Abstract
The performance of fatty acid profiling for strain differentiation of Histoplasma capsulatum was assessed. Total fatty acids were isolated from the yeast-phase cells of seven stock and two previously unreported clinical strains of H. capsulatum var. capsulatum, as well as from one unreported clinical strain and one stock strain of H. capsulatum var. duboisii, and one strain of each of three other dimorphic zoopathogenic fungal species, Blastomyces dermatitidis, Paracoccidioides brasiliensis and Sporothrix schenckii. Different colony morphology and pigmentation types of the H. capsulatum strains were also included. The most frequently occurring fatty acids were oleic, palmitic, stearic and linoleic acids. There were variations in the relative percentage fatty acid contents of H. capsulatum strains that could be used for strain identification and discrimination. Differentiation between H. capsulatum strains was achieved by the comparison of detected fatty acids accompanied by principal component analysis using calculated Varimax-rotated principal component loadings. Statistical analysis yielded three major principal components that explained over 94% of total variance in the data. All the strains of H. capsulatum var. capsulatum RFLP classes II and III were grouped into two distinct clusters: the heterogenic RFLP class I formed a large, but also well-defined group, whereas the outgroup strains of H. capsulatum var. duboisii, B. dermatitidis, P. brasiliensis and S. schenckii were shifted away. These data suggest that fatty acid profiling can be used in H. capsulatum strain classification and epidemiological studies that require strain differentiation at the intraspecies level.
Introduction
Histoplasma capsulatum is the aetiologic agent of histoplasmosis, the most common pulmonary mycosis of humans and other mammals. It is estimated that over 40 million people in the USA have become infected with H. capsulatum, with approximately 500 000 new cases each year. Approximately 95% of histoplasmosis cases are subclinical or self-limiting. Only a small percentage of chronic H. capsulatum infections may turn into a progressive, serious and often fatal systemic disease, especially in immunocompromised patients with AIDS or the lymphoma–leukaemia–Hodgkin's group of diseases, or those on steroid therapy or other immunosuppressive agents (Rippon, 1980; Wheat, 2003). Under these circumstances, H. capsulatum can cause frequent opportunistic infections, and for this reason, along with its worldwide distribution, this fungus is considered an important human pathogen.
The accurate diagnosis of histoplasmosis has classically relied on direct visualization of H. capsulatum in tissue or body fluids and/or its isolation by culturing (Wheat, 2003). The efficiency and sensitivity of these approaches vary with the extent and severity of the infection, and many cases may be undetected or misidentified. These cases might in all probability be properly diagnosed using modern molecular biology techniques. H. capsulatum and other dimorphic fungi can be identified using various commercially available assays, including exoantigen reagents (DiSalvo et al., 1980, 1981; Standard & Kaufman, 1982; Denys et al., 1983; Sekhon et al., 1984, 1986; Body et al., 1988; Wheat et al., 1989, 1991, 1992; Padhye et al., 1992; Sandin et al., 1993; Sutton et al., 1997; Lacaz et al., 1999) and acridinium ester-labelled chemiluminescent DNA probes (Accuprobe, Gen-Probe) (Hall et al., 1992; Padhye et al., 1992; Huffnagle & Gander, 1993; Sandin et al., 1993; Sutton et al., 1997; Chemaly et al., 2001; Brandt et al., 2005). These two methods are accurate and commonly used for H. capsulatum diagnosis (Wheat, 2003).
The use of genetic methods in the diagnosis of histoplasmosis offers not only precise identification, but also the ability to distinguish particular types of H. capsulatum strains. Because of the natural diversity and differences in relative virulence among strains, the application of various nucleic acid-based analytical techniques contributes to efficient strain typing, the determination of biology and infection mechanisms, and epidemiological studies. Initially, most applications were directed above the species level; however, this situation no longer applies, and advances in molecular biology make it possible to distinguish individual pathogenic fungi at the inter- and intrageneric levels. There are also several other strategies, such as grouping based on RFLP analysis of either mitochondrial DNA (Vincent et al., 1986; Spitzer et al., 1989) and rDNA (Vincent et al., 1986; Spitzer et al., 1989; Jiang et al., 2000; Kasuga et al., 1999; Ueda et al., 2003), or Hc-specific protein-coding genes such as yps3 (Keath et al., 1992), arf, ole, tub1 (Kasuga et al., 1999; Taylor et al., 2005), H-anti (Kasuga et al., 1999; Bracca et al., 2003; Taylor et al., 2005) and M-anti (Guedes et al., 2003). H. capsulatum isolates of different types and origins can also be classified by PCR-based random amplification of polymorphic DNA (RAPD) (Kersulyte et al., 1992; Poonwan et al., 1998; Reyes-Montes et al., 1999; Muniz et al., 2001; Taylor et al., 2000, 2005; Zancope-Oliveira et al., 2005), a method sometimes combined with single-strand conformational polymorphism (SSCP) (Carter et al., 1996, 1997), or by repetitive-sequence-based PCR (rep-PCR) (Pounder et al., 2006), or finally by electrophoretic karyotyping and chromosome-length polymorphism analysis (Canteros et al., 2005).
Despite the abundance of already-developed molecular techniques and those still being developed, not all these typing methods are equally effective, and some can lead to misinterpretation. In the case of H. capsulatum and other dimorphic fungal pathogens, no single approach has evolved as dominant over other methods, and for that reason various additional biotyping tests, such as carbohydrate assimilation/fermentation or isoenzyme analysis, are also concurrently in use (Soll, 2000). The chemical composition of cells is a characteristic trait that can also be exploited for the identification and differentiation of various microbial species. Fatty acid profiling [commercially available as the Microbial Identification System (MIS; MIDI, Inc.)] is one such method that is exploited in mycological laboratories, where it has successfully been used for the identification of various yeast species (e.g. Augustyn et al., 1990; El Menyawi et al., 2000; Hinton et al., 2002; Jeennor et al., 2006), including those of clinical importance (Gunasekaran & Hugh, 1980; Kellogg et al., 1998; Peltroche-Llacsahuanga et al., 2000). In comparison with bacteria, yeasts possess a relatively limited range of cellular fatty acids, but enough complexity for chemotaxonomic classification (El Menyawi et al., 2000).
The current study was undertaken to examine total fatty acid patterns of a number of H. capsulatum var. capsulatum strains and to evaluate the usefulness of fatty acid analysis in the differentiation of this pathogen at the intraspecies level. We also examined the fatty acid composition of H. capsulatum var. duboisii and three other dimorphic fungal species, Blastomyces dermatitidis, Paracoccidioides brasiliensis and Sporothrix schenckii. This new approach provides not only an alternative tool for H. capsulatum strain polymorphism studies, but also a means to resolve questions concerning the epidemiology of H. capsulatum infections.
