Abstract
The bulky octahedral complex, Rh(bpy)2chrysi3+ (chrysi = 5, 6- chrysenequinone diimine), binds single base mismatches in a DNA duplex with micromolar binding affinities and high selectivity. Here we present an NMR solution study to characterize the binding mode of this bulky metal complex with its target CC mismatch in the oligonucleotide duplex (5′-CGGACTCCG-3′)2. Both NOESY and COSY studies indicate that Rh(bpy)2chrysi3+ inserts deeply in the DNA at the mismatch site via the minor groove and with ejection of both destabilized cytosines into the opposite major groove. The insertion only minimally distorts the conformation of the oligonucleotide local to the binding site. Both flanking, well-matched base pairs remain tightly hydrogen-bonded to each other, and 2D DQF-COSY experiments indicate that all sugars maintain their original C2′ endo conformation. Remarkably, 31P NMR reveals that opening of the phosphate angles from a BI to a BII conformation is sufficient for insertion of the bulky metal complex. These results corroborate those obtained crystallographically and, importantly, provide structural evidence for this specific insertion mode in solution.
INTRODUCTION
Octahedral metal complexes containing an extended bidentate aromatic ligand have been designed to target single base mismatches in duplex DNA. 1–3 Rh(bpy)2chrysi3+ (chrysi = 5,6-chrysenequinone diimine, Figure 1), is a sterically bulky DNA intercalator that binds specifically in the destabilized regions near DNA base mismatches and, upon photoactivation, cleaves the DNA backbone. The complex is both a general and remarkably specific mismatch recognition agent. 3 Specific DNA cleavage is observed at over 80% of mismatch sites irrespective of the sequence context around the mispaired bases. Furthermore, the complex binds and with photoactivation cleaves at a single base mismatch in a 2725 base pair linearized plasmid heteroduplex. This mismatch-specific targeting is based upon the thermodynamic destabilization associated with the base mispair. The metal complex contains a bulky aromatic bidentate ligand that is difficult to stack within well-matched DNA, but the extended complex can insert more easily within the DNA duplex at the destabilized mismatched site.2
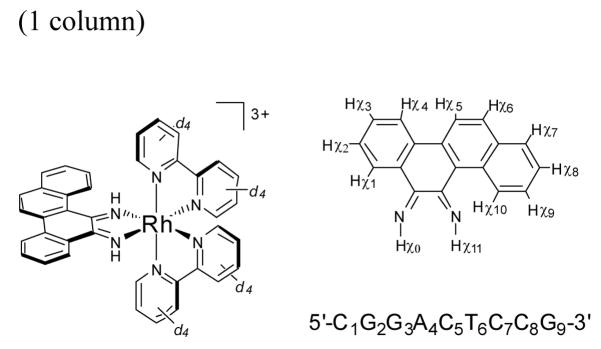
Figure 1.
Chemical structure of Δ-Rh(bpy-d8)2chrysi3+ with numbering scheme for the protons of the chrysi ligand and sequence and numbering scheme for the oligonucleotide.
The uniquely high specificity in targeting mismatches has led to many applications for these bulky metal complexes. The mismatch-targeting agents have been fruitful in developing new methods for the discovery of single nucleotide polymorphisms. 4 Pooling and annealing together DNA samples from a test population generates mismatches at the polymorphic sites, and these can be marked by photocleavage with the mismatch-specific complex. Additionally, since there is an association between deficiencies in mismatch repair and cancerous transformation, 5–8 these complexes may provide a route to novel diagnostics for cancer. Measurements of the abundance of mismatches provides an early report on deficiencies in mismatch repair. 9 Towards that end, we have developed luminescent analogues as probes for mismatches. 10, 11 Bifunctional complexes have also been designed to target alkylators and platinating agents to mismatched sites. 12, 13 Moreover, we have found that these bulky rhodium intercalators can differentially inhibit cellular proliferation in mismatch repair-deficient cells compared with cells that are mismatch repair-proficient. 14 Significantly, then, targeting DNA mismatches may provide a new cell-selective strategy for chemotherapeutic design.14–16
Given the interest in applying these complexes for a range of biological applications, a structural understanding of their interactions with mismatched DNA becomes critically important. The crystal structure of Δ-Rh(bpy)2chrysi3+ bound to a CA mismatch within a DNA oligonucleotide was recently determined. 17 The structure revealed that the metal complex recognizes the thermodynamically destabilized site via a novel binding mode: insertion into the double helix with ejection of both mismatched bases. Importantly, this insertion mode differs from the well characterized intercalation mode18–20 in that the DNA does not unwind to enable introduction of a new smaller ligand in its base stack, but replaces and ejects the faulty mispair using the extended ligand. Indeed, given the width of the chrysi ligand (11.3 Å), wider than the span of a base pair (10.8 Å), insertion of the metal complex to a destabilized site is favored over intercalation in stable, matched DNA. The crystal structure thus corroborates the direct correlation observed between the binding affinity of the metalloinsertor for a given mismatch and the amount by which that mismatch destabilizes DNA.2
The crystal structure revealed many distinctive aspects of this binding mode, (i) insertion into the duplex stack without increasing the base pair rise, (ii) ejection of both mismatched bases, and (iii) binding to the DNA from the minor groove side. This mode is clearly distinguished from the structurally characterized intercalation of metal complexes which has been found to give a doubling of base pair rise at the intercalation site, no base pair opening, and binding from the major groove side. 18–20 In fact, in the crystal structure of the mismatched DNA, an additional Rh complex is bound at the center of the helix through the characteristic intercalative mode. 17 We attributed this extra complex to be bound as a result of crystal packing forces.
Given this observation, it is important also to obtain structural information regarding these binding interactions in solution. Are the bases fully ejected in solution as in the solid state, where additional stacking interactions help to stabilize the ejected mismatched bases? Does the interaction arise from the minor or major groove side? Are there significant perturbations in the sugar-phosphate backbone and are they local to the site or extended over a portion of the duplex? Moreover, it is most important to ascertain not only whether the observations made for the crystal hold as well in solution, but also that the observations can be extended to other mismatches and other duplex sites. Thus we describe here, using 1H and 31P-NMR, a solution study of the insertion of Δ-Rh(bpy)2chrysi3+ into its target CC mismatch within an oligonucleotide DNA duplex.
RESULTS AND DISCUSSION
Choice of oligonucleotide
Rh(bpy)2chrysi3+ binds single base mismatches with affinities of 105–107 M−1 depending on the thermodynamic destabilization associated with the mismatch. Our initial study thus concentrated on characterizing the insertion of the bulky metal complex into the sites to which it binds most tightly, the most destabilizing CC and CA mismatches.2 Several oligonucleotides containing either 1 or 2 mismatches and varying both in length and % GC content were thus evaluated. In most cases, titration of 1 equivalent of Rh complex/mismatch results in broad NMR spectra, characteristic of an intermediate exchange process, and not useful for structure determination (Figure S1, supporting information). Although not sufficient for a detailed structural analysis, sharper spectra are obtained with a palindromic 9-mer containing a central CC mismatch: 5′-C1G2G3A4C5T6C7C8G9-3′. It was therefore this sequence that was used for this NMR study. It should be noted that changing the ionic strength or the pH between 6 and 8 of the DNA/Rh complex solution does not sharpen the spectra, nor does addition of spermine. Furthermore, since the melting point of the oligonucleotide in the absence of the metalloinsertor is barely 18 °C, all NMR experiments, both in the presence and absence of Rh(bpy)2chrysi3+, were thus performed at or below 10 °C so as to ensure that the DNA maintains a fully duplex form.
