Abstract
Active-site inhibitors of HIV-1 PR (protease) block viral replication by preventing viral maturation. However, HIV-1 often develops resistance to active-site inhibitors through multiple mutations in PR and therefore recent efforts have focused on inhibiting PR dimerization as an alternative approach. Dimerization inhibitors have been identified using kinetic analysis, but additional characterization of the effect of these inhibitors on PR by physical methods has been difficult. In the present study, we identified a PRMDR (multi-drug-resistant HIV-1 PR) that was highly resistant to autoproteolysis. Using this PR and a novel size-exclusion chromatographic approach that incorporated fluorescence and MS detection, we were able to demonstrate inhibition of dimerization using P27 (peptide 27), a peptide dimerization inhibitor of PR previously identified on the basis of kinetic analysis. Incubation of PRMDR with P27, or other dimerization inhibitors, led to a dose- and time-dependent formation of PR monomers based on the change in elution time by size exclusion and its similar elution time to engineered forms of monomeric PR, namely PRT26A and glutathionylated PR. In contrast, incubation of PRMDR with a potent active-site inhibitor did not change the elution time for the PRMDR dimer. The monomeric PR induced by P27 had fluorescent characteristics which were consistent with unfolded PR. Structure–activity studies identified the active regions of P27 and experiments were performed to examine the effect of other dimerization inhibitors on PR. The present study is the first characterization of dimerization inhibition of PRMDR, a prime target for these inhibitors, using a novel size-exclusion chromatographic approach.
Keywords: autoproteolysis, dimerization inhibitor, gel filtration, HIV-1 protease (PR), monomer, peptide
INTRODUCTION
HIV-1 PR (protease) is an obligate dimer consisting of two identical 99-amino-acid subunits. PR is essential for viral maturation, as it is required for processing HIV-1 Gag and Gag–Pol polyproteins [1,2]. PR is encoded within the Gag–Pol polyprotein, and Gag–Pol undergoes several self-processing steps through the action of the embedded PR to ultimately yield mature dimeric PR [3]. Inhibition of Gag and Gag–Pol processing with active-site PR inhibitors leads to the formation of immature virions that are released from infected cells but remain non-infectious even if the inhibitor is subsequently removed [4,5]. Although significant progress has been made in the development of active-site inhibitors of mature PR for use in the treatment of HIV-1 infection, the virus eventually develops resistance to these inhibitors as a result of multiple mutations both in and outside of the active-site region of PR [6–15]. These resistance mutations often provide cross-resistance to other active-site inhibitors, making changes in therapy more challenging. Since HIV-1 PR is only active in its dimeric state, several groups have pursued the development of dimerization inhibitors to increase the arsenal for the treatment of HIV-1 infection [16–23]. Although several agents that inhibit dimerization in vitro have been identified based primarily on kinetic analysis, more research is necessary to corroborate these findings and provide a further understanding of how these inhibitors affect PR structure leading to inhibition of activity.
Size-exclusion chromatography is often used to examine subunit structure; however, to date it has not been successfully applied to assess the effects of dimerization inhibitors on PR. PRWT (wild-type HIV-1 PR) readily undergoes autoproteolysis resulting in PR fragmentation, and this has hindered the analysis of PR structure by various biophysical methods, such as NMR and analytical ultracentrifugation [24–27]. This obstacle was overcome by using engineered HIV-1 PR mutants that were demonstrated to be highly resistant to autoproteolysis, making it possible to solve the NMR structures for dimeric and monomeric forms of PR [28,29]. Ishima et al. [30] developed a NMR method which made it possible to analyse an engineered monomeric PR at concentrations as low as 20 μM, and this method may ultimately yield insights into the interactions of dimerization inhibitors with the PR monomer. However, using NMR [30], it was not possible to confirm the inhibition of PR dimerization with certain peptides that were previously reported to do so based primarily on kinetic data [17,31]. Recently, Giralt and colleagues have provided the first NMR evidence for the interaction of a peptide dimerization inhibitor (Ac-SEYL-OH) with both monomeric and dimeric forms of PR using 13C labelling of the tryptophan residues of PR [21].
In order to corroborate the kinetic and NMR results obtained with dimerization inhibitors and further characterize their effect on PR structure, we developed a novel size-exclusion chromatographic approach that couples the separation of PR with fluorescence and MS detection. Using this approach, we investigated the effects of the dimerization inhibitor peptide P27 (peptide 27) on PRMDR (multi-drug-resistant HIV-1 PR). P27 was previously shown to inhibit PRMDR activity [20] and to block PR dimerization within infected cells using a fluorescence resonance energy transfer assay [32]. The sequence for PRMDR was derived from an HIV-1-infected patient on antiviral therapy. PRMDR contains eight drug-resistant related mutations that often arise in patients on antiviral therapy [14,20]. However, none of these mutations reside in the N- or C-terminal regions that make up the dimerization interface, suggesting that PRMDR could be susceptible to dimerization inhibitors that have been found to be effective on PRWT. During our attempts to analyse PR by size-exclusion chromatography, PRWT readily underwent auto-proteolysis, whereas PRMDR was highly resistant to autoproteolysis under the same conditions. Thus PRMDR was ideal for studies involving dimerization inhibition, as these types of studies can require extended incubation times due to the PR monomer–dimer system being characterized by low dissociation equilibrium constants [33,34] with presumably slow dissociation rate constants. By using MS detection, it was possible to selectively obtain the elution times for intact PR eluting as a dimer or as a monomer. In the present study, we characterize the inhibition of PRMDR using the dimerization inhibitor P27 and other dimerization inhibitors with a novel size-exclusion chromatographic approach that combines fluorescence and MS detection.
EXPERIMENTAL
PRs, peptides and reagents
PRWT and PRMDR were prepared and refolded as described previously [20,35] and stored as stock solutions at concentrations between 20–50 μM in 20 mM HCl. Unmodified and glutathionylated autoproteolysis-resistant PRKIIA (autoproteolysis-resistant HIV-1 PR) was prepared as described previously [36] and dialysed into running buffer [150 mM ammonium acetate buffer (pH 5.4)] prior to analysis. PRT26A (autoproteolysis-resistant and monomeric HIV-1 PR) T26A (N15-labelled) monomeric PR was a gift from Dr John Louis [NIDDK (National Institute of Diabetes and Digestive and Kidney Diseases), NIH (National Institutes of Health), Bethesda, MD, U.S.A.] and refolded as described previously [30]. Unmodified HIV-2 PR and PRHIV-2-OX (oxidized HIV-2 PR) were prepared as described previously [37], as were both unmodified and modified PRKIIA, an engineered form of PR that is autoproteolysis-resistant [36]. The concentrations of the purified PRs were determined spectrophotometrically (ε280 = 12 300 M−1 · cm−1 for HIV-1 PRs and ε280 = 9150 M−1 · cm−1 for HIV-2 PR). To prepare samples for size-exclusion chromatography, PRs were diluted to a final monomeric concentration of 0.25–2 μM (1 μM is approximately 11 μg · ml−1) into running buffer containing a final concentration of 0.1 mg · ml−1 BSA (also prepared in running buffer). The inclusion of BSA improved PR recovery, stabilized activity and was useful as an internal standard for column performance. Peptides, including P27 (PQITLRKKRRQRRRPPQVSFNFCTLNF), were obtained as described previously [20] and prepared as 1 mM stocks in running buffer [150 mM ammonium acetate buffer (pH 5.4)]. The active-site inhibitor JE-2147, which is effective against PRMDR, a gift from Dr Hiroaki Mitsuya [HIV and AIDS Malignancy Branch, NCI (National Cancer Institute), NIH (National Institutes of Health), Bethesda, MD, U.S.A.], and compound 10 [38] were prepared as 1 mM stock solutions in 100 % DMSO. In experiments with these compounds, the final concentration of DMSO did not exceed 1 %, and 1 % DMSO alone was used as a control. After treatment, the PR samples were incubated at 37 °C for the indicated times and then analysed (8 μl) by size-exclusion chromatography. PR activity was measured using the fluorescent PR substrate as described previously [20].