Methods
Fungal strains and growth conditions
The strains used in this study are listed in Table 1. Fungi were routinely grown in liquid Histoplasma-macrophage medium (HMM) (Woods et al., 1998). Cultures were aerated by rotary shaking (150 r.p.m.) at 37 °C in a 5% CO2/95% air atmosphere for 4 days in 20 ml HMM in 50 ml Erlenmeyer flasks. For all experiments, glass was initially treated with a solution of CHCl3/MeOH (1 : 1, v/v), dried and then thoroughly washed with double-distilled water. The experiment was repeated five times for laboratory stock strains and three times for the clinical ones, and each culture was extracted as described below in the lipid-extraction section. In addition, to examine how fatty acid profiles vary during culture growth, six selected strains (Downs, UCLA 531S, G217B, G222B, G186AS, RV26821S) were grown for 3, 4 and 5 days in time-course experiments.
Table 1.
Fungal strains used in this study
| Strain* | RFLP class† |
|---|---|
| H. capsulatum | |
| var. capsulatum | |
| Downs (ATCC 38904) | I |
| UCLA 531S | I |
| UCLA 531E | I |
| UCLA 531R | I |
| G217A (ATCC 26031) | II |
| G217B (ATCC 26032) | II |
| G222B (ATCC 26034) | II |
| G184AS | III |
| G184AE | III |
| G184AR (ATCC 26027) | III |
| G186AS | III |
| G186AE | III |
| G186AR (ATCC 26029) | III |
| G186B (ATCC 26030) | III |
| var. duboisii | |
| RV26821S | |
| RV26821E | |
| RV26821R (ATCC 32281) | |
| Clinical isolates | |
| UWclin01‡ | I |
| UWclin02‡ | III |
| UWclin03‡ | var. duboisii |
| Other dimorphic fungal species | |
| B. dermatitidis ATCC 26199 | |
| P. brasiliensis ATCC 32071 | |
| S. schenckii ATCC 10212 |
Morphological colony and pigment forms: R, rough (virulent); S, smooth (avirulent); E, epithelial variant (avirulent) (Eissenberg et al., 1991); A, albino; B, brown (Berliner, 1973).
Strain classification according to RFLP analysis (Spitzer et al., 1989).
For the purpose of this study, the clinical isolates were identified based on RFLP profiles after digestion of genomic DNA with HindIII.
Lipid extraction
Yeast cells were harvested by centrifugation of 1 ml culture aliquots (3000 g, 5 min) and washed twice with sterile distilled water. The pellet was resuspended in 0.5 ml 0.97% KCl, and transferred into a 2 ml screw-topped plastic tube containing 0.5 ml of acid- and chloroform-washed glass beads (106 mm and finer, Sigma). Cells were disrupted using a Mini-BeadBeater-8 (Biospec Products) for three 1 min periods interspersed with 1 min periods of cooling on ice. Afterwards, cell homogenates were transferred into glass tubes with Teflon seals and 1.5 ml of a CHCl3/MeOH mixture (2 : 1, v/v) was added to each sample. Tubes were thoroughly vortexed for 3 min and lipids were extracted for 2 h at 4 °C. Next, 1.0 ml 0.97% KCl was added to each tube, mixed and then left to stand for 1 h. To separate organic and aqueous phases, the tubes were centrifuged (300 g, 5 min) and the top aqueous layer and the interphase were removed with a Pasteur pipette. The organic layer containing extracted lipids was washed with 1.0 ml 0.97% KCl and the top-separated phase was again removed. In order to achieve complete extraction, the above procedure was repeated twice, and the extracts were pooled in a clean glass test tube and the solvent was completely removed under nitrogen. Fatty acids were converted into corresponding methyl ester derivatives in the presence of 14% BF3 in MeOH (Sigma). Prepared fatty acid methyl esters (FAMEs) were extracted with n-hexane. All solvents contained 5 mg l−1 butylated hydroxytoluene (BHT) as antioxidant.
GC analysis
FAMEs were identified by GC using a Hewlett Packard 5890 gas chromatograph equipped with a capillary column coated with DB-225 (30 m length, 0.25 mm external diameter, 0.25 μm internal diameter; Agilent Technologies). Column temperature was kept at 70 °C for 1 min, increased to 180 °C at a rate of 20 °C min−1 and then to 220 °C at a rate of 3 °C min−1. The temperature was kept at 220 °C for 15 min. Injector and detector temperature were set at 250 °C, and the injection port temperature was set at 300 °C. Peaks were identified by comparison of retention times with those of a set of authentic fatty acid standards (Supelco). The abundance of fatty acids was calculated from relative peak areas.
Statistical analysis
Six major fatty acids detected in these fungi [myristic (14 : 0), palmitic (16 : 0), palmitoleic (16 : 1), stearic (18 : 0), oleic (18 : 1) and linoleic (18 : 2) acids] were examined as descriptors for principal component analysis (PCA). PCA is a common technique for finding a set of weighted linear composites, principal components (PCs), of original variables such that each PC is uncorrelated with the others (Hotelling, 1933). The first PC (PC 1) is a weighted linear composite of the original variables with weights chosen such that the composite accounts for the maximum variation in the original data. The second PC (PC 2) accounts for the maximum variation that is not accounted for by the first. The third PC (PC 3) likewise accounts for the maximum, given the first two PCs, and so on. The technique is thus a useful device for representing a set of a large number of variables by a much smaller set of PCs that account for much of the variance among the set of original data. A more detailed description of the use of PCA is given elsewhere (Zarnowski et al., 2004).
In this study, PCA was used to classify the strains into corresponding groups, and a dendrogram was constructed using calculated Varimax-rotated PC loadings. The proper matrices were constructed on the basis of total fatty acid composition in these fungal strains and the data were processed using Statistica for Windows version 5.1 (StatSoft).
Influence of prolonged culture growth on the fatty acid composition
To examine how fatty acid profiles vary during culture growth, six selected strains (Downs, UCLA 531S, G217B, G222B, G186AS, RV26821S) were grown for 3, 4 and 5 days in time-course experiments. Afterwards, fungal cells were harvested and processed for FAME analysis, as described above.
Results
Composition of fatty acids
Fatty acid patterns found in the fungi studied here are shown in Table 2. In addition, data for a set of selected strains are shown in Fig. 1. In all cases, at least six fatty acids could be detected. The most abundant fatty acids were palmitic (16 : 0), oleic (18 : 1) and linoleic (18 : 2) acids, which formed over 90% of the total fatty acid pool. Myristic (14 : 0), palmitoleic (16 : 1) and stearic (18 : 0) acids were present in considerably smaller amounts. Other fatty acids identified in this study, including linolenic (18 : 3), eicosanoic (20 : 0), eicosaenoic (20 : 1), eicosatetraenoic (20 : 4) and docosanoic (22 : 0) acids, were detected in spurious amounts only and for that reason were excluded from further statistical analyses as nonsignificant variables (data not shown). An example of a typical gas chromatogram obtained after chromatographic separation of fatty acids isolated from fungal cells is shown in Fig. 2. Despite this simplicity in fatty acid composition, certain variations between strains and species were detected.
Table 2.