Binding of Δ-Rh(bpy-d8)2chrysi3+ to the CC mismatch
Previous studies with octahedral tris- chelate complexes demonstrated their binding preference for DNA of matching chirality.21 In each case, it is the Δ enantiomer that preferentially intercalates into right-handed B-DNA. 21–25 In the case of metalloinsertors, this chiral discrimination is even more dramatic. Only the Δ enantiomer of Rh(bpy)2chrysi3+ effectively binds DNA, and minimal interaction is observed with the Λ enantiomer. 1, 14 For the purpose of this study, the Δ and Λ enantiomers were thus separated by chiral chromatography using antimonyl tartrate as an eluent and only the Δ enantiomer was titrated with the DNA. 25 Furthermore, in order to simplify the aromatic region of the NMR spectra and avoid overlap between the resonances of the protons on the ancillary ligands and those of the bases, the metalloinsertor was synthesized with deuterated bipyridines.
As commonly observed with complexes that stack within the duplex, insertion of Δ-Rh(bpy- d8)2chrysi3+ in the oligonucleotide containing a CC mismatch significantly stabilizes the DNA duplex by 19 °C (Figure S2, supporting information), consistent with insertion of the chrysi ligand inside the DNA base stack. Furthermore, photocleavage experiments on the 9-mer followed by MALDI-TOF mass spectrometry indicate a single cleaving point neighboring T6, consistent with insertion of the Rh complex at the CC mismatch (Figure S3, supporting information).26 Thus, independent of the NMR results, photocleavage experiments and melting temperature studies indicate that Δ-Rh(bpy-d8)2chrysi3+ does insert in the oligonucleotide selectively at the CC mismatch.
Assignments and structure of the mismatched DNA without metal Complex
The free, unbound oligonucleotide 5′-CGGACTCCG-3′ was characterized by NOESY, HOHAHA, DQF-COSY and 31P NMR spectroscopy. Concomitant assignment of the NOESY, HOHAHA, and DQF-COSY spectra confirmed that the mismatch-containing oligonucleotide is in a duplex form and adopts the regular B conformation. 27, 28 The two self-complementary strands are related by C2 symmetry. All bases maintain the standard anti conformation. The pattern of the H1′-H2′ DQF-COSY crosspeaks, together with the absence of H3′-H2″ correlations and the relative intensities of the crosspeaks in the NOESY spectrum all confirm that every sugar pucker is predominantly in the C2′-endo conformation (Figure S4, supporting information). Note that the H2′ and H2″ chemical shifts are inverted for the terminal G9 and the mismatch compared to the other nucleotides of the sequence. Furthermore, the 31P 1D NMR spectrum indicates that the CC mismatch does not significantly distort the backbone of the DNA, as all phosphodiester junctions are in the canonical BI conformation (vide infra). Importantly, there is no break in nOe connectivity along the strand (Figure 2). All base pairs are therefore canonically stacked, including the mismatched cytosines. Insertion of the bulky metal complex into the mismatched site is thus not a matter of finding a “hole” in the DNA base stack, but of breaking apart weakly paired bases and ejecting them out of the double helix.
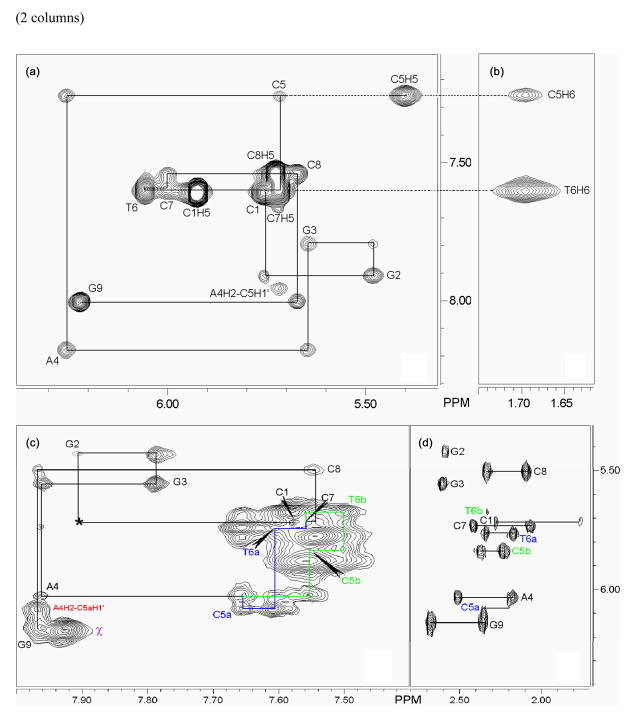
Figure 2.
NOESY data for the free oligonucleotide and in the presence of metal complex. (a) (F2 × F1: H1′ × aromatic) and (b) (F2 × F1: Me × aromatic) NOESY sub-spectra of the free DNA showing the sequential walk of the nOes along the full strand (a). No break in the correlation network confirms the intrahelical stacking of the two mismatched cytosines inside the double helix. Experimental conditions: D2O, 10 °C, 300 ms mixing time. (c) NOESY sub-spectrum of Δ-Rh(bpy-d8)2chrysi3+ inserted in DNA and sequential walk of the nOes along the full strand. (F2 × F1: aromatic × H1′). The loss of the C2 symmetry in the A4pC5pT6 step of the DNA is highlighted by two chains of connectivities displayed in dark blue (strand a) and light green (strand b). The nOe between G2 and C1 (marked by *) is observed at lower signal to noise ratio. Intramolecular nOe correlations between chrysi protons (χ3 -χ4) is labeled χ. Experimental conditions: D2O, 10 °C, 300 ms mixing time. (d) TOCSY sub-spectrum of Δ-Rh(bpy-d8)2chrysi3+ inserted in DNA. (F2 × F1: H2′-H2″ × H1′). Experimental conditions: D2O, 10 °C, 100 ms mixing time.
The NOESY spectrum recorded in H2O at 4 °C enables the assignment of the exchangeable imino protons of the bases (Figure S5, supporting information). All matched bases are paired in the normal Watson-Crick mode. We were unable to identify the amino protons of the CC mismatch, however. This observation was also previously reported for a CC mismatch studied under slightly acidic conditions.29–31 Since the two cytosines adopt the anti conformation, they may be paired according to a Wobble-type of conformation in which a single amino hydrogen bonds the two cytosines. Since this hydrogen bond would be in rapid equilibrium between two conformations, its exchange with bulk solvent is facilitated and its signal is thereby reduced.