Analysis of HIV PR by size-exclusion chromatography
Chromatography on the different forms of PR was carried out using a BioSep SEC3000 column (300 mm × 4.6 mm; Phenomenex, Torrance, CA, U.S.A.) with 150 mM ammonium acetate (pH 5.4) running buffer on a 1100 series HPLC–MS system (Agilent, Santa Clara, CA, U.S.A.). A range of pH values and ionic strengths were evaluated and the final conditions used in this report represent those found to be most optimal for chromatographic performance, activity and reproducible detection of HIV-1 PR. The isocratic flow rate was 0.35 ml · min−1. Prior to use, each new column and pre-column was conditioned with several injections of 1 mg · ml−1 BSA and PRMDR (1 μM) in running buffer to first reduce non-specific irreversible binding of these proteins to the column. Proteins eluting from the column were monitored using an Agilent 1100 series fluorescent detector connected in series with an Agilent 1100 series MS detector. Elution times obtained by MS detection were therefore slightly later than those obtained by fluorescence detection. Fluorescence emission spectra for the PR was obtained between 305 and 400 nm with the following settings: step size of 2 nm, a peak width of > 0.4 min (8s, slow) and gain of 15. To allow for efficient ionization of PR eluting from the column, the flow exiting the fluorescence detector was coupled to a mixing tee that provided a mixture of acetonitrile, 0.01 %(v/v) TFA (trifluoroacetic acid) and 0.0075 % (v/v) FA (formic acid) from a second pump set at 0.4 ml · min−1. The mixture entered an Agilent 1100 mass spectrometer set to monitor specific PR molecular ions set in SIM (selective ion monitoring) mode.
Evaluation of PR stability and measurements of autoproteolysis
To study the stability of PRWT and PRMDR for extended incubation times, each PR was incubated in 150 mM ammonium acetate buffer (pH 5.4) at the indicated concentrations in the absence or presence of 1 mM TCEP [tris-(2-carboxyethyl) phosphine] (Calbiochem) and incubated for 16–24 h. Following incubation, the activity was determined using the fluorescent PR substrate as described previously [20] and the extent of proteolysis was assessed by SDS/PAGE (10 % Bis/Tris gels) or analysed by RP-HPLC (reverse-phase HPLC) analysis. To examine the extent of autoproteolysis by RP-HPLC following incubation at 37 °C, enzyme activity was terminated with the addition of solid guanidine hydrochloride to a final concentration of 6 M. Each sample was then separated on a Vydac C18 column using a water/acetonitrile gradient to assess the extent of autoproteolysis. Solvent A was deionized water containing 0.05 %TFA and solvent B was acetonitrile containing 0.05 %FA and 0.025 % TFA. Starting conditions were 5 % (v/v) solvent B and this was linearly increased to 25 % (v/v) in the first 2.5 min. Solvent B was then increased at a rate of 2 % (v/v) per min for 35 min to 95 % (v/v) solvent B and then returned to starting conditions. Elution of PR and PR fragments was monitored at 205 nm and 276 nm and electrospray MS in scan mode.
Evaluation of PRs by MS and quantification
To obtain the most abundant charged species for each PR for use in PR detection during separation and detection in SIM mode, the PRs were first diluted to a a concentration of 10 μM in running buffer (without BSA) and analysed directly by MS in scan mode via direct flow injection at 0.35 ml · min−1 running buffer and 0.4 ml · ml−1 acetontrile/TFA/FA post-column buffer using a mixing tee. The two most abundant mass-over-charge (m/z) PR ions obtained from the ion scan spectrum (the m/z values corresponding to the 10+ and 11+ ions of the PR subunit) were chosen to monitor the elution of PR. A maximum of four selective ions could be monitored per run and the specific m/z values chosen for analysis on the different PRs are shown in Table 1. Using SIM mode, we could obtain specific and sensitive detection of the full-length PR, regardless of its elution time, as the ions monitored are specific for the intact mass of the PR subunit. This obviated interference from inhibitors or PR fragments, if present. In order to quantify the amount of PR dimer and monomer eluting from the column, we used the peak area obtained by MS. In order to do this, it was first necessary to determine the relative response of the mass spectrometer to a known concentration of eluting dimer or monomer. This was done by injection of 2 μM PR dimer (PRMDR treated with the active-site inhibitor JE-2147 to ensure the dimeric form) or monomer (generated by treatment with the dimerization inhibitor peptide CTLNF [16]) and obtaining the area at 276 nm for each form. This was then equated to the area obtained by the mass spectrometer for the 10+ and 11+ PR ions. The amount of eluting PR in pmoles could then be obtained from the area at 276 nm (obtained with a UV diode array detector) using the formula mol=area (276) × flow/εM × path length (1 cm) using the PR molar absorption coefficient ε276 = 12 300 M−1 · cm−1, as described previously [39], and then equated to the area obtained by MS for the same sample using the m/z of 987.6 and 1086.2 (corresponding to the 11+ and 10+ ions of the PRMDR subunit respectively) for each peak. It was found that 1 pmol of dimeric PR typically gave areas of 2.8 × 10−6 and 3.38 × 10−6 for the 10+ and 11+ PR ions respectively. The monomer gave an area of 5.0 × 10−6 and 2.4 × 10−6 for the 10+ and 11+ PR ions respectively.
Table 1.
Specific m/z (10+ and 11+ molecular ions for the PR subunit) used to monitor, by MS analysis, the different HIV PRs used in the present study
| PR | Predicted subunit mass (Da) | Selective ion (10+) (m/z) | Selective ion (11+) (m/z) |
|---|---|---|---|
| PRMDR | 10 852 | 1086 | 987.6 |
| PRWT | 10 790 | 1080 | 981.6 |
| PRKIIA | 10 745 | 1075.5 | 977.6 |
| PRKIIA-Glut | 11 049 | 1105.9 | 1005.6 |
| PRT26A (N15) | 10 818 | 1082.8 | 984.5 |
| PRHIV-2 | 10 720 | 1072.9 | 975.5 |
| PRHIV-2-OX | 10 751 | 1076.1 | 978.4 |
RESULTS
Comparison of PRMDR and PRWT stability following prolonged incubation in assay buffer
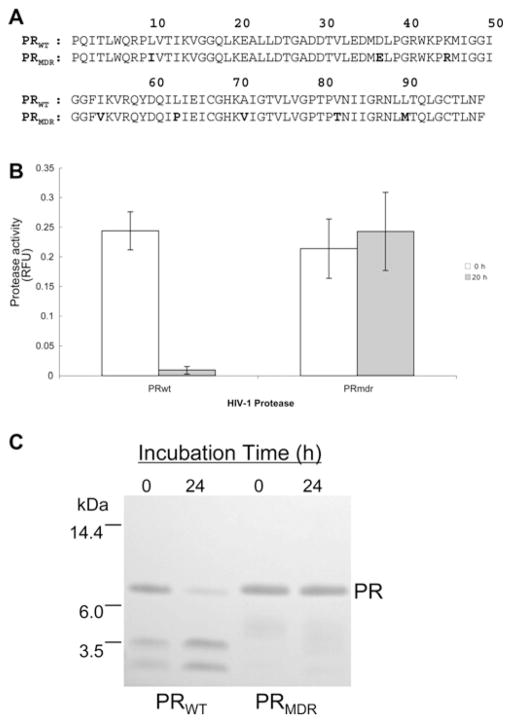
The aligned 99-amino-acid sequences for PRWT and PRMDR are shown in Figure 1(A). The PRWT and PRMDR sequences were derived from primary HIV-1 isolated from a patient prior to and following extensive treatment with antiviral therapy, which included several PR inhibitors (indinavir, ritonavir, saquinavir and amprenavir) and reverse transcriptase inhibitors [14]. PRMDR contains multiple drug-resistant mutations (L10I, K45R, I54V, L63P, A71V, V82T, L90M and I93L) and is highly resistant to a number of active-site inhibitors, although it remains sensitive to the experimental active-site inhibitor JE-2147 [14]. In addition, PRMDR activity is sensitive to the peptide dimerization inhibitor P27 [20]. Since HIV-1 PR is known to undergo autoproteolysis [24,26], the stability of PRWT and PRMDR was examined under the buffer conditions to be used for assessing dimerization inhibition by size-exclusion chromatography. Following a 20 h incubation at 37 °C in 150 mM ammonium acetate buffer (pH 5.4), more than 95 % of the activity of PRWT was lost, whereas PRMDR activity remained unaffected (Figure 1B). To determine if the loss of PRWT activity could be attributed to autoproteolysis, the PRs were examined by SDS/PAGE after incubation. As expected, PRWT undergoes extensive autoproteolysis, whereas PRMDR remained intact (Figure 1C). Further examination of the PRs by RP-HPLC–MS indicated that PRWT had undergone substantial autoproteolysis, as indicated by the presence of the expected autoproteolytic fragments (amino acids 1–33, 34–99 and 64–99) [25], whereas PRMDR remained intact (results not shown). Even within 4 h of incubation, PRWT underwent substantial (approx. 40 %) autoproteolysis.