Fatty acid composition in lipid extracts from dimorphic fungi
Values are mean±SD for five experimental repeats.
| Strain | Fatty acid (percentage of total) | |||||
|---|---|---|---|---|---|---|
| Myristic (14 : 0) | Palmitic (16 : 0) | Palmitoleic (16 : 1) | Stearic (18 : 0) | Oleic (18 : 1) | Linoleic (18 : 2) | |
| H. capsulatum: | ||||||
| Downs | 0.5 ± 0.1 | 30.6 ± 0.4 | 1.0 ± 0.1 | 4.5 ± 0.1 | 27.9 ± 0.5 | 35.5 ± 0.8 |
| UCLA 531S | 0.5 ± 0.1 | 32.7 ± 0.9 | 3.2 ± 0.1 | 2.4 ± 0.5 | 43.5 ± 0.8 | 17.6 ± 0.6 |
| UCLA 531E | 0.6 ± 0.1 | 32.5 ± 0.4 | 2.4 ± 0.0 | 2.5 ± 0.2 | 41.5 ± 0.4 | 20.4 ± 0.5 |
| UCLA 531R | 0.6 ± 0.0 | 33.5 ± 0.6 | 2.8 ± 0.2 | 2.5 ± 0.2 | 41.7 ± 0.3 | 19.0 ± 0.9 |
| G217A | 0.4 ± 0.1 | 34.5 ± 0.1 | 1.6 ± 0.1 | 2.5 ± 0.1 | 23.1 ± 0.1 | 37.9 ± 0.2 |
| G217B | 0.3 ± 0.0 | 34.0 ± 0.9 | 2.1 ± 0.1 | 2.3 ± 0.3 | 23.3 ± 0.8 | 38.1 ± 0.7 |
| G222B | 0.3 ± 0.0 | 34.4 ± 0.7 | 1.7 ± 0.1 | 2.4 ± 0.3 | 22.4 ± 0.8 | 38.7 ± 0.4 |
| G184AS | 0.5 ± 0.0 | 36.0 ± 0.1 | 2.7 ± 0.1 | 2.5 ± 0.1 | 23.5 ± 0.1 | 34.7 ± 0.0 |
| G184AE | 0.5 ± 0.1 | 37.7 ± 0.5 | 1.8 ± 0.2 | 2.6 ± 0.3 | 23.4 ± 0.4 | 33.9 ± 0.3 |
| G184AR | 0.4 ± 0.0 | 36.4 ± 0.3 | 1.6 ± 0.0 | 2.5 ± 0.1 | 23.0 ± 0.5 | 34.0 ± 0.2 |
| G186AS | 0.5 ± 0.1 | 35.4 ± 0.2 | 2.3 ± 0.2 | 2.8 ± 0.4 | 24.2 ± 0.2 | 34.8 ± 0.4 |
| G186AE | 0.4 ± 0.0 | 36.8 ± 0.2 | 1.9 ± 0.2 | 2.6 ± 0.2 | 24.0 ± 0.7 | 34.3 ± 0.6 |
| G186AR | 0.5 ± 0.0 | 35.9 ± 0.7 | 2.1 ± 0.1 | 2.4 ± 0.2 | 24.8 ± 1.2 | 34.3 ± 0.5 |
| G186B | 0.5 ± 0.0 | 36.3 ± 1.1 | 1.9 ± 0.2 | 2.7 ± 0.2 | 24.1 ± 0.5 | 34.5 ± 1.2 |
| UWclin01 | 0.6 ± 0.1 | 33.6 ± 0.3 | 2.6 ± 0.2 | 2.8 ± 0.4 | 33.6 ± 0.3 | 26.8 ± 0.8 |
| UWclin02 | 0.7 ± 0.1 | 36.1 ± 0.8 | 1.5 ± 0.0 | 2.5 ± 0.4 | 24.1 ± 0.2 | 35.2 ± 0.9 |
| UWclin03 | 0.6 ± 0.0 | 41.6 ± 0.8 | 1.2 ± 0.0 | 3.7 ± 0.2 | 10.9 + 0.5 | 42.0 ± 0.7 |
| RV26821S | 0.6 ± 0.1 | 40.2 ± 0.4 | 1.4 ± 0.1 | 3.4 ± 0.4 | 12.1 ± 0.3 | 42.3 ± 0.5 |
| RV26821E | 0.5 ± 0.1 | 40.2 ± 0.5 | 1.5 ± 0.0 | 3.3 ± 0.5 | 12.0 ± 1.0 | 42.5 ± 0.1 |
| RV26821R | 0.6 ± 0.1 | 42.1 ± 1.0 | 1.1 ± 0.1 | 3.7 ± 0.2 | 10.4 ± 0.6 | 42.1 ± 0.4 |
| B. dermatitidis | 0.3 ± 0.0 | 23.2 ± 0.4 | 1.4 ± 0.0 | 1.7 ± 0.1 | 7.6 ± 0.2 | 65.8 ± 0.5 |
| P. brasiliensis | 0.5 ± 0.1 | 37.9 ± 0.3 | 0.7 ± 0.0 | 7.4 ± 0.1 | 18.8 ± 0.2 | 34.8 ± 0.1 |
| S. schenckii | 1.5 ± 0.1 | 32.7 ± 0.6 | 1.8 ± 0.1 | 5.6 ± 0.4 | 11.1 ± 0.2 | 47.3 ± 0.9 |
Fig. 1.
Fatty acid composition of lipid extracts from selected dimorphic fungi. (a–h) H. capsulatum strains: (a) Downs (class I); (b) UCLA 531S (class I); (c) G217B (class II); (d) G186B (class III); (e) UWclin01 (class I); (f) UWclin02 (class III); (g) UWclin03 (var. duboisii); (h) RV26821R (var. duboisii). (i–k) The remaining three dimorphic fungal species: (i) B. dermatitidis; (j) P. brasiliensis; (k) S. schenckii.
Fig. 2.
Typical gas chromatogram obtained after separation of total cellular fatty acids isolated from H. capsulatum cells. The conditions for fatty acid analysis were as described in Methods. RSI, relative signal intensity.
In H. capsulatum var. capsulatum strains of RFLP class III, palmitic and linoleic acids were predominant, and were encountered in concentrations ranging between 35.4 and 37.7%, and 33.9 and 34.7%, respectively. Oleic acid was also detected in notable amounts in those strains, and its concentration varied between 23.4 and 24.8%. Strains of H. capsulatum RFLP class II contained higher amounts of linoleic acid (37.9–38.7%), whereas palmitic and oleic acids were present in slightly lower concentrations (34.0–34.5 and 22.4–23.1%, respectively). The greatest discrepancies were observed with respect to fatty acid patterns of H. capsulatum strains of RFLP class I. The level of palmitic acid in Downs and UCLA 531 variants was similar (30.6 versus 32.5–33.5%, respectively), but concentrations of oleic and linoleic acids varied in those strains considerably. Similar discrepancies were also observed in the content of palmitoleic and stearic acids (Fig. 1a, b).