NMR characterization of the insertion
Titration of the metalloinsertor into the oligonucleotide results in significant line broadening (Figure S1, supporting information). Nonetheless, most protons can be assigned by a combination of NOESY, HOHAHA, and 2D DQF COSY starting from the resonance of the H1′. Significantly, the 1D 1H NMR spectra for the titration clearly indicate that the aromatic protons of the chrysi ligand shift significantly upfield upon addition of the DNA, consistent with its insertion and stacking inside the DNA base stack.32
Intraduplex 1H-correlations
Notably, both the NOESY and the HOHAHA spectra recorded in D2O clearly indicate the loss of the C2 symmetry in the central part of the oligonucleotide upon insertion of the rhodium complex (Figures 2 and 3). For instance, two thymine H6/Me correlations and five cytosine H6/H5 correlations are observed in the presence of Δ-Rh(bpy-d8)2chrysi3+, as opposed to only one and four respectively for free DNA (Figure 3). Two strands, labeled a and b, are thus clearly distinguished in the central part of the oligonucleotide comprising the A4pC5pT6 steps. Accordingly, in the NOESY spectra, two sequential NOESY walks, labeled a and b, were built along the central part of the DNA (Figure 2b). These NOESY data confirm that the double helix maintains its original B-conformation upon insertion of the chrysi ligand despite its extended width.
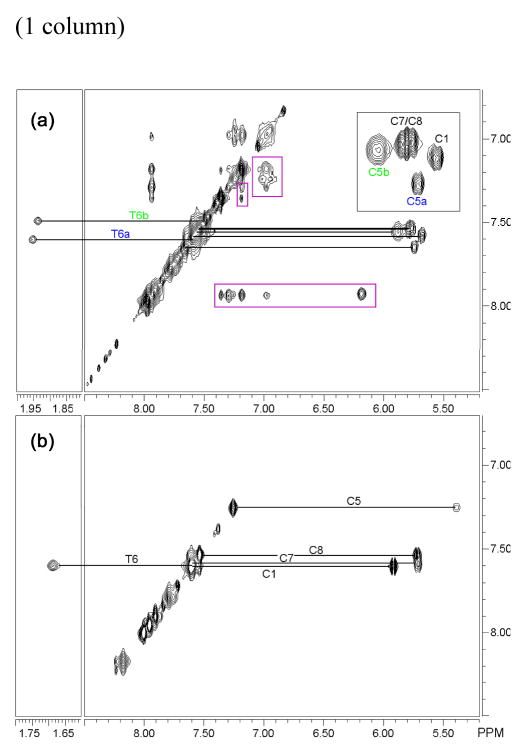
Figure 3.
HOHAHA of the Rh complex inserted into the DNA (a) versus the free oligonucleotide (b). The thymine H6/Me and the cytosine H6/H5 resonances are connected by solid lines. Insert: expansion of the cytosine correlations. The loss of C2 symmetry in the central part of the oligonucleotide is clearly apparent with the two ejected mismatched cytosines, C5a and C5b, which are no longer chemically equivalent. Also observable are correlations between the chrysi protons, χ (purple box). Experimental conditions: D2O, 10 °C, 100 ms mixing time.
Importantly, no nOes are observed in either strand between T6 Me and C5 H6. These nOes are, however, clearly present in the free oligonucleotide. Furthermore, the nOes observed between A4 and C5H6 clearly indicate that, in both strands, the mismatched cytosines are still close to their neighboring adenosines. Together, these observations are consistent with ejection of the mismatched cytosines from the double helix asymmetrically in such a way that they remain closer to the purines than the pyrimidines. Furthermore, the crosspeaks assigned to the C5 from strand b are broader than those from strand a. This suggests that one of the ejected cytosines is more flexible and less constrained than the other.
No anomalous intra nOe crosspeaks for the bases H6 and H8 are detected suggesting that, even after insertion, all bases maintain their anti conformation. Furthermore, the patterns of the 2D DQF- COSY crosspeaks, although more difficult to observe in the Rh-bound DNA than in the free oligonucleotide, suggest that all sugars, including the ejected cytosines, maintain their C2′-endo puckering (Figure 4). This is further supported by the absence of correlations between H3′ and H2″.
Figure 4.
2D DQF-COSY sub-spectrum of Δ-Rh(bpy-d8)2chrysi3+ inserted in DNA at 10 °C (F2 × F1: H1′ × H2′-H2″). Only the cross-peaks associated with H1′-H2′ correlations are visible. Those corresponding to H1′-H2″ correlations are only observed with lower signal to noise ratio. The cross-peak patterns indicate that all sugars, including the ejected cytosines, maintain the C2′-endo puckering.
Again, recording NMR spectra in H2O at 4 °C enables the assignment of the exchangeable imino protons (Figures S6 and S7, supporting information). The T6a and T6b imino protons were assigned according to their dipolar correlations with T6aMe and T6bMe respectively, as well as with A4a and A4b to which they are paired. These T6 imino protons markedly shift upfield upon insertion of the chrysi ligand. Again, no correlations are observed between the imino protons of the mismatched cytosines. Yet, in this instance, the lack of contact between C5a and C5b is more likely due to their ejection from the DNA base stack than to any Wobble-type pairing between them.
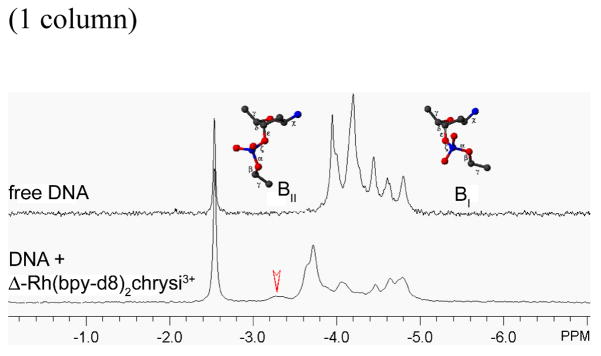
31P-NMR results
The more flexible part of the DNA, namely the phosphodiester backbone, undergoes significant distorsion upon insertion. Addition of Δ-Rh(bpy-d8)2chrysi3+ significantly changes the 1D 31P NMR spectrum of the oligonucleotide: at least one peak shifts significantly downfield from the others (Figure 5). Gorenstein and coworkers previously reported that the torsion angle difference (ε-ξ) can be directly calculated from the chemical shifts of the 31P NMR spectrum.33 The resulting (ε-ξ) values directly indicates whether the phosphodiester backbone is in the BI or BII conformation. In the case of the free oligonucleotide, all phosphodiester junctions are in the normal BI conformation. However, in the Rh-bound DNA, while most of the phosphates have chemical shifts that clearly indicate torsion angles for the BI conformation, one of the phosphates, that which is shifted downfield, has shifts that reflect adoption of the more open BII conformation.
Figure 5.