Figure 1. Sequence alignment for PRWT and PRMDR and analysis of PR stability.
(A) Amino acid sequence for PRWT and PRMDR. The amino acid mutations from PRWT are indicated in bold typeface in the PRMDR sequence. (B) PR activity for PRWT and PRMDR (2 μM) following a 20 h incubation in 150 mM ammonium acetate buffer (pH 5.4) at 37 °C. Results are means ± S.D. (n = 3). (C) Coomassie-stained SDS/PAGE of PRWT and PRMDR following a 20 h incubation at 40 μM in 150 mM ammonium acetate buffer (pH 5.4) at 20 °C. RFU, relative fluorescent units.
MS dose–response profile of PRMDR by size-exclusion chromatography in the absence and presence of an active-site inhibitor
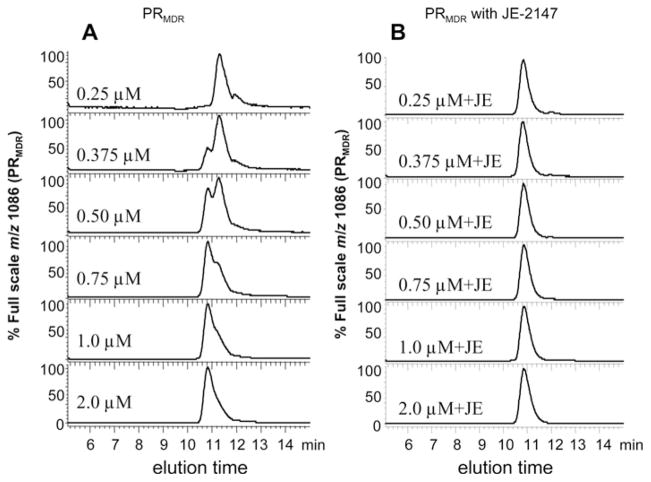
PRMDR is a clinically significant target for next generation PR inhibitors, and since it was found to be highly stable it was used to further examine the effects of dimerization inhibitors on PR structure. To detect PRMDR by size-exclusion chromatography, two PRMDR specific molecular ions were monitored by MS in SIM mode. This allowed for sensitive and specific detection of full-length unmodified PR. In addition, the intrinsic tryptophan fluorescence of PRMDR was also monitored. The PRMDR-specific molecular ions were identified by determining the positive charged molecular ion profile for PRMDR by MS analysis in scan mode (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/419/bj4190497add.htm). The two most abundant PRMDR-specific ions generated corresponded to m/z values of 1086 and 987.6 (the 10+ and 11+ ions for the PR subunit respectively), so these were monitored during chromatography. The same procedure was used to obtain characteristic m/z values for the other forms of HIV PR used in the present study. The charged species that were chosen to detect the various PRs are shown in Table 1. PRMDR was analysed by size-exclusion chromatography at PR subunit concentrations of 0.25–2 μM under the conditions to be used for dimerization inhibitor studies. PRMDR was detectable by MS at concentrations of 0.25 μM and above (Figure 2A, chromatograms shown normalized to full scale to aid comparison) and by fluorescence at 0.75 μM and above (results not shown). The peak elution time for PRMDR was concentration dependent (Figure 2A). These results are consistent with a concentration-dependent formation of PRMDR dimer. At a concentration of 0.5 μM, PRMDR eluted in both forms (monomer and dimer) at essentially equal proportions. This indicated an approximate apparent Kd for dimer dissociation of 0.5 μM (0.25 μM dimeric concentration) under these conditions. On the basis of a standard curve obtained with several different proteins of known molecular mass (BSA, carbonic anhydrase, ribonuclease and insulin), the elution times for the earlier and later forms for PRMDR corresponded to apparent molecular masses of 12 and 6 kDa respectively (expected molecular masses of 22 and 11 kDa) (results not shown). This was consistent with the presence of dimeric and monomeric forms of PR. Although the calculated molecular masses are lower than expected, their relative positions are consistent with the earlier peak representing the dimeric form of PR and with the later-eluting peak representing the monomeric form. The later than predicted elution time for both forms of PR based on the standards may be due to their relatively high pI, since lysozyme, which also has a high pI, was found to elute on this column with a calculated molecular mass which was significantly lower than expected based on the other standards (results not shown). The detection of the earlier and later-eluting forms of PR by MS confirmed that these peaks consisted of intact PR, eliminating the possibility that these peaks were autoproteolytic fragments. PRMDR was also analysed following incubation with the active-site inhibitor JE-2147 (structure shown in Supplementary Figure S2 at http://www.BiochemJ.org/bj/419/bj4190497add.htm). At all of the concentrations tested, PRMDR eluted as the earlier form (dimer) in the presence of the active-site inhibitor JE-2147 (Figure 2B), consistent with the elution of dimeric PR bound to JE-2147. Although the addition of the inhibitor did not alter the peak elution time of 2 μM PRMDR, it did noticeably sharpen the peak shape obtained for the eluting PR, indicating that in the absence of inhibitor, some monomer may exist even at 2 μM (Figure 2, chromatograms shown normalized to full scale for clarity). Since active-site inhibitors of PR are known to bind to and stabilize the PR dimer [28,40,41], these results further substantiate the earlier-eluting form of PRMDR as the dimeric PR [14,42].
Figure 2. Elution profile for PRMDR at increasing concentrations in the absence or presence of the active-site inhibitor JE-2147.
(A) PRMDR (final monomeric protein concentrations of 0.25, 0.375, 0.5, 0.75, 1.0 and 2.0 μM) was incubated for 16 h at 37 °C in 150 mM ammonium acetate buffer containing 100 μg · ml−1 BSA without JE-2147 or (B) with 1 μM JE-2147 (+JE). The column effluent was monitored for PRMDR-specific ions (m/z 1086, 10+) and (m/z 987.6, 11+) in SIM mode. The m/z 1086 is shown. The earlier- and later-eluting PRMDR peaks had approximate elution times of 10.8 and 11.3 min respectively. This experiment was repeated three times with similar results and a representative experiment is shown.
Analysis of the eluting PRMDR by monitoring the intrinsic tryptophan fluorescence reveals the elution profile of the PR as well as BSA (see Supplementary Figure S3 at http://www.BiochemJ.org/bj/419/bj4190497add.htm), which is included in the analysis to improve recovery of PRMDR during chromatography and to help stabilize PRMDR activity during the incubation, as described previously [43]. When PRMDR was analysed in the presence of JE-2147, the elution profile was nearly identical to that seen in the absence of inhibitor (see Supplementary Figure S3). However, the peak was sharper in the presence of inhibitor. We also monitored the molecular ion of JE-2147 by MS and it was found to elute at the same time as that for PRMDR (elution times for both inhibitor and PRMDR were 10.8 min), indicating that the eluting PR was an inhibitor-bound form (see Supplementary Figure S3). In dose–response studies with PRMDR the molecular ion for JE-2147 was detected co-eluting with PRMDR at all concentrations examined (results not shown). There was a strong correlation between the area for JE-2147 by MS and the area for PRMDR by MS (correlation coefficient of 0.997), further suggesting that the dimeric PR elutes as an inhibitor-bound form of the dimer. In addition, there was a good correlation between the amount of PRMDR injected and the peak area obtained when analysing the PR in the presence of inhibitor (correlation coefficient of 0.959). However, the correlation was not as good in the absence of inhibitor (correlation coefficient of 0.889). This is probably due to a decrease in the amount of PR recovered for samples at concentrations below 1 μM and may indicate that PR at lower concentrations is less stable in solution during analysis in the absence of added inhibitor. Other active-site inhibitors (ritonavir, saquinavir and KNI-272) that are not effective inhibitors of the drug-resistant PR did not elute with PRMDR, although they were seen to co-elute with PRWT (results not shown). It is important to note that active-site inhibitors of PRWT and PRMDR were not detected if they were analysed in the absence of PR. This was due to a loss of the inhibitor to the column material, as it was found that the inhibitors could be flushed from the column with injections of 100 % DMSO (results not shown). This suggests that the inhibitor eluting from the column in the presence of PR is specifically bound to the PR rather than co-eluting with the PR as an unbound species.