The pattern of fatty acids isolated from H. capsulatum var. duboisii was substantially different from those observed for H. capsulatum var. capsulatum. In this case, the major fatty acids (palmitic and linoleic) formed over 80% of the total fatty acid pool, whereas the content of oleic acid was significantly reduced to only 10–12.1% (Fig. 1h).
Three previously unreported clinical specimens from human histoplasmosis cases were also subjected to fatty acid analysis. The UWclin01 strain contained equal amounts of palmitic and oleic acids (33.6% of each), whereas the level of linoleic acid was lower than those observed in RFLP classes II and III (only 27.4%) (Fig. 1e). The UWclin02 strain contained similar amounts of palmitic and linoleic acids (36.1 and 35.2%, respectively), but the concentration of oleic acid was considerably lower (24.1%) (Fig. 1f). The UWclin03 strain was found to possess a different pattern of fatty acids. In this case, the most abundant fatty acids were palmitic and linoleic acids, which were present in high amounts (41.6 and 42.0%, respectively) (Fig. 1g).
Individual strains of three other dimorphic fungi, B. dermatitidis, P. brasiliensis and S. schenckii, possessed significantly distinct fatty acid profiles. Interestingly, the content of linoleic acid in lipid extracts from B. dermatitidis cells was very high (65.8%), whereas oleic acid was considerably less abundant (only 7.6%) (Fig. 1i). In P. brasiliensis and S. schenckii, the level of stearic acid was elevated (7.4 and 5.6%, respectively) (Fig. 1j, k). The content of myristic acid in S. schenckii cells was also slightly higher (1.5%) than in other strains examined in this work, in which this acid was present in constant, but very small amounts, not exceeding 0.6%.
Statistical analysis
PCA yielded four PCs, of which the major three, PC1, PC2 and PC3, explained over 94% of total variance in the data. We calculated PCs in such a manner that the first PC accounted for as much of the variability in the original dataset as possible, the second PC accounted for as much of the remaining variability in the data as possible, and so on. The majority of this variance was explained by the first and second PCs (58.2 and 35.8%, respectively). Fig. 3 shows a delineation of the examined H. capsulatum strains based on the combination of the first three calculated PCs, which resulted in the formation of three RFLP-related clusters. The strains of H. capsulatum var. capsulatum RFLP classes II and III were grouped into two distinct clusters, whereas the heterogenic RFLP class I formed a large, but also well-defined group. As a result of different levels of major fatty acids as reflected by PC 1, H. capsulatum var. duboisii, together with the three other dimorphic fungal species, B. dermatitidis, P. brasiliensis and S. schenckii (included in this study as the outgroup), was shifted away from other strains of H. capsulatum var. capsulatum.
Fig. 3.
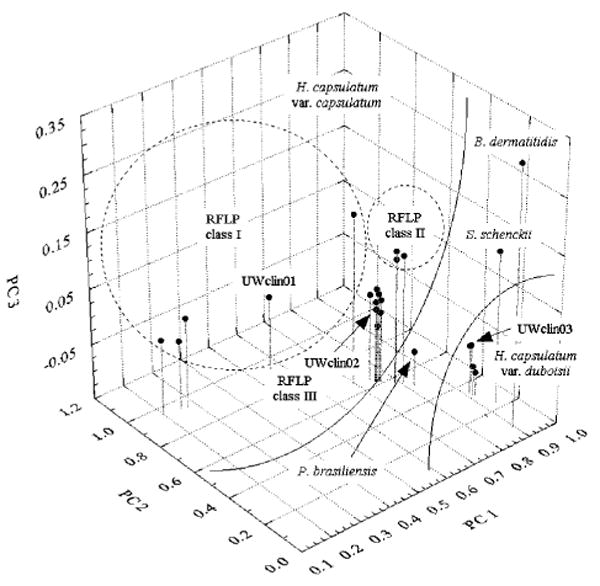
Total fatty acid profile-based grouping of H. capsulatum and other dimorphic fungi using PCA. The 3D score plot was created based on calculated Varimax-rotated PC loadings, PC 1, PC 2 and PC 3, that explained over 94% of total variance in the original dataset.
Overall, the classification pattern observed also resulted from differences in the contents of myristic and 18-carbon carboxylic acids (PC 2), and palmitic and stearic acids (PC 3).
The pattern of classification of the three clinical H. capsulatum isolates tested herein was found to be highly comparable to that of the RFLP-based classification. It is noteworthy that the clinical H. capsulatum strains were novel and had not previously been classified by any genetic method or fatty acid profiling. The latter approach that we describe in this work put UWclin01 into a broad cluster that grouped heterogeneous H. capsulatum var. capsulatum strains of RFLP class I. The UWclin02 strain was grouped within the H. capsulatum var. capsulatum RFLP class III cluster, and the UWclin03 strain was placed along with all strains of H. capsulatum var. duboisii (Fig. 3). The same strain-typing results were obtained by RFLP profiling after genomic DNA digestion with HindIII (data not shown).
Influence of prolonged culture growth on the fatty acid composition
As the differences observed in fatty acid patterns might have been the result of differences in culture and/or metabolism of the fungi at the time of harvest, we examined fatty acids isolated from cells of six selected fungal strains (Downs, UCLA 531S, G217B, G222B, G186AS, RV26821S) after 3, 4 and 5 days consecutive growth in HMM (data not shown). These time points roughly correspond to early exponential, exponential and stationary growth phases of the G217B strain. The variability in fatty acid patterns was mainly observed in young cultures, whereas cells in the exponential and stationary growth phases had highly comparable profiles (data not shown). The high stability in fatty acid patterns is probably due to an appropriately chosen culture medium. As HMM is a rich medium, fungi grown in this medium do not quickly face nutrient exhaustion. Based on these observations, we assume that the recommended incubation time for fatty acid profiling should be standardized to a minimum of 4 days of culture growth.
Discussion
Fatty acid profiling has been widely used in the identification of micro-organisms. Taxa are distinguishable by the fatty acids produced and their relative ratios. In comparison with bacteria, the spectrum of fatty acids produced by fungi is more limited, e.g. yeasts mainly produce fatty acids with 16 and 18 carbon atoms. For that reason, fungal fatty acids have been considered to have very little value in taxonomic studies of this group of organisms. Regardless of such simplicity in fatty acid patterns, this method has gained recognition as being suitable for the reliable identification of clinically and industrially important species. Indeed, many reports have shown that fatty acids can be successfully used to identify and delineate yeasts and yeast-like fungi at different taxonomical levels (Westhuizen et al., 1987; Brondz et al., 1989; Stahl & Klug, 1996; Kellogg et al., 1998; El Menyawi et al., 2000; Peltroche-Llacsahuanga et al., 2000). Many of these studies employed the automated Microbial Identification System (MIS; MIDI, Inc.), which is equipped with a database for the identification of certain micro-organisms. However, this system also has many limitations. A major drawback still is its inability to discriminate between various fungal species. Another important disadvantage of MIS is the fact that many clinically relevant species, such as dimorphic fungal pathogens, are not included in the databases.