31P 1D NMR of the free oligonucleotide (upper trace) and the Δ-Rh(bpy-d8)2chrysi3+ inserted in DNA (lower trace). Chemical shifts are referenced to an external standard of TMP (δ = 0.00 ppm). The intense peak at δ = −2.54 ppm corresponds to the phosphate buffer. All phosphate-ester junctions in the free oligonucleotide are in the canonical BI conformation. In the Rh complex-bound DNA, a new peak appears at δ = −3.25 ppm (red arrow) corresponding to a (ε–ξ) of 14.7 °. This value reflects a phosphodiester linkage in the more open BII conformation. Experimental conditions: D2O, 10 °C.
1H-correlations between the DNA and metal Complex
The 1H NMR signals of the free Δ-Rh(bpy-d8)2chrysi3+ were first assigned by the combination of its NOESY and COSY spectra (Figure S8, supporting information). As a reminder, since the ancillary bipyridines are deuterated, they remain silent during the 1H NMR experiments. The two exchangeable χ0 and χ11 protons give rise to an additional signal in H2O (4 °C) at δ = 8.70 ppm.
Intermolecular correlations between the chrysi ligand and the central part of the oligonucleotide are also observed and give insights into the insertion binding mode. About twenty specific contacts are detected between the chrysi ligand and the oligonucleotide at the site of insertion (Figure 6 and Table S4, supporting information), principally with the ejected C5 and T6. The chrysi protons advantageously shift upfield upon insertion in a frequency window without strong overlap, such that many contacts with sugar protons can be assigned. However, the A4H2′ (δ = 7.97 ppm) which would have been a valuable indicator to probe the minor groove occupancy by the metalloinsertor, overlaps significantly with some chrysi protons (δ = 7.93 ppm), such that the corresponding intermolecular contacts are masked by the strong intra-chrysi correlations.
Figure 6.
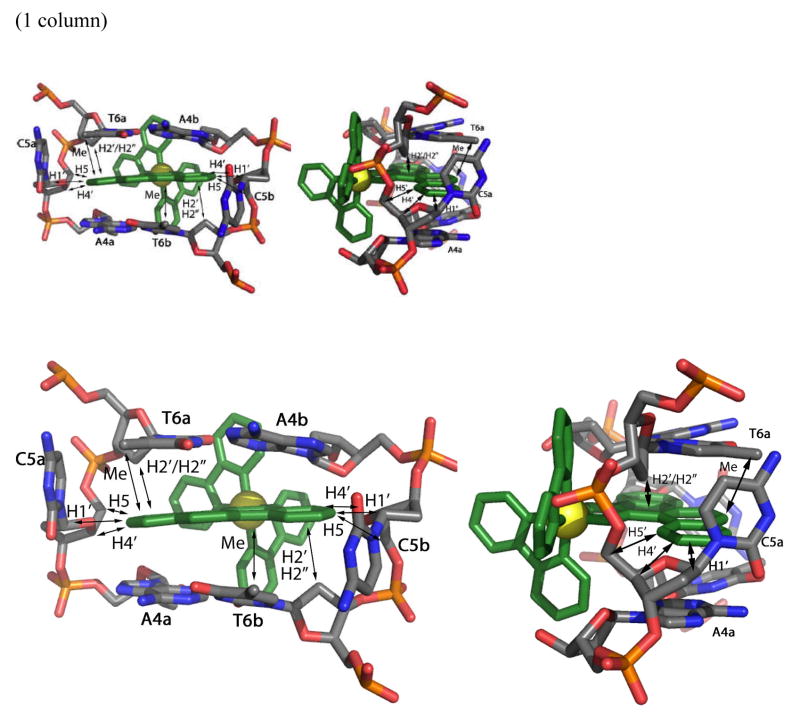
Structural model, adapted from the crystal structure,17 illustrating the insertion of the chrysi complex from the minor groove with ejection of the mismatched cytosines and showing metal complex/DNA nOe’s. Observed nOe contacts are indicated with arrows.
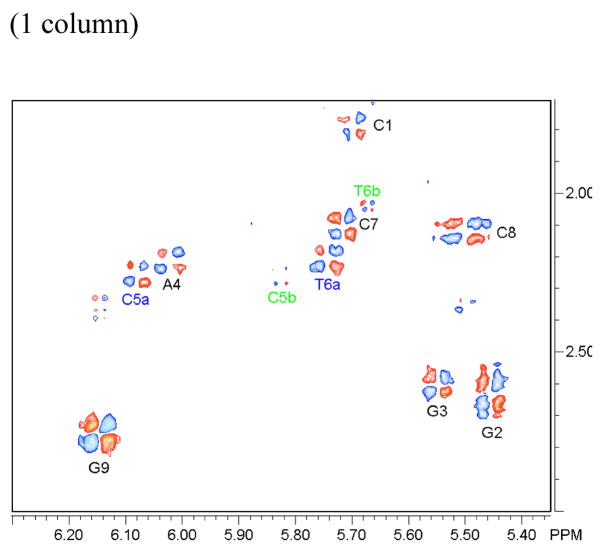
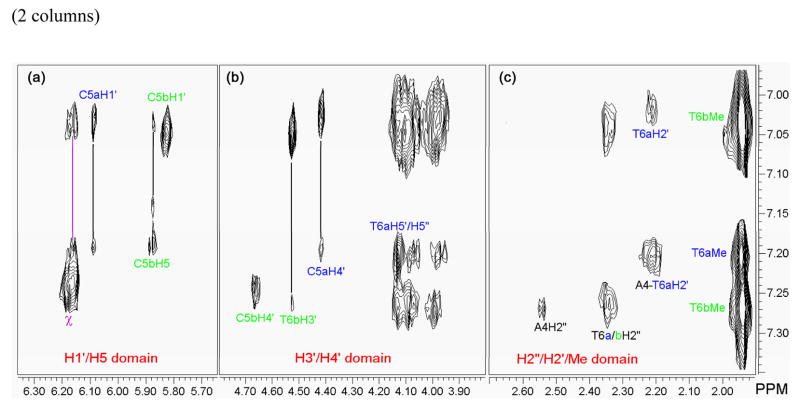
The intra-DNA nOes observed between A4H2 and C5H1′ of strand a are consistent with ejection of the mismatched C5a in the major groove (Figure 2). The homologous contact with C5b was however not detected even at lower signal to noise ratio. Nonetheless, the position of the second mismatched cytosine, C5b, in the major groove could be ascertained by intermolecular contacts with the chrysi ligand (Figure 7). Indeed, the correlation observed between the sugar protons of C5b located in the minor groove (H1′ and to a lesser extend, H4′) and the aromatic chrysi proton clearly also position C5b in the major groove. Additional nOe correlations between the chrysi ligand and the H5′, H5″ and H2′ of T6 of both strands as well as the lack of nOe between T6Me and C5H6 further support insertion of the rhodium complex via the minor groove and ejection of both mismatched cytosines in the opposite major groove. The strong dipolar crosspeaks observed between the chrysi ligand and the T6Me confirm the deep insertion of Rh(bpy-d8)2chrysi3+.
Figure 7.