Characterization of monomeric HIV PRs by size-exclusion chromatography using fluorescence and MS detection
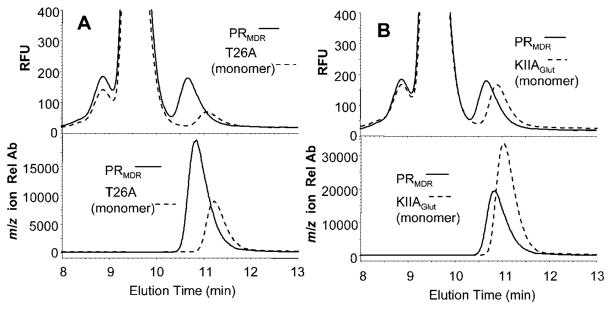
To further confirm that the later-eluting form of PRMDR represented the monomeric form of PRMDR, we analysed the elution profile of two previously identified monomeric forms of HIV PR. The first monomeric form PRT26A (a gift from Dr John Louis) was created by Ishima et al. [30]. PRT26A is resistant to autoproteolysis and has a dissociation constant (Kd) of greater than 1 mM. As demonstrated by NMR analysis, PRT26A behaves exclusively as a monomer at micromolar concentrations [30]. The elution time for the PRT26A monomer was 11.2 min, or approx. 0.4 min later than the PRMDR dimer (10.8 min) (Figure 3A), and is essentially the same as that seen for the later-eluting form of PRMDR. The large peak detected by fluorescence prior to the PR is that of BSA, which is present in the sample. Additionally, we analysed another form of HIV-1 PR, PRKIIA-Glut (PRKIIA with glutathionylation of Cys95), that was made monomeric by glutathionylation of Cys95, an amino acid located near the dimer interface. It was previously shown that glutathionylation of this cysteine residue leads to reversible inactivation of the PR and induces PR to behave as a monomer [36]. PRKIIA-Glut ran slower than dimeric PRMDR with an elution time of 11.0 min (Figure 3B). The earlier elution time for this monomer as compared to PRT26A may be due to the addition of the glutathione moiety that alters both the size and charge characteristics of the protein. To further investigate this, we analysed the elution times for increasing concentrations of PRKIIA-Glut and compared them to the elution times for the deglutathionylated form, PRKIIA, created by treatment with thioltransferase [44]. The elution time for PRKIIA-Glut remained constant at all concentrations analysed (0.06–4 μM), suggesting that glutathionylation prevented the formation of PR dimer (see Supplementary Figure S4 at http://www.BiochemJ.org/bj/419/bj4190497add.htm). However, if the glutathione moiety of PRKIIA-Glut was first removed by treatment with thioltransferase, yielding PRKIIA, then the elution time for PRKIIA became concentration dependent (see Supplementary Figure S4). HIV-2 PR contains a methionine residue at position 95, and oxidation of this residue also leads to reversible inactivation of PR [37]. This inhibition of activity is thought to be due to prevention of PR dimerization. Analysis of PRHIV-2-OX as a result of treatment with hydrogen peroxide as described previously [37] led to an elution time for PRHIV-2-OX that was consistent with a monomeric form of the PR (results not shown).
Figure 3. Elution profile for PRT26A and PRKIIA-Glut monomeric PRs as compared to PRMDR.
(A) PRMDR (1 μM, solid lines) and PRT26A (2 μM, dashed lines) were incubated at 37 °C for 60 min, and then 8 μl of each PR was separated by size-exclusion chromatography and detected by fluorescence (upper panel) or MS (lower panel) in SIM mode for the 10+ m/z molecular ion for each PR (see Table 1 for details). (B) PRMDR (1 μM, solid lines) and PRKIIA-Glut (1 μM, dashed lines) were incubated at 37 °C for 60 min, and then 8 μl of each PR was separated by size-exclusion chromatography and PRs were detected by fluorescence (upper panel) or MS (lower panel) in SIM mode for the PR m/z (10+) molecular ions. The elution times as determined by MS for PRMDR, PRT26A and PRKIIAGlut were 10.8, 11.0 and 11.2 min respectively. The major peak observed in the fluorescence profile is that of BSA. Rel Ab, relative ion abundance; RFU, relative fluorescent units.
Effect of the dimerization inhibitor P27 on PRMDR as analysed by size-exclusion chromatography
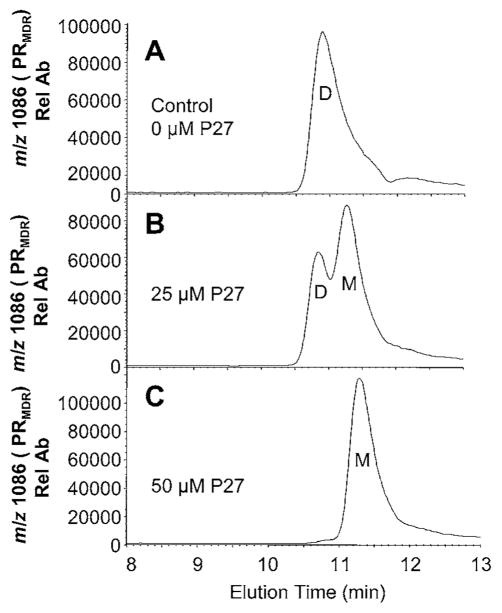
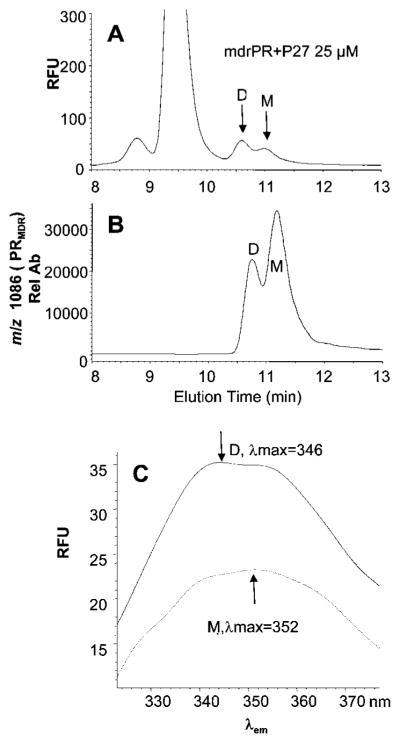
To assess the effect of the dimerization inhibitor P27 on PRMDR, the PR was incubated for 16 h with 0, 25 μM or 50 μM P27 and then analysed by size-exclusion chromatography (Figure 4). Treatment of PRMDR with 25 μM P27 induced the formation of a later-eluting PRMDR species with an elution time consistent with monomeric PR (Figure 4B). Treatment with 50 μM P27 converted all of the detectable PRMDR into the monomeric form of PRMDR (Figure 4C). In this experiment, approx. 20 % of the total PRMDR was not recovered after treatment with P27 as compared to untreated PRMDR. Decreases in recoverable PR were probably due to the unstable nature of monomeric PR that may tend to aggregate and precipitate as described previously for monomeric forms of PR [29]. PRMDR enzyme activity determined in parallel decreased by 27 %and 99 %following treatment with 25 μM and 50 μM P27 respectively. The substantial increase in inhibition observed for 50 μM P27 is surprising. This behaviour may be due to the presence of N- and C-terminal sequences in P27 that could self-associate, leading to the formation of varying amounts of active and inactive forms of P27 depending on the concentration used. The extent of monomer formation was also assessed over time in the absence and presence of 50 μM P27. Over a 12 h period, the percentage of monomer in the untreated control remained relatively constant, whereas in the presence of P27 there was clear evidence of monomer formation within 4 h, although the maximum effect was not obtained until 10–12 h after treatment (see Supplementary Figure S5 at http://www.BiochemJ.org/bj/419/bj4190497add.htm). It was found that the absolute amount of detectable monomer peaked within 6–8 h after treatment and thereafter steadily decreased, suggesting that PRMDR was unstable in its monomeric form (results not shown), similar to that described for engineered monomeric PR [29]. As expected, the percentage of PR converted to monomer with a given concentration of P27 was inversely proportional to the concentration of PRMDR. Treatment of PRMDR at 1 μM with 25 μM P27 induced approx. 40 % monomer, whereas treatment of 0.5 μM induced 80 % monomer. To further explore the physical characteristics of PRMDR following treatment with P27, we compared the emission spectra obtained for the dimer to that of the monomer induced by P27. As shown in Figure 5, the monomeric PRMDR had less intrinsic fluorescence than the dimer (Figure 5A), based on the amount of each form quantified by MS (Figure 5B). In addition, P27 treatment induced a red-shift in the emission maximum for the monomer (shifted from λmax = 346 to λmax = 352 nm) as compared to the dimeric PR (Figure 5C). These changes in the fluorescent characteristics of the PR in its monomeric form are consistent with those seen for an unfolded (urea treated) monomeric form of the PR as compared to the folded dimer, as described previously [45]. Treating PRMDR with 8 M urea also led to a red-shift and decrease in fluorescence intensity (results not shown). Taken together, the results suggest that treatment of PRMDR with P27 leads to greater solvent exposure of PRMDR tryptophan residues and may ultimately lead to unfolding of the PR monomer.