This study was aimed at improving the identification and differentiation of the yeast phase of H. capsulatum var. capsulatum by the establishment of an appropriate fatty acid profile database. In this study, we have analysed the fatty acid composition of two clinical isolates and seven laboratory stock strains that included different colony and pigment types of H. capsulatum. One clinical and one laboratory stock strain of H. capsulatum var. duboisii, and one strain of each of three other dimorphic zoopathogenic fungal species, B. dermatitidis, P. brasiliensis and S. schenckii, were included in this work as the outgroup. The most frequently occurring fatty acids were oleic, palmitic, stearic and linoleic acids. There were certain variations in the relative percentage fatty acid content of H. capsulatum that could be successfully applied to discriminate this fungus at the intraspecies level. The differences in fatty acid patterns among the strains demonstrated here are in good agreement with other phenotypic and/or molecular studies. This indicates that fatty acid profiling is a reliable tool for the identification/differentiation of H. capsulatum. It is also noteworthy that to the best of our knowledge, this is the first comparative study of fatty acid profiles obtained from various H. capsulatum strains that is taxonomically relevant.
Our qualitative data are basically in agreement with those of Domer & Hamilton (1971), who demonstrated the presence of the same major fatty acid homologues in H. capsulatum. However, our data differ quite drastically when the relative percentage of fatty acids is compared. These discrepancies might be related to different culture conditions as well as to different types of fatty acid isolation/preparation techniques. From a classification viewpoint, other investigations of H. capsulatum lipids provide limited data (Al-Doory, 1960; Nielsen, 1966; Domer & Hamilton, 1971).
We show a strong correlation between the fatty acid composition and the RFLP type of H. capsulatum var. capsulatum strains. In RFLP class III, the predominant fatty acids were palmitic (mean 36.6%) and linoleic (mean 34.3%) acids, followed by oleic acid (mean 24.1%). Strains of H. capsulatum RFLP class II contained higher amounts of linoleic acid (mean 38.3%), whereas palmitic and oleic acids were present in slightly lower concentrations (mean 34.3 and 22.8%, respectively). These two RFLP-related clusters are statistically different. This classification pattern, which is determined by the applied statistical algorithm, can be explained not only by differences in the abundance of the above-mentioned fatty acids, but also by their relative ratios, particularly between saturated and unsaturated fatty acids.
The greatest discrepancies were observed with the fatty acid patterns of H. capsulatum strains of RFLP class I. This class is highly heterogeneous and consists of strains that were obtained from miscellaneous and somewhat unusual patients. The Downs strain was isolated from an 80-year-old woman, who had disseminated disease with extensive joint and vaginal involvement (Gass & Kobayashi, 1969). UCLA 531R was isolated from a 13-year-old boy in a region of California, in which this fungus is not endemic (Stone et al., 1990; Eissenberg et al., 1991). RFLP class I isolates have been found to be less virulent than those of RFLP class II and III, and possess morphological and physiological traits substantially different from those of the other classes (Medoff et al., 1986; Eissenberg & Goldman, 1991). In the RFLP class I strains studied in this work, the level of palmitic acid was relatively constant; however, concentrations of both oleic and linoleic acids varied considerably in these strains. Such discrepancies were also observed in the contents of palmitoleic and stearic acids. For that reason, the RFLP class I strains were sequestered from other H. capsulatum clusters.
The pattern of fatty acids isolated from H. capsulatum var. duboisii was substantially different from that observed for H. capsulatum var. capsulatum. In this case, the major fatty acids (palmitic and linoleic) formed over 80% of the total fatty acid pool, whereas the content of oleic acid was significantly reduced to a mean of only 11.1%.
One characteristic of H. capsulatum strains other than those of RFLP class II is the generation of spontaneous avirulent variants (Eissenberg et al., 1991; Eissenberg & Goldman, 1991). When plated on solid medium, these smooth-colony type variants (S and E) can be easily distinguished from rough wild-type colonies (R). The major difference in this rough–smooth variation lies in the cell wall polysaccharides. From the data presented in Table 2, no differences in fatty acid patterns between these virulent (R) and avirulent (S and E) morphology type variants of H. capsulatum were detected. This observation concurs with an earlier hypothesis of Nielsen (1966), who suggested a lack of any relationship between extractable lipids and virulence.
There have also been relatively few published investigations of lipids and/or fatty acids of B. dermatitidis (DiSalvo & Denton, 1963; Domer & Hamilton, 1971), P. brasiliensis (Kanetsuna et al., 1969; San-Blas et al., 1977; Manocha, 1980) and S. schenckii (De Bieuvre & Mariat, 1975; Dart, 1976; Dart & Stretton, 1976; Stretton & Dart, 1976; Yamada, 1990). Published data show oleic acid as the predominant fatty acid and linoleic acid detected in negligible amounts or completely absent. In contrast, our study demonstrated the latter in substantially higher amounts in all examined species and strains. Linoleic acid contains two double bonds in its carbon side chain, and for that reason, this molecule is relatively unstable. However, linoleic acid (as well as other unsaturated compounds) can be protected from degradation by adding small amounts of antioxidants. In this study, we successfully isolated linoleic acid using BHT, while no application of any antioxidants by other investigators is mentioned.
The ratios of major fatty acids in the three other dimorphic fungi involved in this study, B. dermatitidis, P. brasiliensis and S. schenckii, were substantially different from those found in both variants of H. capsulatum. Cells of B. dermatitidis contained elevated amounts of linoleic acid (over 65%), and this increase was compensated by decreases in the content of palmitic and oleic acids. In P. brasiliensis and S. schenckii, the level of stearic acid was elevated (∼7.4 and 5.6%, respectively). Additionally, the content of myristic acid in S. schenckii cells was also slightly higher (1.5%), whereas in other strains this compound was present in constant but very small amounts, and its concentration did not exceed 0.6%.
We conclude that the high discriminative power of fatty acid profiling allows the differentiation of the yeast phase of H. capsulatum strains at the intraspecies level. Our preliminary results demonstrate fatty acid profiling to be a reliable method that could help to discriminate between H. capsulatum var. capsulatum and other variants within this species (such as H. capsulatum var. duboisii), or even between other closely related dimorphic fungal species. However, in this study, we examined only one strain each of B. dermatitidis, P. brasiliensis and S. schenckii, and more work is needed to test additional strains of these fungal species. To recapitulate, the proposed method is a valuable alternative tool that will be useful for further studies of H. capsulatum population biology and epidemiology, and possibly for other issues in fungal diagnostics and therapeutics.
Acknowledgments
We thank Drs David Andes (University of Wisconsin), George Deepe, Jr (University of Cincinnati), Carol Spiegel (University of Wisconsin) and Robert Striker (University of Wisconsin) for providing clinical isolates, and Dr Megan Bohse for critical reading of the manuscript. This work was supported by grants NIH R01s AI52303 and HL55949 (to J. P. W.), NIH R37 AI42747 (to George Deepe, Jr) and NIH R01 DK62388 (to J. M. N.).
