tris, 10mM DTT, and 0 or 1 mM MgCl2.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Richmond T, Davey C. The structure of DNA in the nucleosome core. Nature. 2003;423:145–50. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 2.Robinson PJ, Rhodes D. Structure of the ‘30 nm’ chromatin fibre: a key role for the linker histone. Curr Opin Struct Biol. 2006;16:336–43. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bühler M, Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol. 2007;14:1041–1048. doi: 10.1038/nsmb1315. [DOI] [PubMed] [Google Scholar]
- 5.Fousteri M, van Hoffen A, Vargova H, Mullenders LH. Repair of DNA lesions in chromosomal DNA impact of chromatin structure and Cockayne syndrome proteins. DNA Repair (Amst) 2005;4:919–25. doi: 10.1016/j.dnarep.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 6.Topp CN, Dawe RK. Reinterpreting pericentromeric heterochromatin. Curr Opin Plant Biol. 2006;9:647–53. doi: 10.1016/j.pbi.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Brutlag DL. Molecular arrangement and evolution of heterochromatic DNA. Annu Rev Genet. 1980;14:121–44. doi: 10.1146/annurev.ge.14.120180.001005. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Widom J. Mechanism of transcriptional silencing in yeast. Cell. 2005;120:37–48. doi: 10.1016/j.cell.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 9.Chen D, et al. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. The Journal of Cell Biology. 2005;168:41–54. doi: 10.1083/jcb.200407182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheutin T, et al. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–5. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 11.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. Journal of Molecular Biology. 1995;254:130–49. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 12.Li G, Widom J. Nucleosomes facilitate their own invasion. Nat Struct Mol Biol. 2004;11:763–9. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 13.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 14.Bucceri A, Kapitza K, Thoma F. Rapid accessibility of nucleosomal DNA in yeast on a second time scale. EMBO J. 2006;25:3123–32. doi: 10.1038/sj.emboj.7601196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein BE, Liu C, Humphrey EL, Perlstein EO, Schreiber S. Global nucleosome occupancy in yeast. Genome Biol. 2004;5:R62. doi: 10.1186/gb-2004-5-9-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Research. 2008;18:1084–91. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams CC, Workman JL. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Molecular and Cellular Biology. 1995;15:1405–21. doi: 10.1128/mcb.15.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polach KJ, Widom J. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. Journal of Molecular Biology. 1996;258:800–12. doi: 10.1006/jmbi.1996.0288. [DOI] [PubMed] [Google Scholar]
- 19.Miller JA, Widom J. Collaborative competition mechanism for gene activation in vivo. Molecular and Cellular Biology. 2003;23:1623–32. doi: 10.1128/MCB.23.5.1623-1632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vashee S, Willie J, Kodadek T. Synergistic activation of transcription by physiologically unrelated transcription factors through cooperative DNA-binding. Biochem Biophys Res Commun. 1998;247:530–5. doi: 10.1006/bbrc.1998.8820. [DOI] [PubMed] [Google Scholar]
- 21.Luger K, Hansen JC. Nucleosome and chromatin fiber dynamics. Curr Opin Struct Biol. 2005;15:188–96. doi: 10.1016/j.sbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 22.Widom J. Toward a unified model of chromatin folding. Annual review of biophysics and biophysical chemistry. 1989;18:365–95. doi: 10.1146/annurev.bb.18.060189.002053. [DOI] [PubMed] [Google Scholar]
- 23.Poirier MG, Bussiek M, Langowski J, Widom J. Spontaneous access to DNA target sites in folded chromatin fibers. Journal of Molecular Biology. 2008;379:772–86. doi: 10.1016/j.jmb.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clegg RM. Fluorescence resonance energy transfer and nucleic acids. Meth Enzymol. 1992;211:353–88. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 25.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. Journal of Molecular Biology. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 26.Schalch T, Duda S, Sargent D, Richmond T. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–41. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 27.Dorigo B, et al. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–3. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 28.Routh A, Sandin S, Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc Natl Acad Sci USA. 2008;105:8872–7. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dorigo B, Schalch T, Bystricky K, Richmond T. Chromatin Fiber Folding: Requirement for the Histone H4 N-terminal Tail. Journal of Molecular Biology. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 30.Bussiek M, Tóth K, Schwarz N, Langowski J. Trinucleosome compaction studied by fluorescence energy transfer and scanning force microscopy. Biochemistry. 2006;45:10838–46. doi: 10.1021/bi060807p. [DOI] [PubMed] [Google Scholar]