NOESY sub-spectra and assignments of intermolecular contacts between the DNA and the chrysi protons. (a) (F2 × F1: H1′/H5′ ×χ), (b) (F2 × F1: H3′/H4′ ×χ), (c) (F2 × F1: H2′-H2″/Me × χ). The intramolecular contacts between the chrysi protons are noted χ (purple line). Contacts between the chrysi ligand and strand a (dark blue) and strand b (light green) of the oligonucleotide are clearly separated from each other. Experimental conditions: D2O, 10 °C, 300 ms mixing time.
Changes in chemical shift associated with binding
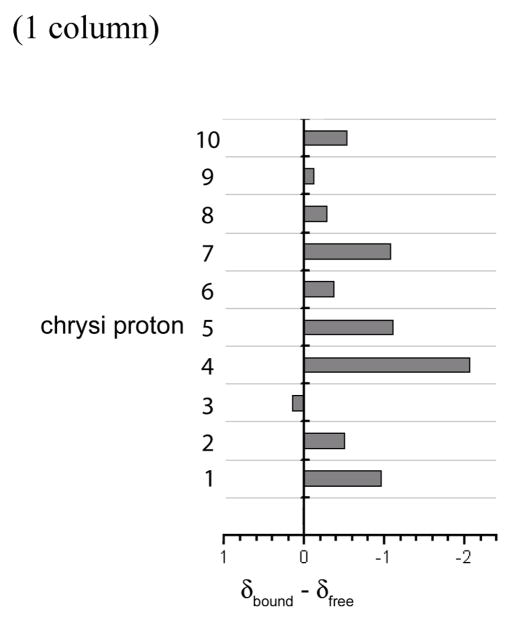
Changes in chemical shifts also give valuable information on the binding of the bulky metal complex to DNA. Upon insertion in the oligonucleotide, most of the protons of the chrysi ligand shift upfield by as much as −2.05 ppm (Figure 8), consistent with insertion and π-stacking of the bulky ligand inside the DNA base stack. 32, 34–37 Notably, one of the chrysi protons, χ3, shifts slightly downfield, while its neighbor, χ4, shifts significantly upfield. These two unusual shifts may be explained by insertion of the rhodium complex from the minor groove. Indeed, this minor groove orientation places χ3 directly below the carbonyl of T6a which may perturb the ring current, while χ4 in the ortho positions is directly placed under the ring current and thus undergoes a significant upfield shift.
Figure 8.
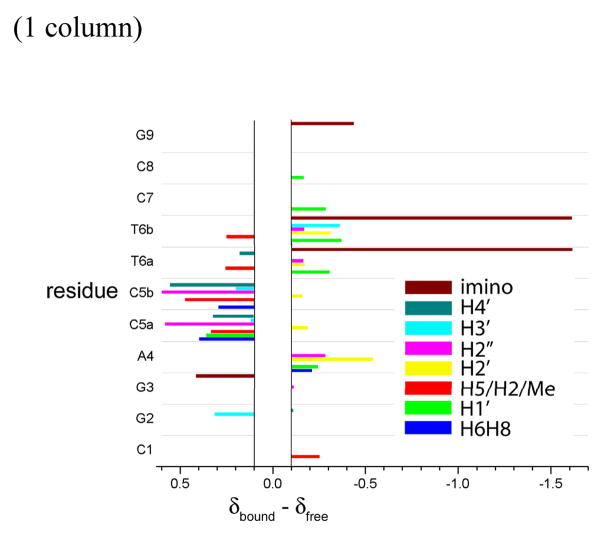
Variation of the chemical shifts, δbound –δfree, of the chrysi protons upon insertion in the DNA. A positive value corresponds to a downfield shift and a negative one to an upfield shift. See Figure 1 for numbering of the chrysi protons.
Just as the metalloinsertor undergoes significant chemical shifts upon insertion, so does the DNA (Figure 9). For clarification, only those protons which shift more than 0.1 ppm are considered; also the H5′ and H5″ are not considered since they strongly overlap with each other. Nonetheless, as is evident from the bar graph, the central part of the oligonucleotide marking the site of insertion, namely A4pC5pT6, undergoes a significantly greater shift, reflecting a greater change in magnetic surrounding, than does the rest of the DNA. Importantly, the protons of both mismatched cytosines (C5a and C5b) shift downfield, consistent with their ejection from the base stack. The exception of C5H2′ could be explained by the fact that the H2′ and H2″ of C5 are inverted in the free oligonucleotide. Similarly, most protons of the flanking A4·T6 base pairs are shifted upfield due to their efficient π-stacking with the inserted chrysi ligand. Significantly, the H1′ of the flanking A4·T6 base pairs shift upfield, consistent with insertion of Δ-Rh(bpy-d8)2chrysi3+ via the minor groove (Δδ = −0.24 ppm, −0.31 ppm, and −0.37 ppm for A4, T6a and T6b respectively).
Figure 9.
Variation of the chemical shifts, δbound– δfree, of the DNA protons upon insertion of the Rh complex. A positive value corresponds to a downfield shift and a negative one to an upfield shift. Only significant changes (± 0.1 ppm) are considered.
The exchangeable imino protons of the flanking T6 also undergo significant upfield shifting (Δδ = −1.61 ppm) and this dramatic shift is another strong indicator of the deep insertion of the chrysi ligand. This thymine is still tightly hydrogen bonded to its adenine; no changes are observed for the base pairs flanking the extended chrysi ligand. Indeed, analysis of the amino protons of the adenosines indicate that the chemical shift differences between the free and bonded amino,Δδamino are 1.46 ppm and and 1.12 ppm for A4 of strands a and b respectively. This Δδamino is a direct indicator of the strength of the base pairing and is comparable to that of the triply hydrogen-bonded C1, C7 and C8 (Δδamino = 1.43 ppm, 1.52 ppm and 1.53 ppm respectively).
Notably, homologous protons in strand a and b of the oligonucleotide are perturbed differently, resulting in the break in the C2 symmetry at the site of insertion. Strand b of the DNA undergoes greater changes in chemical shift than does strand a, suggesting that the wider part of the chrysi ligand (corresponding to the protons χ7–10) is pointed toward strand b.
Insertion mode
Figure 10 displays the basic characteristics associated with insertion of the bulky metal complex into the mismatched DNA that are evident from the NMR study. Consistent with the chemical shift changes seen both for the metal complex and for the DNA as well as the NOESY data, the metal complex is clearly bound at the central mismatched site by insertion and stacking of the chrysi ligand. This leaves sufficient room for the ancillary ligands of the Δ-isomer but not the Λ-isomer to insert into the duplex, if there is no increase in base pair rise at the insertion site. Based upon the NOESY data, the metal complex binds from the minor groove side and inserts deeply into the duplex so that the chrysi protons can interact with major groove protons. The cytosines are ejected out into the major groove, in an asymmetric, likely more flexible, fashion; no stacking or hydrogen bonding by the mismatched cytosines occur. The insertion mode is, furthermore, local to the mismatched site, and the remainder of the duplex remains in a hydrogen-bonded and stacked B-conformation.