Figure 4. Effect of P27 on PRMDR elution profile and PR activity.
PRMDR (1 μM) was incubated for 16 h at 37 °C in 150 mM ammonium acetate buffer containing 100 μg · ml−1 BSA. Samples (8 μl) were separated by size-exclusion chromatography and elution was monitored by MS for the PRMDR-specific ion (m/z 1086, 10+). (A) Treatment with 0 μM P27 (elution time 10.7 min), (B) 25 μM P27 (elution times 10.8 and 11.3 min) or (C) 50 μM P27 (elution time 11.3 min). The dimeric (D) and monomeric (M) forms of PRMDR are indicated in the Figures. PR activity of each sample was also measured following the incubation. PR activity was (A) 2.6 RFU min−1 · μg−1 (where RFU is relative fluorescence units) (B) 1.85 RFU min−1 · μg−1 and (C) 0.0 RFU min−1 · μg−1 PRMDR. Rel Ab, relative ion abundance.
Figure 5. Size-exclusion chromatography of PRMDR treated with P27 and the corresponding fluorescent spectra for the dimeric and monomeric forms.
PRMDR (1 μM final concentration) was incubated for 16 h at 37 °C in 150 mM ammonium acetate buffer containing 100 μg · ml−1 BSA with P27 at 25 μM. Following incubation, 8 μl of the PR was sampled and analysed by size-exclusion chromatography. (A) PRMDR was detected by fluorescence by excitation at 280 nm and emission at 350 nm and (B) by MS in SIM mode for the PRMDR-specific ion (m/z 1086, 10+). (C) The fluorescent emission spectra λem from 325–375 nm were obtained at the peak apex for each peak using reference spectra at 10.2 and 12 min. The spectra for the dimer (D) (solid line, λmax = 346 nm) and the monomer (M) (dashed line, λmax = 352 nm) are shown. Rel Ab, relative ion abundance; RFU, relative fluorescence units.
Structure–function analysis of P27 and related peptides on the ability to induce PRMDR monomer and inhibit PR activity
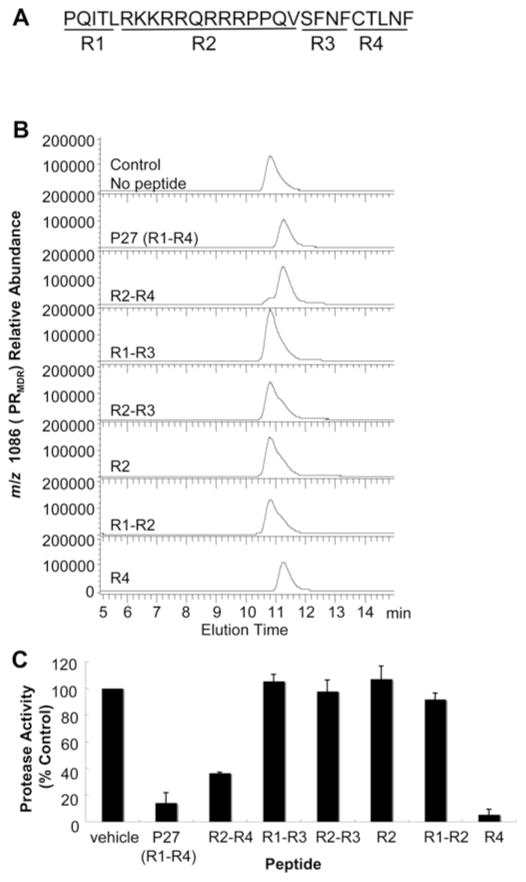
P27 is made up of four distinct peptide regions (Figure 6A). R1 (region 1) consists of the first five amino acids found in the N-terminus of PR, R2 (region 2) consists of a 13-aminoacid peptide containing the Tat membrane-permeable domain, R3 (region 3) contains the last four amino acids found in the transframe domain in Gag–Pol, and R4 (region 4) consists of the last five amino acids in the C-terminus of PR. To determine which of these regions were important for inhibition of PRMDR dimerization, we examined combinations of these regions for activity against PRMDR by size-exclusion chromatography. As shown in Figure 6(B), treatment of PRMDR with 50 μM P27 (R1–R4) converted all PRMDR to monomer. The peptide lacking the R1 domain (R2–R4) was slightly less active and converted most of PRMDR to monomer as well (Figure 6B). However, all peptides lacking the C-terminal domain (CTLNF, R4) were relatively inactive (Figure 6B; R1–R3, R2–R3, R2 and R1–R2). The CTLNF domain used alone, however, was effective at inducing monomer formation (Figure 6B; R4). Thus the majority of monomer-inducing activity resides in the R4 (CTLNF) domain of P27. This activity of CTLNF is consistent with a previous report of enzyme inhibition by this peptide [16]. Interestingly, changing the cysteine residue in P27 to aspartic acid eliminated its ability to induce monomerization (results not shown). As shown in Figure 6(C), the inhibition of activity by the various peptides, under these conditions correlated closely to the ability of the peptides to induce monomer formation. The dimerization inhibitor peptide [Ac-TLNF (acetylated-TLNF)] previously reported by Zhang et al. [31] also had some activity (50 μM) as an inhibitor of dimerization, but was noticeably weaker than Ac-CTLNF (acetylated-CTLNF) and CTLNF peptides (results not shown). On the basis of dose–response data obtained with these various peptides, the relative potencies of the different peptides at inducing monomer formation were P27 > CTLNF > Ac-CTLNF > Ac-TLNF (results not shown). We also tested the ability of a previously reported dimerization inhibitor, compound 10 (see Supplementary Figure S6 at http://www.BiochemJ.org/bj/419/bj4190497add.htm for structure of compound 10), with a Ki of 2.3 μM, to induce monomer formation of PRMDR [38]. PRMDR treated overnight with increasing concentrations of compound 10 induced the formation of monomer in a dose-dependent fashion, whereas the DMSO vehicle alone induced the formation of a low level of monomer (see Supplementary Figure S7 at http://www.BiochemJ.org/bj/419/bj4190497add.htm). There was a clear decrease in the total PRMDR recovered following treatment with compound 10, suggesting that this dimerization inhibitor leads to PRMDR instability and loss during chromatography, perhaps through aggregation of the resulting monomer. At the highest concentration tested (10 μM), compound 10 induced up to 50 % monomer within 4 h after treatment (results not shown). Taken together, these results suggest that this approach could be useful as a means to screen for potential dimerization inhibitors of HIV PR and be used to differentiate active-site inhibitors from dimerization inhibitors.
Figure 6. Effect of P27 and P27-related peptides on PRMDR elution profile.
PRMDR was incubated at 1 μM for 16 h at 37 °C with 50 μM of each peptide. Samples were then analysed by size-exclusion chromatography. (A) P27 sequence with the four major peptide regions (R1–R4) indicated. (B) The elution profile for PRMDR by MS of the PRMDR-specific ion (m/z 1086, 10+) is shown for each sample The total area corresponding to PRMDR was: no peptide, 6.1 × 106; R1–R4, 1.6 × 106; R2–R4, 2.9 × 106; R1–R3, 6.5 × 106; R2–R3, 6.9 × 106; R2, 7.1 × 106; R1–R2, 7.7 × 106; and R4, 3.9 × 106. Peak elution times were 10.8 min for control, R1–R3, R2–R3, R2 and R1–R2, and 11.3 min for R1–R4, R2–R4 and R4. (C) Corresponding PR activity for each sample following peptide treatment. Results are means ± S.D. (n = 3).