Abbreviations
- BHT
butylated hydroxytoluene
- FAME
fatty acid methyl ester
- PC
principal component
- PCA
principal component analysis
References
- Al-Doory Y. Free lipids and phospholipid phosphorus of Histoplasma capsulatum and other pathogenic fungi. J Bacteriol. 1960;80:565–566. doi: 10.1128/jb.80.4.565-566.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustyn OPH, Kock JFL, Ferriera D. Differentiation between yeast species and strains within a species by cellular fatty acid analysis. 3. Saccharomyces sensu lato, Arxiozyma and Pachytichospora. Syst Appl Microbiol. 1990;13:44–55. [Google Scholar]
- Berliner MD. Histoplasma capsulatum: vital staining for the differentiation of the albino and brown phenotypes in vitro. Sabouraudia. 1973;11:271–273. doi: 10.1080/00362177385190551. [DOI] [PubMed] [Google Scholar]
- Body BA, Spicer A, Burgwyn CM. Immuno-identification of Histoplasma capsulatum and Blastomyces dermatitidis with commercial exoantigen reagents. Effect of culture age. Arch Pathol Lab Med. 1988;112:519–522. [PubMed] [Google Scholar]
- Bracca A, Tosello ME, Girardini JE, Amigot SL, Gomez C, Serra E. Molecular detection of Histoplasma capsulatum var. capsulatum in human clinical samples. J Clin Microbiol. 2003;41:1753–1755. doi: 10.1128/JCM.41.4.1753-1755.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt ME, Gaunt D, Iqbal N, McClinton S, Hambleton S, Sigler L. False-positive Histoplasma capsulatum Gen-Probe chemiluminescent test result caused by a Chrysosporium species. J Clin Microbiol. 2005;43:1456–1458. doi: 10.1128/JCM.43.3.1456-1458.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondz I, Olsen I, Sjostrom M. Gas chromatographic assessment of alcoholized fatty acids from yeasts: a new chemotaxonomic method. J Clin Microbiol. 1989;27:2815–2819. doi: 10.1128/jcm.27.12.2815-2819.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canteros CE, Zuiani MF, Ritacco VD, Perrotta E, Reyes-Montes MR, Granados J, Zuniga G, Taylor ML, Davel G. Electrophoresis karyotype and chromosome-length polymorphism of Histoplasma capsulatum clinical isolates from Latin America. FEMS Immunol Med Microbiol. 2005;45:423–428. doi: 10.1016/j.femsim.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Carter DA, Burt A, Taylor JW, Koenig GL, White TJ. Clinical isolates of Histoplasma capsulatum from Indianapolis, Indiana, have a recombining population structure. J Clin Microbiol. 1996;34:2577–2584. doi: 10.1128/jcm.34.10.2577-2584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter DA, Burt A, Taylor JW, Koenig GL, Dechairo BM, White TJ. A set of electrophoretic molecular markers for strain typing and population genetic studies of Histoplasma capsulatum. Electrophoresis. 1997;18:1047–1053. doi: 10.1002/elps.1150180703. [DOI] [PubMed] [Google Scholar]
- Chemaly RF, Tomford JW, Hall GS, Sholtis M, Chua JD, Procop GW. Rapid diagnosis of Histoplasma capsulatum endocarditis using the AccuProbe on an excised valve. J Clin Microbiol. 2001;39:2640–2641. doi: 10.1128/JCM.39.7.2640-2641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dart RK. Effect of temperature on the fatty-acid composition of Sporotrichum thermophile. Trans Br Mycol Soc. 1976;66:532–533. [Google Scholar]
- Dart RK, Stretton RJ. Fatty acid composition of Sporotrichum species. Trans Br Mycol Soc. 1976;66:529–532. [Google Scholar]
- De Bieuvre C, Mariat F. Composition en acides gras des lipides polaires et neutres de Sporothrix schenckii et de. Ceratocystis stenoceras Sabouraudia. 1975;13:226–230. in French. [PubMed] [Google Scholar]
- Denys GA, Newman MA, Standard PG. Evaluation of a commercial exoantigen test system for the rapid identification of systemic fungal pathogens. Am J Clin Pathol. 1983;79:379–381. doi: 10.1093/ajcp/79.3.379. [DOI] [PubMed] [Google Scholar]
- DiSalvo AF, Denton JF. Lipid content of four strains of Blastomyces dermatitidis of different mouse virulence. J Bacteriol. 1963;85:927–931. doi: 10.1128/jb.85.4.927-931.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiSalvo AF, Sekhon AS, Land GA, Fleming WH. Evaluation of the exoantigen test for identification of Histoplasma species and Coccidioides immitis cultures. J Clin Microbiol. 1980;11:238–241. doi: 10.1128/jcm.11.3.238-241.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiSalvo AF, Terreni AA, Wooten AK. Use of the exoantigen test to identify Blastomyces dermatitidis, Coccidioides immitis, and Histoplasma capsulatum in mixed cultures. Am J Clin Pathol. 1981;75:825–826. doi: 10.1093/ajcp/75.6.825. [DOI] [PubMed] [Google Scholar]
- Domer JE, Hamilton JG. The readily extracted lipids of Histoplasma capsulatum and Blastomyces dermatitidis. Biochim Biophys Acta. 1971;231:465–478. doi: 10.1016/0005-2760(71)90114-7. [DOI] [PubMed] [Google Scholar]
- Eissenberg LG, Goldman WE. Histoplasma variation and adaptive strategies for parasitism: new perspectives on histoplasmosis. Clin Microbiol Rev. 1991;4:411–421. doi: 10.1128/cmr.4.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg LG, West JL, Woods JP, Goldman WE. Infection of P388D1 macrophages and respiratory epithelial cells by Histoplasma capsulatum: selection of avirulent variants and their potential role in persistent histoplasmosis. Infect Immun. 1991;59:1639–1646. doi: 10.1128/iai.59.5.1639-1646.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Menyawi I, Wogerbauer M, Sigmund H, Burgmann H, Graninger W. Identification of yeast species by fatty acid profiling as measured by gas-liquid chromatography. J Chromatogr B Biomed Sci Appl. 2000;742:13–24. doi: 10.1016/s0378-4347(00)00044-x. [DOI] [PubMed] [Google Scholar]
- Gass M, Kobayashi GS. Histoplasmosis. An illustrative case with unusual vaginal and joint involvement. Arch Dermatol. 1969;100:724–727. doi: 10.1001/archderm.100.6.724. [DOI] [PubMed] [Google Scholar]
- Guedes HL, Guimaraes AJ, Muniz Mde M, Pizzini CV, Hamilton AJ, Peralta JM, Deepe GS, Jr, Zancope-Oliveira RM. PCR assay for identification of Histoplasma capsulatum based on the nucleotide sequence of the M antigen. J Clin Microbiol. 2003;41:535–539. doi: 10.1128/JCM.41.2.535-539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekaran M, Hugh WT. Gas-liquid chromatography: a rapid method for identification of different species of Candida. Mycologia. 1980;72:505–511. [PubMed] [Google Scholar]