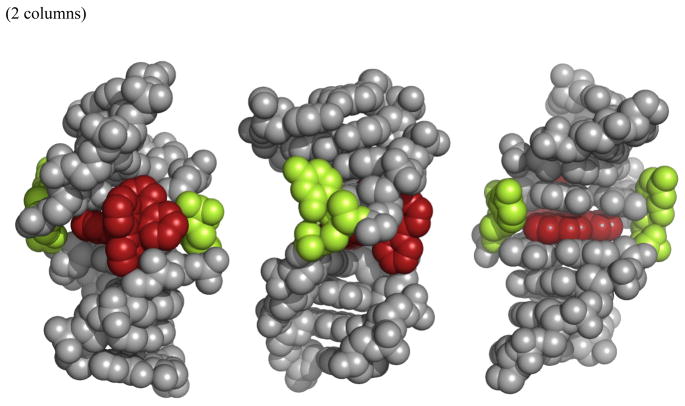
Figure 10.
Structural model for binding of the Rh complex at the mismatched site in the 9-mer duplex based upon the NMR study with views looking into the minor groove (left), from the side (center) and into the major groove (right). The Rh complex (red) is shown bound from the minor groove, with the chrysi ligand inserted deeply into the mismatched site and the mispaired cytosines (green) ejected asymmetrically into the major groove.
Comparison with the crystal structure
Taken together, the NMR solution study and the solid state structure obtained from single crystal X-ray diffraction17 give a clear picture of the novel insertion binding mode of the bulky metal complex with a single base mismatch. Since the crystal structure and the NMR studies were performed on different oligonucleotides containing different mismatches (CA and CC mismatches respectively) and the results are seen to be mutually consistent, it appears that the features seen here reflect insertion of the complex inside any thermodynamically destabilized mismatched duplex.
Significantly, insertion differs from the previously well characterized intercalation22–25, 34–37 in that the DNA does not unwind to enable a ligand to enter the base stack, but rather the incoming ligand simply ejects both bases of the destabilized or weakly bonded pair. The present NMR study confirms that minimal perturbation of the DNA is required for insertion of the bulky ligand and base ejection. Both studies indicate that all sugars maintain their C2′-endo puckering and all bases maintain their anti conformation, including the ejected ones. The oligonucleotide enables insertion of the chrysi ligand by opening its phosphodiester junctions from a BI to a more open BII conformation. The present NMR study also clearly confirms that, even in solution, the base pairs flanking the chrysi ligand remain paired, and this despite the extended length of chrysi, 0.5 Å wider than the span of a base pair.
Thus Δ-Rh(bpy-d8)2chrysi3+ can discriminate single base mismatches effectively. The present NMR solution study and the crystal structure17 illustrate this ejection of destabilized bases by the bulky rhodium complex without perturbing well-paired neighboring sites. Both studies demonstrate that there is sufficiently deep insertion of the chrysi ligand so that it protrudes through the base stack and into the opposite major groove. Both studies also reveal the break in symmetry upon insertion of the chiral metal complex. Importantly, this results in two ejected bases of different nature. Indeed, both studies indicate that one of the ejected bases is significantly more flexible than the other. In the crystal structure, the more flexible base (in that case an adenosine) is able to fold back into the minor groove where it is π-stacked with both an ejected purine and an ancillary bpy ligand of a rhodium complex inserted in a nearby crystallographically related oligonucleotide; such an interwoven conformation is unlikely in solution. Indeed, the present study illustrates that in solution, although more flexible, the second mismatched base is still positioned in the major groove.
Remarkably, this difference in flexibility between the two ejected bases is in accordance with the photoactivated cleavage experiments performed with this class of compounds.26 In all cases, only one strand of the DNA is cleaved upon photoactivation of the inserted rhodium complex. This corresponds, both in solution and in the solid state, to the strand with the more rigid ejected base. Furthermore, both studies place the sugar protons of the base flanking the mismatch closer to the chrysi ligand than the sugar of the ejected base, thereby explaining why photoactivation of the inserted rhodium complex cleaves the base one away from the mismatch and not at the mismatch itself.
It should be noted that there is, however, one clear difference between the present solution study and the crystal structure. In the crystal structure, an additional Rh complex is bound by intercalation from the major groove side at the central 5′AT-3′ step.17 Here, while there is significiant broadening of all the resonances in the bound form, there is no direct indication of any specific perturbation of the well-matched DNA sites, only binding at the mismatched site. This lack of evidence for binding at a well-matched site in solution is consistent, in the crystal, with binding at the matched site being a result of crystal packing forces. Also consistent with this difference between solid state and solution, we had observed photoactivated cleavage at the matched site as well as the mismatched sites in the crystal but only at the mismatched site in solution.
CONCLUSION
The NMR solution study of Δ-Rh(bpy-d8)2chrysi3+ bound to its target CC mismatch in DNA reveals the key features associated with insertion into a mismatched DNA site. Bulky metalloinsertors can target destabilized single-base mismatches, replacing the weakly bonded bases and ejecting them out of the DNA base stack. The insertor enters the DNA via the minor groove, thereby pushing the ejected bases into the opposite major groove. This observation corroborates the direct correlation between the binding affinity of Δ-Rh(bpy-d8)2chrysi3+ for mismatches and the extent to which these mismatches destabilize DNA. 1–3 The C2 symmetry of the palindromic oligonucleotide is broken upon insertion of the rhodium complex resulting in two distinct strands. Consequently, the two ejected bases are no longer equivalent: one of the two mismatched cytosines is significantly more flexible than the other. This result is in accordance with previous photoactivated cleavage experiments. Notably, insertion of the bulky metalloinsertor also only minimally and locally distorts the structure of the DNA duplex. All bases maintain their anti conformation and the sugars maintain their regular C2′-endo puckering, including the ejected ones; insertion of the bulky ligand is accomodated only by a small change in the phosphodiester junction.
EXPERIMENTAL SECTION
Materials
Unless otherwise noted, starting materials were obtained from commercial suppliers and used without further purification. Phosphoramidites, reagents and solid supports for DNA synthesis were obtained from Glen Research. Trimethylphosphate (TMP) and bipyridine-d8 were obtained from Aldrich. RhCl3 was obtained from Pressure Chemicals. D2O (99.96 %) and 2,2-dimethyl-2-silanepentane-5-sulfonate sodium (1% DSS-d6 in 99.99% D2O) were obtained from Cambridge Isotopes Labs and Itotec respectively. Water was deionized and further purified by a Millipore cartridge system (resistivity 18 × 106 Ω).
Rh(bpy-d8)2chrysi3+ was synthesized according to literature procedure using bpy-d8.38 The two enantiomers were separated as previously described using chromatography with the chiral eluant, antimonyl tartrate. 1, 25 Only the Δ enantiomer was titrated into the DNA. The self-complementary oligonucleotide, 5′-CGGACTCCG-3′ was synthesized on an Applied Biosystems 394 automatic DNA synthesizer and purified according to standard protocols.