DISCUSSION
Several groups, including our own, have developed peptides, peptide mimetics or lipopeptides that can disrupt the dimerization of HIV-1 PR [19,20,22,23,31,46–48]. These inhibitors have been shown to inhibit dimerization based primarily on the results on dimerization kinetics by Zhang et al. [31]. However, few studies are available to corroborate these findings and provide further insights into how these inhibitors affect PR structure. Until the present study, size-exclusion chromatography has not been used as a primary means to study or screen the effects of potential inhibitors on HIV-1 PR structure. One obstacle has been the propensity of retroviral PRs to undergo substantial autoproteolysis under native conditions [24–26], making structural analysis by biophysical methods difficult. To overcome autoproteolysis, some laboratories have utilized engineered autoproteolysis-resistant forms of PR for structural studies [21,29]. Autoproteolysis is of particular concern when studying the effects of dimerization inhibitors on PR, since the monomeric or unfolded species of the enzyme is expected to be highly susceptible to autoproteolysis by even low levels of active PR dimer [25]. In the present study, we have identified PRMDR, a multi-drug-resistant PR derived from HIV-1 isolated from a patient heavily treated for HIV-1 infection. This PR was found to be highly resistant to autoproteolysis. This prime target for dimerization inhibitors was ideal for studies using size-exclusion chromatography. In addition, by taking advantage of the recent advances that have allowed for the use of MS as a detector during liquid chromatography [49], it was possible to attain both sensitive and specific detection of the intact full-length PR at relatively low concentrations. These two advances have made it possible to further our understanding of the effect of dimerization inhibitors on drug-resistant PR using size-exclusion chromatography on low concentrations of PR.
Several criteria were used to differentiate the PRMDR dimer from monomer in the present study. The dimeric form of PRMDR was identified based on its elution time in the presence of a potent active-site inhibitor (JE-2147) known to bind only to dimeric forms of PRMDR, and by the co-elution of this inhibitor with PRMDR. This earlier-eluting form of PRMDR had an apparent molecular mass which was twice that of the later-eluting PRMDR. The identification of the later-eluting form as monomeric PRMDR was performed on the basis of its elution time as compared to two known monomeric forms of HIV PR (PRT26A and PRKIIA-Glut), the appearance of this form at low PR concentrations, and its identification as an intact PR by MS. Also, treatment of PRMDR with P27 led to a time- and dose-dependent conversion of PRMDR dimer to monomer and the PRMDR species generated following P27 treatment had an elution time similar to known monomeric forms of HIV-1 PR, namely PRT26A [30] and PRKIIA-Glut [36]. In addition, the later-eluting form of PRMDR induced by treatment with P27 had a lower intrinsic tryptophan fluorescence and red-shifted, both characteristics of unfolded monomeric PR [45]. With this approach, we were able to dissect the functional regions of P27 that contributed to the inhibition of dimerization and confirm the effect of another previously identified dimerization inhibitor (compound 10) [38] on PRMDR. Finally, it was possible to demonstrate that HIV PRs reversibly inactivated by oxidation of the residue at position 95, namely PRKIIA-Glut and HIV-2 PR, were monomeric in nature [36,50].
It has been described previously by others that certain peptides identified as dimerization inhibitors of HIV-1 PR, based on kinetic data, were not able to induce monomeric PR based on NMR analysis [30]. However, more recently, Frutos et al. [21] have provided NMR evidence for the interaction of a dimerization inhibitor peptide (Ac-SYEL-OH) with an engineered autoproteolysis-resistant PR by using selective tryptophan side chain labelling with PR at 200 μM. In the present study, we were able to demonstrate the formation of PRMDR monomer with peptides and peptide mimetics that were previously characterized as dimerization inhibitors based on the dimerization kinetics of Zhang et al. [31]. Our approach required significantly lower concentrations of PR (1 μM) as compared to that required for NMR. Not surprisingly, the most important region of P27 for inhibition of dimerization was the five C-terminal amino acids CTLNF. Although previous studies demonstrated that this peptide had an IC50 of 150 μM, this was determined with only a 5 min pre-incubation step with ≤25 nM PRWT [16]. Using 50 μM P27, it took 10–12 h for monomer formation to reach a plateau at a PR concentration of 1 μM. Indeed, the extent of monomer formation induced by P27 increased as the PR concentration was decreased, as expected for an effect that is dependent on the dissociation of PR. These results are consistent with these inhibitors being dependent on PR dissociation for activity [40] and emphasize the importance of PR concentration and incubation time when testing these inhibitors in various biophysical assays. Our results also suggest that monomer induced by P27 may be relatively unstable, as the absolute amount of PR detected decreased after reaching a peak at between 6–8 h. The monomeric PRMDR may be lost to aggregation over time like that observed with engineered monomeric forms of the PR when incubated under native conditions [29].
Although it was possible to detect an active-site inhibitor co-eluting with dimeric PRMDR by MS, we did not detect the peptides co-eluting with PRMDR monomer. This may be due to relatively weak associations of these inhibitors with monomeric PR or to poor ionization of the peptides that are bound to PR. Another possibility is that the interaction of dimerization-inhibitor peptides with PR affects the proper folding of PR, ultimately resulting in PR unfolding. Such a mechanism has been suggested as an alternative approach to disrupting PR dimerization [18,51]. Addition of JE-2147 to samples containing monomer induced by P27 led to a partial reversal of the monomeric PR to dimer (results not shown), indicating that inhibition by P27 is at the level of protein–protein dimerization. Nevertheless, this approach should be useful in identifying and optimizing conditions suitable for studying the structure and formation of PR monomer in the presence of inhibitors and allow one to rapidly screen for more potent dimerization inhibitors of multi-drug-resistant PR.
As dimerization inhibitors could be useful in cases where HIV-1 develops resistance to standard therapy, their activity against multi-drug-resistant PRs is of particular interest. It has been suggested that dimerization inhibitors that target the conserved anti-parallel β-sheet making up the dimerization interface may be useful in overcoming drug resistance to active-site inhibitors [52]. There are no amino acid differences between the N- and C-termini which make up the dimer interface of PRWT and PRMDR, so it is not surprising that they are both susceptible to these inhibitors. Our previous results on PRs that were inactivated by oxidative modification [36,50] and the new data provided in the present study further demonstrate that simple oxidation of a single residue of PR monomer is sufficient to eliminate dimerization and activity. It is interesting that such small changes in the residues either near the active site, like that seen in PRT26A, or within the dimer interface, like that in PRHIV-2-OX, can completely disrupt dimerization. Future studies utilizing this very sensitive and specific chromatographic approach could reveal the important structural requirements needed to disrupt PR dimerization by candidate drugs and provide a means to easily differentiate active-site inhibitors from inhibitors of PR dimerization.
Supplementary Material
Acknowledgments
We thank Dr Hiroaki Mitsuya [HIV and AIDS Malignancy Branch, NCI (National Cancer Institute), NIH (National Institutes of Health), Bethesda, MD, U.S.A.] for providing the PR active-site inhibitors.