- 31.Du Q, Smith C, Shiffeldrim N, Vologodskaia M, Vologodskii A. Cyclization of short DNA fragments and bending fluctuations of the double helix. Proc Natl Acad Sci USA. 2005;102:5397–402. doi: 10.1073/pnas.0500983102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iqbal A, et al. Orientation dependence in fluorescent energy transfer between Cy3 and Cy5 terminally attached to double-stranded nucleic acids. Proc Natl Acad Sci USA. 2008;105:11176–81. doi: 10.1073/pnas.0801707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bao Y, White CL, Luger K. Nucleosome core particles containing a poly(dA.dT) sequence element exhibit a locally distorted DNA structure. Journal of Molecular Biology. 2006;361:617–24. doi: 10.1016/j.jmb.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 34.Yao J, Lowary PT, Widom J. Direct detection of linker DNA bending in defined-length oligomers of chromatin. Proc Natl Acad Sci USA. 1990;87:7603–7. doi: 10.1073/pnas.87.19.7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson PJ, Fairall L, Huynh VA, Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc Natl Acad Sci USA. 2006;103:6506–11. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–7. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 37.Widom J. Chromatin structure: linking structure to function with histone H1. Curr Biol. 1998;8:R788–91. doi: 10.1016/s0960-9822(07)00500-3. [DOI] [PubMed] [Google Scholar]
- 38.Ushinsky SC, et al. Histone H1 in Saccharomyces cerevisiae. Yeast. 1997;13:151–61. doi: 10.1002/(SICI)1097-0061(199702)13:2<151::AID-YEA94>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 39.Bates DL, Thomas JO. Histones H1 and H5: one or two molecules per nucleosome? Nucleic Acids Research. 1981;9:5883–94. doi: 10.1093/nar/9.22.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan Y, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 41.Anderson JD, Thåström A, Widom J. Spontaneous access of proteins to buried nucleosomal DNA target sites occurs via a mechanism that is distinct from nucleosome translocation. Molecular and Cellular Biology. 2002;22:7147–57. doi: 10.1128/MCB.22.20.7147-7157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson JD, Widom J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. Journal of Molecular Biology. 2000;296:979–87. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- 43.Polach KJ, Widom J. Restriction enzymes as probes of nucleosome stability and dynamics. Meth Enzymol. 1999;304:278–98. doi: 10.1016/s0076-6879(99)04017-3. [DOI] [PubMed] [Google Scholar]
- 44.Thåström A, Bingham LM, Widom J. Nucleosomal locations of dominant DNA sequence motifs for histone-DNA interactions and nucleosome positioning. Journal of Molecular Biology. 2004;338:695–709. doi: 10.1016/j.jmb.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 45.Feng HP, Scherl DS, Widom J. Lifetime of the histone octamer studied by continuous-flow quasielastic light scattering: test of a model for nucleosome transcription. Biochemistry. 1993;32:7824–31. doi: 10.1021/bi00081a030. [DOI] [PubMed] [Google Scholar]
- 46.Little JW, et al. Cleavage of LexA repressor. Meth Enzymol. 1994;244:266–84. doi: 10.1016/0076-6879(94)44022-0. [DOI] [PubMed] [Google Scholar]
- 47.Thåström A, Lowary PT, Widom J. Measurement of histone-DNA interaction free energy in nucleosomes. Methods. 2004;33:33–44. doi: 10.1016/j.ymeth.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 48.Bonnet G, Krichevsky O, Libchaber A. Kinetics of conformational fluctuations in DNA hairpin-loops. Proc Natl Acad Sci USA. 1998;95:8602–6. doi: 10.1073/pnas.95.15.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krichevsky O, Bonnet G. Reports on Progress in Physics. 2002. Fluorescence correlation spectroscopy: the technique and its applications. [Google Scholar]
- 50.Hess ST, Huang S, Heikal AA, Webb W. Biological and chemical applications of fluorescence correlation spectroscopy: a review. Biochemistry. 2002;41:697–705. doi: 10.1021/bi0118512. [DOI] [PubMed] [Google Scholar]
- 51.Elson EL, Webb WW. Concentration correlation spectroscopy: a new biophysical probe based on occupation number fluctuations. Annu Rev Biophys Bioeng. 1975;4:311–34. doi: 10.1146/annurev.bb.04.060175.001523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.