The DNA concentrations of all samples were determined by UV-visible spectroscopy at 70 °C on a Beckman DU 7400 diode array spectrophotometer equipped with a Peltier heating sample holder. An extinction coefficient for the single stranded oligonucleotide of ε260 ssDNA = 88,200 M−1cm−1 was used. The melting profiles were similarly measured with a heating and cooling rate of 0.5 °C/min between 5 °C and 50 °C.
Photocleavage experiments
Photoactivated cleavage was performed as previously described26 using the following concentrations: 7.5 μM dsDNA, 7.5 μM Δ-Rh(bpy-d8)2chrysi3+ (1 equivalent/mismatch), 10 mM Na phosphate, 50 mM NaCl, pH 7.0. Irradiation was carried out at 313 nm using a 1000 W Oriel Hg/Xe arc lamp with a monochromator fitted with a 300 nm cutoff filter and IR filter. MALDI-TOF mass spectra were recorded on a PerSeptive Biosystem Voyager-DE Pro.
NMR sample preparations
The NMR sample of the free oligonucleotide was prepared by dissolving the lyophilized 9-mer in 500 μL of 50 mM Na phosphate buffered at pH = 6.10 containing 20 mM NaCl.39 The final concentration of duplex DNA was 2.33 mM. For spectra measured in D2O, the solvent was lyophilized from the aqueous buffer, then twice redissolved in 99.9 % D2O and lyophilized, and finally redissolved in 500 μL of 99.96 % D2O. A drop of 1 % DSS-d6 in D2O was added. The DSS- d6 methyl signal was used as internal reference (δ = 0.000 ppm). For investigation of the exchangeable protons, the D2O sample was lyophilized and redissolved in H2O/D2O, 90/10.
The NMR sample of Rh-bound DNA was similarly prepared with a final concentration of 1.62 mM duplex DNA, 1.62 mM Δ-Rh(bpy-d8)2chrysi3+ (1 equivalent/mismatch), 50 mM Na phosphate buffered at pH = 6.10, 20 mM NaCl. Likewise, the NMR sample of the free metalloinsertor contained 1.62 mM Δ-Rh(bpy-d8)2chrysi3+, 50 mM Na phosphate buffered at pH = 6.10, 20 mM NaCl.
NMR Measurements
H NMR spectra were collected on a Varian Unity-Plus 600 spectrometer equipped with a variable temperature unit and pulse-field gradients in three dimensions. 31P NMR spectra were recorded at 242 MHz on a Varian 300 MHz instrument. 1H and 31P NMR spectra were recorded using a dual (1H, 13C) and a quadra (1H, 13C, 15N, 31P) probes respectively. NMR data were processed using the MestReC software (version 4.8.6.0).
The non-exchangeable (D2O) and exchangeable (H2O/D2O: 90/10) proton spectra were recorded at 10 °C and 4 °C respectively. NOESY spectra in D2O of the free DNA and Δ-Rh(bpy- d8)2chrysi3+ inserted in DNA were collected with mixing times of 150 and 300 ms respectively (12 ppm sweep width, TPPI, 2048 complex points, 560 t1 blocks, 64 scans per t1 block, 2.5 s relaxation delay, water suppression by presaturation of the residual signal during relaxation time and mixing time). WaterGATE NOESY spectra in H2O/D2O (90/10) were recorded with a mixing time of 300 ms for the free DNA and Δ-Rh(bpy-d8)2chrysi3+ inserted in DNA (20 ppm sweep width, TPPI, 4096 complex points,560 t1 blocks, 96 scans per t1 block, 2 s relaxation delay). HOHAHA experiments were collected with a mixing time of 110 ms so as to optimize the coherence transfers from the H1′ to H2′ and H2″ protons40,41 (12 ppm sweep width, hyper-complex mode, TPPI, 2048 complex points, 380 t1 blocks, 64 scans per t1 block, 1.5 s relaxation delay, water suppression by presaturation of the residual signal). DQF-COSY experiments42 were collected with 512 and 680 t1 blocks for the free DNA and the Rh-bound DNA respectively (12 ppm sweep width, TPPI, 4096 complex points, 64 scans per t1 block, 1.5 s relaxation delay, water suppression by presaturation of the residual signal). 1D 31P NMR experiments were recorded at 10 °C and referenced to an external standard of TMP (0.1 M in D2O, δ = 0.000 ppm, 20 ppm sweep width, 4096 complex points, 250 and 800 scans, 1.0 s relaxation delay).
For the free Rh(bpy-d8)2chrysi3+ an additional set of COSY and NOESY experiments were performed in D2O at 20 °C (COSY: 9 ppm sweep width, 2048 complex points, 512 t1 blocks, 16 scans per t1 block, 1.0 s relaxation delay, water suppression by presaturation of the residual signal; NOESY: 9 ppm sweep width, 2048 complex points, 640 t1 blocks, 16 scans per t1 block, water suppression by presaturation of the residual signal during 1.0 s relaxation time and 0.8 s mixing time). Exchangeable protons of the Rh complex were assigned in H2O/D2O: 90/10 at 4 °C by 1D NMR (12 ppm sweep width, 32 K complex points, 16 scans, 1.0 s relaxation delay).
Supplementary Material
Acknowledgments
We are grateful to the National Institutes of Health (GM33309) for their financial support. We also thank the Universite D. Diderot (Paris 7) for sabbatical support to C. C. Additionally we thank P. K. Bhattacharya, K. Crowhurst, M. Shahgholi, and S. Ross for helpful discussions.
