The annual outbreak of influenza in Scotland is monitored by sentinel general practices, which report influenza-like illness. We piloted real time virological surveillance to investigate whether polymerase chain reaction (PCR)1,2 is useful for monitoring an outbreak while it is evolving; to compare PCR with two standard techniques—culture and serology; and to compare two media for submitting samples.
Methods and results
Six practices took part. Influenza-like illness was defined by using standard criteria. Combined nose and throat swabs were submitted in both lysis buffer3 and viral transport medium. Two serum samples were taken a minimum of three weeks apart. All samples were posted to the laboratory. Influenza A and B reverse transcription PCR was performed on both media.3 Primary rhesus monkey kidney cells (Biowhittaker, Wokingham) were used to isolate virus. Influenza A and B antibodies were measured using the complement fixation test.
Patients were aged 17 to 72 years (mean 50.5 years), comprising 104 women and 64 men. Samples were taken 1-21 (mean 5.3) days after onset of illness, although 84% of samples were taken within seven days of onset.
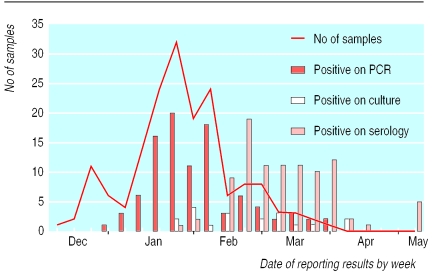
PCR results were available within 36 hours of sample arrival, culture took at least a week, and serology took a minimum of three weeks in this study (figure). Overall, 112 (67%) patients had influenza infection that was confirmed by the laboratory. Of 168 samples, 97 were positive for PCR (57% overall): 84 for influenza A and 13 for influenza B. Nineteen of these also had positive results by culture. Of 153 patients tested serologically, 94 (61%) showed a rising or high (⩾128) titre. Fifteen patients with positive serology had negative results with PCR; nine of these had their swabs taken eight or more days after onset of illness. Excluding samples taken after eight days, the sensitivity of PCR compared with any positive diagnosis (PCR, serology, and culture) was 94.2%. Conversely, 12 patients had positive results with PCR, but had negative results with serology. Thus, serology sensitivity compared with any positive result was 88.7%.
An additional 10% of samples were positive by PCR in lysis buffer alone.
Five of 13 samples that were negative for influenza and that were submitted in the first two weeks of the study were later confirmed by PCR to be rhinovirus infections.
Comment
Real time surveillance using PCR with a rapid turnaround time confirms that influenza is circulating. As the PCR results were faxed back the next day, there was a stimulus to send in further samples. Results of serology and PCR correlated well, although serology took three weeks longer (figure). No false positives were generated by PCR. Culture was insensitive and slow because of the variation between batches of the primary cell line used in this laboratory. The time since onset of illness is critical for the sensitivity of virus isolation and PCR. In the late phase of illness, when results of culture and PCR were negative, there was already a high antibody titre.
Although culture is required to accumulate virus isolates for antigenic characterisation of the circulating viruses, PCR should now be the front line assay for diagnosis of influenza, even in non-specialist laboratories after initial training. It is clear that additional pathogens cause influenza-like illness, and the introduction of a multiplex PCR to test for a wider number of pathogens2,4 will considerably improve surveillance of the winter respiratory burden. New treatments for influenza strengthen the case for improved virological surveillance to alert clinicians to the cause of influenza-like illness and for rapid diagnosis and appropriate treatment of individual cases.
Figure.
Date of arrival of samples (line graph) at the laboratory and dates of reporting results for polymerase chain reaction, virus culture, and serology (bar charts)
Acknowledgments
We acknowledge the hard work performed by the staff of the participating general practices.
Footnotes
Funding: The Scottish Centre for Infection and Environmental Health provided partial financial support.
Competing interests: None declared.
References
- 1.Atmar RL, Baxter BD, Dominguez EA, Taber LH. Comparison of reverse transcription-PCR with tissue culture and other rapid diagnostic assays for detection of type A influenza virus. J Clin Microbiol. 1996;34:2604–2606. doi: 10.1128/jcm.34.10.2604-2606.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stockton J, Ellis JS, Saville M, Clewley JP, Zambon MC. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J Clin Microbiol. 1998;36:2990–2995. doi: 10.1128/jcm.36.10.2990-2995.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace LA, McAulay KA, Douglas JDM, Elder AG, Stott DJ, Carman WF. Influenza diagnosis: from dark isolation into the molecular light. J Infect. 1999;39:221–226. doi: 10.1016/s0163-4453(99)90053-1. [DOI] [PubMed] [Google Scholar]
- 4.Grondahl B, Puppe W, Hoppe A, Kuhne I, Weigl JA, Schmitt HJ. Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J Clin Microbiol. 1999;37:1–7. doi: 10.1128/jcm.37.1.1-7.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]