- Hall GS, Pratt-Rippin K, Washington JA. Evaluation of a chemiluminescent probe assay for identification of Histoplasma capsulatum isolates. J Clin Microbiol. 1992;30:3003–3004. doi: 10.1128/jcm.30.11.3003-3004.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton A, Jr, Cason JA, Ingram KD. Enumeration and identification of yeasts associated with commercial poultry processing and spoilage of refrigerated broiler carcasses. J Food Prot. 2002;65:993–998. doi: 10.4315/0362-028x-65.6.993. [DOI] [PubMed] [Google Scholar]
- Hotelling H. Analysis of a complex of statistical variables into principal components. J Educ Psychol. 1933;24:417–441. [Google Scholar]
- Huffnagle KE, Gander RM. Evaluation of Gen-Probe's Histoplasma capsulatum and Cryptococcus neoformans AccuProbes. J Clin Microbiol. 1993;31:419–421. doi: 10.1128/jcm.31.2.419-421.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeennor S, Laoteng K, Tanticharoen M, Cheevadhanarak S. Comparative fatty acid profiling of Mucor rouxii under different stress conditions. FEMS Microbiol Lett. 2006;259:60–66. doi: 10.1111/j.1574-6968.2006.00242.x. [DOI] [PubMed] [Google Scholar]
- Jiang B, Bartlett MS, Allen SD, Smith JW, Wheat LJ, Connolly PA, Lee CH. Typing of Histoplasma capsulatum isolates based on nucleotide sequence variation in the internal transcribed spacer regions of rRNA genes. J Clin Microbiol. 2000;38:241–245. doi: 10.1128/jcm.38.1.241-245.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanetsuna F, Carbonell LM, Moreno RE, Rodriguez J. Cell wall composition of the yeast and mycelial forms of Para-coccidioides brasiliensis. J Bacteriol. 1969;97:1036–1041. doi: 10.1128/jb.97.3.1036-1041.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga T, Taylor JW, White TJ. Phylogenetic relationships of varieties and geographical groups of the human pathogenic fungus Histoplasma capsulatum Darling. J Clin Microbiol. 1999;37:653–663. doi: 10.1128/jcm.37.3.653-663.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keath EJ, Kobayashi GS, Medoff G. Typing of Histoplasma capsulatum by restriction fragment length polymorphisms in a nuclear gene. J Clin Microbiol. 1992;30:2104–2107. doi: 10.1128/jcm.30.8.2104-2107.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg JA, Bankert DA, Chaturvedi V. Limitations of the current microbial identification system for identification of clinical yeast isolates. J Clin Microbiol. 1998;36:1197–1200. doi: 10.1128/jcm.36.5.1197-1200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersulyte D, Woods JP, Keath EJ, Goldman WE, Berg DE. Diversity among clinical isolates of Histoplasma capsulatum detected by polymerase chain reaction with arbitrary primers. J Bacteriol. 1992;174:7075–7079. doi: 10.1128/jb.174.22.7075-7079.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacaz CD, Del Negro GM, Vidal MS, Heins-Vaccari EM, Santos RF, Martins MA, Ozaki MM, Romiti R, Proenca R, Castro LG. Atypical disseminated cutaneous histoplasmosis in an immunocompetent child, caused by an ‘aberrant’ variant of Histoplasma capsulatum var. capsulatum. Rev Inst Med Trop Sao Paulo. 1999;41:195–202. doi: 10.1590/s0036-46651999000300012. [DOI] [PubMed] [Google Scholar]
- Manocha MS. Lipid compostion of Paracoccidioides brasiliensis: comparison between the yeast and mycelial forms. Sabouraudia. 1980;18:281–286. doi: 10.1080/00362178085380481. [DOI] [PubMed] [Google Scholar]
- Medoff G, Maresca B, Lambowitz AM, Kobayashi G, Painter A, Sacco M, Carratu L. Correlation between pathogenicity and temperature sensitivity in different strains of Histoplasma capsulatum. J Clin Invest. 1986;78:1638–1647. doi: 10.1172/JCI112757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz MM, Pizzini CV, Peralta JM, Reiss E, Zancope-Oliveira RM. Genetic diversity of Histoplasma capsulatum strains isolated from soil, animals, and clinical specimens in Rio de Janeiro State, Brazil, by a PCR-based random amplified polymorphic DNA assay. J Clin Microbiol. 2001;39:4487–4494. doi: 10.1128/JCM.39.12.4487-4494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen HS., Jr Variation in lipid content of strains of Histoplasma capsulatum exhibiting different virulence properties for mice. J Bacteriol. 1966;91:273–277. doi: 10.1128/jb.91.1.273-277.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhye AA, Smith G, McLaughlin D, Standard PG, Kaufman L. Comparative evaluation of a chemiluminescent DNA probe and an exoantigen test for rapid identification of Histoplasma capsulatum. J Clin Microbiol. 1992;30:3108–3111. doi: 10.1128/jcm.30.12.3108-3111.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltroche-Llacsahuanga H, Schmidt S, Lutticken R, Haase G. Discriminative power of fatty acid methyl ester (FAME) analysis using the microbial identification system (MIS) for Candida (Torulopsis) glabrata and Saccharomyces cerevisiae. Diagn Microbiol Infect Dis. 2000;38:213–221. doi: 10.1016/s0732-8893(00)00205-4. [DOI] [PubMed] [Google Scholar]
- Poonwan N, Imai T, Mekha N, Yazawa K, Mikami Y, Ando A, Nagata Y. Genetic analysis of Histoplasma capsulatum strains isolated from clinical specimens in Thailand by a PCR-based random amplified polymorphic DNA method. J Clin Microbiol. 1998;36:3073–3076. doi: 10.1128/jcm.36.10.3073-3076.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pounder JI, Hansen D, Woods GL. Identification of Histoplasma capsulatum, Blastomyces dermatitidis, and Coccidioides species by repetitive-sequence-based PCR. J Clin Microbiol. 2006;44:2977–2982. doi: 10.1128/JCM.00687-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Montes MR, Bobadilla-Del Valle M, Martinez-Rivera MA, Rodriguez-Arellanes G, Maravilla E, Sifuentes-Osornio J, Taylor ML. Relatedness analyses of Histoplasma capsulatum isolates from Mexican patients with AIDS-associated histoplasmosis by using histoplasmin electrophoretic profiles and randomly amplified polymorphic DNA patterns. J Clin Microbiol. 1999;37:1404–1408. doi: 10.1128/jcm.37.5.1404-1408.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippon JW. Dimorphism in pathogenic fungi. Crit Rev Microbiol. 1980;8:49–97. doi: 10.3109/10408418009085078. [DOI] [PubMed] [Google Scholar]