FUNDING
This work was supported in part by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
Abbreviations used
- Ac-CTLNF
acetylated-CTLNF
- Ac-TLNF
acetylated-TLNF
- FA
formic acid
- PR
protease
- PRHIV-2-OX
oxidized HIV-2 PR
- PRKIIA
auto-proteolysis-resistant HIV-1 PR
- PRKIIA-Glut
PRKIIA with glutathionylation of Cys95
- PRMDR
multi-drug-resistant HIV-1 PR
- PRWT
wild-type HIV-1 PR
- PRT26A
autoproteolysis-resistant and monomeric HIV-1 PR
- P27
peptide 27
- R1 etc
region 1 etc
- RP-HPLC
reverse-phase HPLC
- SIM
selective ion monitoring
- TFA
trifluoroacetic acid
References
- 1.Kohl NE, Emini EA, Schleif WA, Davis LJ, Heimbach JC, Dixon RAF, Scolnick EM, Sigal IS. Active human immunodificiency virus protease is required for viral infectivity. Proc Natl Acad Sci USA. 1988;85:4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng C, Ho BK, Chang TW, Chang NT. Role of human immunodeficiency virus type 1-specific protease in core protein maturation and viral infectivity. J Virol. 1989;63:2550–2556. doi: 10.1128/jvi.63.6.2550-2556.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pettit SC, Everitt LE, Choudhury S, Dunn BM, Kaplan AH. Initial cleavage of the human immunodeficiency virus type 1 GagPol precursor by its activated protease occurs by an intramolecular mechanism. J Virol. 2004;78:8477–8485. doi: 10.1128/JVI.78.16.8477-8485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Humphrey RW, Ohagen A, Davis DA, Fukazawa T, Hayashi H, Hoglund S, Mitsuya H, Yarchoan R. Removal of HIV-1 protease inhibitors from preparations of immature HIV-1 Virions does not result in an increase in infectivity or the appearance of mature morphology. Antimicrob Agents Chemother. 1997;41:1017–1023. doi: 10.1128/aac.41.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambert DM, Petteway SR, Jr, McDanal CE, Hart TK, Leary JJ, Dreyer GB, Meek TD, Bugelski PJ, Bolognesi DP, Metcalf BW, Matthews TJ. Human immunodeficiency virus type 1 protease inhibitors irreversibly block infectivity of purified virions from chronically infected cells. Antimicrob Agents Chemother. 1992;36:982–988. doi: 10.1128/aac.36.5.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Condra JH. Resistance to HIV protease inhibitors. Haemophilia. 1998;4:610–615. doi: 10.1046/j.1365-2516.1998.440610.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang YM, Imamichi H, Imamichi T, Lane HC, Falloon J, Vasudevachari MB, Salzman NP. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J Virol. 1997;71:6662–6670. doi: 10.1128/jvi.71.9.6662-6670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schock HB, Garsky VM, Kuo LC. Mutational anatomy of an HIV-1 protease variant conferring cross-resistance to protease inhibitors in clinical trials. Compensatory modulations of binding and activity. J Biol Chem. 1996;271:31957–31963. doi: 10.1074/jbc.271.50.31957. [DOI] [PubMed] [Google Scholar]
- 9.Romano L, Venturi G, Giomi S, Pippi L, Valensin PE, Zazzi M. Development and significance of resistance to protease inhibitors in HIV-1-infected adults under triple-drug therapy in clinical practice. J Med Virol. 2002;66:143–150. doi: 10.1002/jmv.2123. [DOI] [PubMed] [Google Scholar]
- 10.Schmit JC, Ruiz L, Clotet B, Raventos A, Tor J, Leonard J, Desmyter J, De Clercq E, Vandamme AM. Resistance-related mutations in the HIV-1 protease gene of patients treated for 1 year with the protease inhibitor ritonavir (ABT-538) AIDS. 1996;10:995–999. doi: 10.1097/00002030-199610090-00010. [DOI] [PubMed] [Google Scholar]
- 11.Svicher V, Ceccherini-Silberstein F, Erba F, Santoro M, Gori C, Bellocchi MC, Giannella S, Trotta MP, Monforte A, Antinori A, Perno CF. Novel human immunodeficiency virus type 1 protease mutations potentially involved in resistance to protease inhibitors. Antimicrob Agents Chemother. 2005;49:2015–2025. doi: 10.1128/AAC.49.5.2015-2025.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ceccherini-Silberstein F, Erba F, Gago F, Bertoli A, Forbici F, Bellocchi MC, Gori C, D’Arrigo R, Marcon L, Balotta C, et al. Identification of the minimal conserved structure of HIV-1 protease in the presence and absence of drug pressure. AIDS. 2004;18:F11–F19. doi: 10.1097/01.aids.0000131394.76221.02. [DOI] [PubMed] [Google Scholar]
- 13.Muzammil S, Ross P, Freire E. A major role for a set of non-active site mutations in the development of HIV-1 protease drug resistance. Biochemistry. 2003;42:631–638. doi: 10.1021/bi027019u. [DOI] [PubMed] [Google Scholar]
- 14.Yoshimura K, Kato R, Yusa K, Kavlick MF, Maroun V, Nguyen A, Mimoto T, Ueno T, Shintani M, Falloon J, et al. JE-2147: a dipeptide protease inhibitor (PI) that potently inhibits multi-PI-resistant HIV-1. Proc Natl Acad Sci USA. 1999;96:8675–8680. doi: 10.1073/pnas.96.15.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ridky T, Leis J. Development of drug resistance to HIV-1 protease inhibitors. J Biol Chem. 1995;270:29621–29623. doi: 10.1074/jbc.270.50.29621. [DOI] [PubMed] [Google Scholar]
- 16.Babé LM, Rose J, Craik CS. Synthetic “interface” peptides alter dimeric assembly of the HIV 1 and 2 proteases. Protein Sci. 1992;1:1244–1253. doi: 10.1002/pro.5560011003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowman MJ, Chmielewski J. Novel strategies for targeting the dimerization interface of HIV protease with cross-linked interfacial peptides. Biopolymers. 2002;66:126–133. doi: 10.1002/bip.10232. [DOI] [PubMed] [Google Scholar]
- 18.Broglia RA, Provasi D, Vasile F, Ottolina G, Longhi R, Tiana G. A folding inhibitor of the HIV-1 protease. Proteins. 2006;62:928–933. doi: 10.1002/prot.20849. [DOI] [PubMed] [Google Scholar]
- 19.Caflisch A, Schramm HJ, Karplus M. Design of dimerization inhibitors of HIV-1 aspartic proteinase: a computer-based combinatorial approach. J Comput Aided Mol Des. 2000;14:161–179. doi: 10.1023/a:1008146201260. [DOI] [PubMed] [Google Scholar]
- 20.Davis DA, Brown CA, Singer KE, Wang V, Kaufman J, Stahl SJ, Wingfield P, Maeda K, Harada S, Yoshimura K, et al. Inhibition of HIV-1 replication by a peptide dimerization inhibitor of HIV-1 protease. Antiviral Res. 2006;72:89–99. doi: 10.1016/j.antiviral.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 21.Frutos S, Rodriguez-Mias RA, Madurga S, Collinet B, Reboud-Ravaux M, Ludevid D, Giralt E. Disruption of the HIV-1 protease dimer with interface peptides: structural studies using NMR spectroscopy combined with [2-(13)C]-Trp selective labeling. Biopolymers. 2007;88:164–173. doi: 10.1002/bip.20685. [DOI] [PubMed] [Google Scholar]
- 22.Schramm HJ, Billich A, Jaeger E, Rucknagel KP, Arnold G, Schramm W. The inhibition of HIV-1 protease by interface peptides. Biochem Biophys Res Commun. 1993;194:595–600. doi: 10.1006/bbrc.1993.1863. [DOI] [PubMed] [Google Scholar]
- 23.Schramm HJ, de Rosny E, Reboud-Ravaux M, Buttner J, Dick A, Schramm W. Lipopeptides as dimerization inhibitors of HIV-1 protease. Biol Chem. 1999;380:593–596. doi: 10.1515/BC.1999.076. [DOI] [PubMed] [Google Scholar]
- 24.Louis JM, Oroszlan S, Tozser J. Stabilization from autoproteolysis and kinetic characterization of the human T-cell leukemia virus type 1 proteinase. J Biol Chem. 1999;274:6660–6666. doi: 10.1074/jbc.274.10.6660. [DOI] [PubMed] [Google Scholar]