Footnotes
Supporting Information Available. Table of nOe contacts of the free and Rh-bound oligonucleotide, chemical shifts of the chrysi ligand in the presence and absence of DNA, list of nOe contacts observed between the chrysi ligand and the DNA, titration of Δ-Rh(bpy-d8)2chrysi3+ to DNA containing CA versus CC mismatches, melting curves of the oligonucleotides with and without the metalloinsertor, mass spectra of photocleavage experiments, COSY and NOESY spectra of free Δ-Rh(bpy-d8)2chrysi3+, 2D DQF-COSY and NOESY subspectra of the free oligonucleotide, NOESY subspectra of the exchangeable protons of the DNA bound to the metalloinsertor. This information is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Jackson BA, Barton JK. J Am Chem Soc. 1997;119:12986–12987. [Google Scholar]
- 2.Jackson BA, Barton JK. Biochemistry. 2000;39:6176–6182. doi: 10.1021/bi9927033. [DOI] [PubMed] [Google Scholar]
- 3.Jackson BA, Alekseyev VY, Barton JK. Biochemistry. 1999;38:4655–4662. doi: 10.1021/bi990255t. [DOI] [PubMed] [Google Scholar]
- 4.Hart JR, Johnson MD, Barton JK. Proc Natl Acad Sci U S A. 2004;101:14040–14044. doi: 10.1073/pnas.0406169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kunkel TA, Erie DA. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 6.Strauss BS. Mutation Research/Reviews in Mutation Research. 1999;437:195–203. doi: 10.1016/s1383-5742(99)00066-6. [DOI] [PubMed] [Google Scholar]
- 7.Arzimanoglou II, Gilbert F, Barber HRK. Cancer. 1998;82:1808–1820. doi: 10.1002/(sici)1097-0142(19980515)82:10<1808::aid-cncr2>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 8.Loeb LA, Loeb KR, Anderson JP. Proc Natl Acad Sci U S A. 2003;100:776–781. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Junicke H, Hart JR, Kisko J, Glebov O, Kirsch IR, Barton JK. Proc Natl Acad Sci U S A. 2003;100:3737–3742. doi: 10.1073/pnas.0537194100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruba E, Hart JR, Barton JK. Inorg Chem. 2004;43:4570–4578. doi: 10.1021/ic0499291. [DOI] [PubMed] [Google Scholar]
- 11.Zeglis BM, Barton JK. J Am Chem Soc. 2006;128:5654–5655. doi: 10.1021/ja061409c. [DOI] [PubMed] [Google Scholar]
- 12.Schatzschneider U, Barton JK. J Am Chem Soc. 2004;126:8630–8631. doi: 10.1021/ja048543m. [DOI] [PubMed] [Google Scholar]
- 13.Petitjean A, Barton JK. J Am Chem Soc. 2004;126:14728–14729. doi: 10.1021/ja047235l. [DOI] [PubMed] [Google Scholar]
- 14.Hart JR, Glebov O, Ernst R, Kirsch IR, Barton JK. Proc Natl Acad Sci U S A. 2006;103:15359–15363. doi: 10.1073/pnas.0607576103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunner J, Barton JK. Biochemistry. 2006;45:12295–12302. doi: 10.1021/bi061198o. [DOI] [PubMed] [Google Scholar]
- 16.Puckett CA, Barton JK. J Am Chem Soc. 2007;123:46–47. doi: 10.1021/ja0677564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierre VC, Kaiser JT, Barton JK. Proc Natl Acad Sci U S A. 2007;104:429–434. doi: 10.1073/pnas.0610170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erkkila KE, Odom DT, Barton JK. Chem Rev. 1999;99:2777– 2795. doi: 10.1021/cr9804341. [DOI] [PubMed] [Google Scholar]
- 19.Sitlani A, Long EC, Pyle AM, Barton JK. J Am Chem Soc. 1992;114:2303–2312. [Google Scholar]
- 20.Kielkopf CL, Erkkila KE, Hudson BP, Barton JK, Rees DC. Nat Struc Biol. 2000;7:117–121. doi: 10.1038/72385. [DOI] [PubMed] [Google Scholar]
- 21.Barton JK. Science. 1986;233:727–734. doi: 10.1126/science.3016894. [DOI] [PubMed] [Google Scholar]
- 22.David SS, Barton JK. J Am Chem Soc. 1993;115:2984–2985. [Google Scholar]
- 23.Sitlani A, Dupureur CM, Barton JK. J Am Chem Soc. 1993;115:12589–12595. [Google Scholar]
- 24.Hiort C, Lincoln P, Norden B. J Am Chem Soc. 1993;115:3448–3454. [Google Scholar]
- 25.(a) Dupureur CM, Barton JK. Inorg Chem. 1997;36:33. [Google Scholar]; (b) Zeglis BM, Barton JK. Nature Prot. 2007;2:357–371. doi: 10.1038/nprot.2007.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunner J, Barton JK. J Am Chem Soc. 2006;128:6772–6773. doi: 10.1021/ja0612753. [DOI] [PubMed] [Google Scholar]
- 27.(a) Wuthrich K. NMR of Proteins and Nucleic Acids. Wiley; New York: 1986. [Google Scholar]; (b) James TL, editor. Nuclear Magnetic Resonance and Nucleic Acids. Vol. 261. Academic Press, Inc; San Diego: 1995. Methods in Enzymology. [Google Scholar]
- 28.Van de Ven FJ, Hilbers CW. Eur J Biochem. 1988;178:1–38. doi: 10.1111/j.1432-1033.1988.tb14425.x. [DOI] [PubMed] [Google Scholar]
- 29.Kouchakdjian M, Li BFL, Swann PF, Patel DJ. J Mol Biol. 1988;202:139–155. doi: 10.1016/0022-2836(88)90526-8. [DOI] [PubMed] [Google Scholar]
- 30.Gray DM, Cui T, Ratliff RL. Nucl Acids Res. 1984;12:7565–7580. doi: 10.1093/nar/12.19.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boulard Y, Cognet JAH, Fazakerley GV. J Mol Biol. 1997;268:331–347. doi: 10.1006/jmbi.1997.0975. [DOI] [PubMed] [Google Scholar]
- 32.(a) Patel DJ, Shapiro L. J Biol Chem. 1986;261:1230. [PubMed] [Google Scholar]; (b) Patel DJ, Shapiro L. Biopolymers. 1986;25:707. doi: 10.1002/bip.360250413. [DOI] [PubMed] [Google Scholar]; (c) Aggarwal A, Islam SA, Kuroda R, Neidle S. Biopolymers. 1984;23:1025–1041. doi: 10.1002/bip.360230605. [DOI] [PubMed] [Google Scholar]; (d) Weiner SJ, Kollman PA, Nguyen DT, Case PA. J Comp Chem. 1986;7:230. doi: 10.1002/jcc.540070216. [DOI] [PubMed] [Google Scholar]; (e) Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alagana G, Piotete S, Weiner P. J Am Chem Soc. 1984;106:765. [Google Scholar]; (f) LaMar G, VanHecke G. Inorg Chem. 1970;9:1546. [Google Scholar]; (g) Huang T, Brewer D. Can J Chem. 1981;59:1689. [Google Scholar]
- 33.Gorenstein DG. Chem Rev. 1994;94:1315–1338. The empirical relation of Gorenstein is (ε–ζ = 254.5 + 72.8 δ 31P) [Google Scholar]
- 34.Franklin SJ, Barton JK. Biochemistry. 1998;37:16093–16105. doi: 10.1021/bi981798q. [DOI] [PubMed] [Google Scholar]
- 35.Dupureur CM, Barton JK. J Am Chem Soc. 1994;116:10286–10287. [Google Scholar]
- 36.Collins JG, Shields TP, Barton JK. J Am Chem Soc. 1994;116:9840–9846. [Google Scholar]
- 37.Hudson BP, Barton JK. J Am Chem Soc. 1998;120:6877–6888. [Google Scholar]
- 38.Murner H, Jackson BA, Barton JK. Inorg Chem. 1998;37:3007–3012. [Google Scholar]
- 39.pD = pH + 0.4 at 25 °C. The electrode (pH) was calibrated with standard buffered solutions. The pH values indicated correspond to the measured and uncorrected value.
- 40.Cavanagh J, Chazin WJ, Rance MJ. J Magn Reson. 1990;87:110–131. [Google Scholar]
- 41.Wijmenga S, Mooren MW, Hilbers C. In: NMR of Macromolecules, a Practical Approach. Roberts GCK, editor. Oxford University Press; Oxford: 1993. pp. 217–288. [Google Scholar]
- 42.Piantini U, Sorensen OW, Ernst RR. J Am Chem Soc. 1982;104:6800–6801. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.