- San-Blas G, San-Blas F, Ormaechea E, Serrano LE. Cell wall analysis of an adenine-requiring mutant of the yeast-like form of Paracoccidioides brasiliensis strain IVIC Pb9. Sabouraudia. 1977;15:297–303. doi: 10.1080/00362177785380121. [DOI] [PubMed] [Google Scholar]
- Sandin RL, Isada CM, Hall GS, Tomford JW, Rutherford I, Rogers AL, Washington JA. Aberrant Histoplasma capsulatum. Confirmation of identity by a chemiluminescence-labeled DNA probe. Diagn Microbiol Infect Dis. 1993;17:235–238. doi: 10.1016/0732-8893(93)90103-e. [DOI] [PubMed] [Google Scholar]
- Sekhon AS, DiSalvo AF, Standard PG, Kaufman L, Terreni AA, Garg AK. Evaluation of commercial reagents to identify the exoantigens of Blastomyces dermatitidis, Coccidioides immitis, and Histoplasma species cultures. Am J Clin Pathol. 1984;82:206–209. doi: 10.1093/ajcp/82.2.206. [DOI] [PubMed] [Google Scholar]
- Sekhon AS, Standard PG, Kaufman L, Garg AK. Reliability of exoantigens for differentiating Blastomyces dermatitidis and Histoplasma capsulatum from Chrysosporium and Geomyces species. Diagn Microbiol Infect Dis. 1986;4:215–221. doi: 10.1016/0732-8893(86)90100-8. [DOI] [PubMed] [Google Scholar]
- Soll DR. The ins and outs of DNA fingerprinting the infectious fungi. Clin Microbiol Rev. 2000;13:332–370. doi: 10.1128/cmr.13.2.332-370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer ED, Lasker BA, Travis SJ, Kobayashi GS, Medoff G. Use of mitochondrial and ribosomal DNA polymorphisms to classify clinical and soil isolates of Histoplasma capsulatum. Infect Immun. 1989;57:1409–1412. doi: 10.1128/iai.57.5.1409-1412.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl PD, Klug MJ. Characterization and differentiation of filamentous fungi based on fatty acid composition. Appl Environ Microbiol. 1996;62:4136–4146. doi: 10.1128/aem.62.11.4136-4146.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standard PG, Kaufman L. Safety considerations in handling exoantigen extracts from pathogenic fungi. J Clin Microbiol. 1982;15:663–667. doi: 10.1128/jcm.15.4.663-667.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone MM, Frenkel LM, Howard DH. Histoplasmosis after multiple trauma. Pediatr Infect Dis J. 1990;9:747–749. [PubMed] [Google Scholar]
- Stretton RJ, Dart RK. Long-chain fatty acids of Sporothrix (Sporotrichum) schenckii. J Clin Microbiol. 1976;3:635–636. doi: 10.1128/jcm.3.6.635-636.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton DA, Padhye AA, Standard PG, Rinaldi MG. An aberrant variant of Histoplasma capsulatum var. capsulatum. J Clin Microbiol. 1997;35:734–735. doi: 10.1128/jcm.35.3.734-735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor ML, Chavez-Tapia CB, Reyes-Montes MR. Molecular typing of Histoplasma capsulatum isolated from infected bats, captured in Mexico. Fungal Genet Biol. 2000;30:207–212. doi: 10.1006/fgbi.2000.1219. [DOI] [PubMed] [Google Scholar]
- Taylor ML, Chavez-Tapia CB, Rojas-Martinez A, del Rocio Reyes-Montes M, del Valle MB, Zuniga G. Geographical distribution of genetic polymorphism of the pathogen Histoplasma capsulatum isolated from infected bats, captured in a central zone of Mexico. FEMS Immunol Med Microbiol. 2005;45:451–458. doi: 10.1016/j.femsim.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Ueda Y, Sano A, Tamura M, Inomata T, Kamei K, Yokoyama K, Kishi F, Ito J, Mikami Y, et al. Diagnosis of histoplasmosis by detection of the internal transcribed spacer region of fungal rRNA gene from a paraffin-embedded skin sample from a dog in Japan. Vet Microbiol. 2003;94:219–224. doi: 10.1016/s0378-1135(03)00104-4. [DOI] [PubMed] [Google Scholar]
- Vincent RD, Goewert R, Goldman WE, Kobayashi GS, Lambowitz AM, Medoff G. Classification of Histoplasma capsulatum isolates by restriction fragment polymorphisms. J Bacteriol. 1986;165:813–818. doi: 10.1128/jb.165.3.813-818.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Westhuizen JPJ, Kock JFL, Smit EJ, Lategan PM. The value of long-chain fatty acid composition in the identification of species representing the basidomycetous genus Rhodosporidium Banno. Syst Appl Microbiol. 1987;10:31–34. [Google Scholar]
- Wheat LJ. Current diagnosis of histoplasmosis. Trends Microbiol. 2003;11:488–494. doi: 10.1016/j.tim.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Wheat LJ, Connolly-Stringfield P, Kohler RB, Frame PT, Gupta MR. Histoplasma capsulatum polysaccharide antigen detection in diagnosis and management of disseminated histoplasmosis in patients with acquired immunodeficiency syndrome. Am J Med. 1989;87:396–400. doi: 10.1016/s0002-9343(89)80820-4. [DOI] [PubMed] [Google Scholar]
- Wheat LJ, Connolly-Stringfield P, Blair R, Connolly K, Garringer T, Katz BP. Histoplasmosis relapse in patients with AIDS: detection using Histoplasma capsulatum variety capsulatum antigen levels. Ann Intern Med. 1991;115:936–941. doi: 10.7326/0003-4819-115-12-936. [DOI] [PubMed] [Google Scholar]
- Wheat LJ, Connolly-Stringfield P, Williams B, Connolly K, Blair R, Bartlett M, Durkin M. Diagnosis of histoplasmosis in patients with the acquired immunodeficiency syndrome by detection of Histoplasma capsulatum polysaccharide antigen in bronchoalveolar lavage fluid. Am Rev Respir Dis. 1992;145:1421–1424. doi: 10.1164/ajrccm/145.6.1421. [DOI] [PubMed] [Google Scholar]
- Woods JP, Heinecke EL, Goldman WE. Electrotransformation and expression of bacterial genes encoding hygromycin phosphotransferase and β-galactosidase in the pathogenic fungus Histoplasma capsulatum. Infect Immun. 1998;66:1697–1707. doi: 10.1128/iai.66.4.1697-1707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K. Unusual fatty acids in Sporothrix schenckii. J Dermatol. 1990;17:647–649. doi: 10.1111/j.1346-8138.1990.tb01710.x. [DOI] [PubMed] [Google Scholar]
- Zancope-Oliveira RM, de Silva Tavares PM, de Medeiros Muniz M. Genetic diversity of Histoplasma capsulatum strains in Brazil. FEMS Immunol Med Microbiol. 2005;45:443–449. doi: 10.1016/j.femsim.2005.05.018. [DOI] [PubMed] [Google Scholar]
- Zarnowski R, Suzuki Y, Pietr SJ. Alkyl- and alkenylresorcinols of wheat grains and their chemotaxonomic significance. Z Naturforsch [C] 2004;59c:190–196. doi: 10.1515/znc-2004-3-411. [DOI] [PubMed] [Google Scholar]