- 25.Mildner AM, Rothrock DJ, Leone JW, Bannow CA, Lull JM, Reardon IM, Sarcich JL, Howe WJ, Tomich CS, Smith CW, et al. The HIV-1 protease as enzyme and substrate: mutagenesis of autolysis sites and generation of a stable mutant with retained kinetic properties. Biochemistry. 1994;33:9405–9413. doi: 10.1021/bi00198a005. [DOI] [PubMed] [Google Scholar]
- 26.Rose JR, Salto R, Craik CS. Regulation of autoproteolysis of the HIV-1 and HIV-2 proteases with engineered amino acid substitutions. J Biol Chem. 1993;268:11939–11945. [PubMed] [Google Scholar]
- 27.Strickler JE, Gorniak J, Dayton B, Meek T, Moore M, Magaard V, Malinowski J, Debouck C. Characterization and autoprocessing of precursor and mature forms of human immunodeficiency virus type 1 (HIV 1) protease purified from Escherichia coli. Proteins. 1989;6:139–154. doi: 10.1002/prot.340060205. [DOI] [PubMed] [Google Scholar]
- 28.Ishima R, Ghirlando R, Tozser J, Gronenborn AM, Torchia DA, Louis JM. Folded monomer of HIV-1 protease. J Biol Chem. 2001;276:49110–49116. doi: 10.1074/jbc.M108136200. [DOI] [PubMed] [Google Scholar]
- 29.Louis JM, Ishima R, Nesheiwat I, Pannell LK, Lynch SM, Torchia DA, Gronenborn AM. Revisiting monomeric HIV-1 protease. Characterization and redesign for improved properties. J Biol Chem. 2003;278:6085–6092. doi: 10.1074/jbc.M209726200. [DOI] [PubMed] [Google Scholar]
- 30.Ishima R, Torchia DA, Louis JM. Mutational and structural studies aimed at characterizing the monomer of HIV-1 protease and its precursor. J Biol Chem. 2007;282:17190–17199. doi: 10.1074/jbc.M701304200. [DOI] [PubMed] [Google Scholar]
- 31.Zhang ZY, Poorman RA, Maggiora LL, Heinrikson RL, Kezdy FJ. Dissociative inhibition of dimeric enzymes. Kinetic characterization of the inhibition of HIV-1 protease by its COOH-terminal tetrapeptide. J Biol Chem. 1991;266:15591–15594. [PubMed] [Google Scholar]
- 32.Koh Y, Matsumi S, Das D, Amano M, Davis DA, Li J, Leschenko S, Baldridge A, Shioda T, Yarchoan R, et al. Potent inhibition of HIV-1 replication by novel non-peptidyl small molecule inhibitors of protease dimerization. J Biol Chem. 2007;282:28709–28720. doi: 10.1074/jbc.M703938200. [DOI] [PubMed] [Google Scholar]
- 33.Darke PL, Jordan SP, Hall DL, Zugay JA, Shafer JA, Kuo LC. Dissociation and association of the HIV-1 protease dimer subunits: equilibria and rates. Biochemistry. 1994;33:98–105. doi: 10.1021/bi00167a013. [DOI] [PubMed] [Google Scholar]
- 34.Jordan SP, Zugay J, Darke PL, Kuo LC. Activity and dimerization of human immunodeficiency virus protease as a function of solvent composition and enzyme concentration. J Biol Chem. 1992;267:20028–20032. [PubMed] [Google Scholar]
- 35.Davis DA, Dorsey K, Wingfield PT, Stahl SJ, Kaufman J, Fales HM, Levine RL. Regulation of HIV-1 protease activity through cysteine modification. Biochemistry. 1996;35:2482–2488. doi: 10.1021/bi951525k. [DOI] [PubMed] [Google Scholar]
- 36.Davis DA, Brown CA, Newcomb FM, Boja ES, Fales HM, Kaufman J, Stahl SJ, Wingfield P, Yarchoan R. Reversible oxidative modification as a mechanism for regulating retroviral protease dimerization and activation. J Virol. 2003;77:3319–3325. doi: 10.1128/JVI.77.5.3319-3325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis DA, Newcomb FM, Moskovitz J, Fales HM, Levine RL, Yarchoan R. Reversible oxidation of HIV-2 protease. In: Sies H, Packer L, editors. Methods in Enzymology. Academic Press; New York: 2002. pp. 249–259. [DOI] [PubMed] [Google Scholar]
- 38.Bowman MJ, Chmielewski J. Sidechain-linked inhibitors of HIV-1 protease dimerization. Bioorg Med Chem. 2008;17:967–976. doi: 10.1016/j.bmc.2008.02.060. [DOI] [PubMed] [Google Scholar]
- 39.Levine RL, Williams JA, Stadtman ER, Shacter E. Carbonyl assays for determination of oxidatively modified proteins. Methods Enzymol. 1994;233:346–357. doi: 10.1016/s0076-6879(94)33040-9. [DOI] [PubMed] [Google Scholar]
- 40.Darke PL, Jordan SP, Hall DL, Zugay JA, Shafer JA, Kuo LC. Dissociation and association of the HIV-1 protease dimer subunits: equilibria and rates. Biochemistry. 1994;33:98–105. doi: 10.1021/bi00167a013. [DOI] [PubMed] [Google Scholar]
- 41.Todd MJ, Freire E. The effect of inhibitor binding on the structural stability and cooperativity of the HIV-1 protease. Proteins. 1999;36:147–156. doi: 10.1002/(sici)1097-0134(19990801)36:2<147::aid-prot2>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 42.Mimoto T, Kato R, Takaku H, Nojima S, Terashima K, Misawa S, Fukazawa T, Ueno T, Sato H, Shintani M, et al. Structure-activity relationship of small-sized HIV protease inhibitors containing allophenylnorstatine. J Med Chem. 1999;42:1789–1802. doi: 10.1021/jm980637h. [DOI] [PubMed] [Google Scholar]
- 43.Jordan SP, Zugay J, Darke PL, Kuo LC. Activity and dimerization of human immunodeficiency virus protease as a function of solvent composition and enzyme concentration. J Biol Chem. 1992;267:20028–20032. [PubMed] [Google Scholar]
- 44.Davis DA, Newcomb FM, Starke DW, Ott DE, Mieyal JM, Yarchoan R. Thioltransferase (glutaredoxin) is detected within HIV-1 and can regulate the activity of glutathionylated HIV-1 protease in vitro. J Biol Chem. 1997;272:25935–25940. doi: 10.1074/jbc.272.41.25935. [DOI] [PubMed] [Google Scholar]
- 45.Grant SK, Deckman IC, Culp JS, Minnich MD, Brooks IS, Hensley P, Debouck C, Meek TD. Use of protein unfolding studies to determine the conformational and dimeric stabilities of HIV-1 and SIV proteases. Biochemistry. 1992;31:9491–9501. doi: 10.1021/bi00154a023. [DOI] [PubMed] [Google Scholar]
- 46.Shultz MD, Ham YW, Lee SG, Davis DA, Brown C, Chmielewski J. Small-molecule dimerization inhibitors of wild-type and mutant HIV protease: a focused library approach. J Am Chem Soc. 2004;126:9886–9887. doi: 10.1021/ja048139n. [DOI] [PubMed] [Google Scholar]
- 47.Shultz MD, Chmielewski J. Probing the role of interfacial residues in a dimerization inhibitor of HIV-1 protease. Bioorg Med Chem Lett. 1999;9:2431–2436. doi: 10.1016/s0960-894x(99)00400-x. [DOI] [PubMed] [Google Scholar]
- 48.Dumond J, Boggetto N, Schramm HJ, Schramm W, Takahashi M, Reboud-Ravaux M. Thyroxine-derivatives of lipopeptides: bifunctional dimerization inhibitors of human immunodeficiency virus-1 protease. Biochem Pharmacol. 2003;65:1097–1102. doi: 10.1016/s0006-2952(02)01622-2. [DOI] [PubMed] [Google Scholar]
- 49.Fligge TA, Bruns K, Przybylski M. Analytical development of electrospray and nanoelectrospray mass spectrometry in combination with liquid chromatography for the characterization of proteins. J Chromatogr. 1998;706:91–100. doi: 10.1016/s0378-4347(97)00535-5. [DOI] [PubMed] [Google Scholar]
- 50.Davis DA, Newcomb FM, Moskovitz J, Wingfield PT, Stahl SJ, Kaufman J, Fales HM, Levine RL, Yarchoan R. HIV-2 protease is inactivated after oxidation at the dimer interface and activity can be partly restored with methionine sulphoxide reductase. Biochem J. 2000;346:305–311. [PMC free article] [PubMed] [Google Scholar]
- 51.Broglia R, Levy Y, Tiana G. HIV-1 protease folding and the design of drugs which do not create resistance. Curr Opin Struct Biol. 2008;18:60–66. doi: 10.1016/j.sbi.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 52.Boggetto N, Reboud-Ravaux M. Dimerization inhibitors of HIV-1 protease. Biol Chem. 2002;383:1321–1324. doi: 10.1515/BC.2002.150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.