Abstract
L-Leucine aminopeptidases (LAPs) are implicated in the progress of many pathological disorders, and play some regulatory roles in tumor cell proliferation, invasion and/or angiogenesis. Thus, LAPs could become not only new diagnostic or prognostic biomarkers, but also have potential as novel molecular targets for the treatment of several cancers. Highly sensitive assays are critical for early detection of changes in LAP activity and for screening potent LAP inhibitors. In this study, we developed a novel and highly sensitive fluorescent assay for LAPs based on substituted aminopyridines as fluorescent reporters. This assay was at least 100 and 20 fold more sensitive than commercial colorimetric or fluorescent LAP substrates, respectively. We also showed that this assay was a useful tool to monitor LAP activities in extracts from cancer cell lines, as well as for the high throughput screening of inhibitors, which could lead to new cancer treatments.
Keywords: metallopeptidases, N-terminal hydrolysis, fluorescent assay, HTS assay
Introduction
L-Leucine aminopeptidases (LAPs; EC 3.4.11.1) are exopeptidases that catalyze the hydrolysis of leucine residues from the amino-termini of protein or peptide substrates [1]. LAPs are zinc containing enzymes, of the M1 and M17 peptidase families. They play diverse biological and physiological roles in mammals, microbes and plants by either degrading bioactive peptides, or interacting with peptide-dependent signaling and DNA [2][3]. Thus, changes in LAP expression pattern and/or catalytic function result in altered peptide activation leading to changes in tumor cell proliferation, invasion and/or angiogenesis. For example, puromycin-insensitive leucyl-specific aminopeptidase (PI-LAP) plays a crucial role in the cell cycle progression of endothelial cells and angiogenesis via the binding and modification of phosphatidylinositol-dependent kinase [4]. Placental LAP (P-LAP) is the biomarker for the evaluation of ovarian epithelial malignancy and a target of molecular therapy [3]. P-LAP is also involved in the promotion and progression of breast cancer through oxytocin and/or vasopressin mis-regulation [5], and plays a significant role in insulin regulation of Glut4 receptors in diabetic patients [6][7]. In addition, adipocyte-derived LAP (A-LAP) mediates endometrial cancer cell growth and differentiation, therefore, the assessment of A-LAP status provides clinically useful prognostic information in patients with endometrial carcinoma [8]. Thus, LAPs may become not only new diagnostic or prognostic biomarkers, but also have potential as novel molecular targets for the treatment of several cancers. Selective chemical inhibitors of LAPs may be useful probes to determine LAP functions in addition to being possible cancer therapeutics.
In order to develop a potent enzyme inhibitor, it is important to have reliable, sensitive, and specific assays that can be used for both measuring enzyme activity and inhibitor potency. Several assays have been developed for LAPs. Briefly, these assays are based on substrates containing an amide functionality formed with (L)-leucine as the acid and either a common colorimetric or fluorescent reporter, such as β-naphthylamine, p-nitroaniline or 7-amino-4-methylcoumarin, as the amine [9]-[11]. While widely used, these assays are not very sensitive for LAPs, with limits of detection (L.O.D.) > 5 μg/mL for the colorimetric assay [11] and > 1 μg/mL for the fluorescent assay [9]. Furthermore, the background chemical hydrolysis of these assays is relatively high. Taken together, the currently available assays are less than suitable for high throughput screening.
Recently, we have discovered a series of novel fluorescent reporters (i.e., amino-pyridine derivatives) [12]. These are small molecules with high quantum yields and large Stokes’ shift. These properties potentially make these compounds excellent reporters for aminopeptidases such as LAPs. However, these reporters have near-UV excitation (280-300 nm) and emission (380-400 nm) wavelengths. These properties suggest that their fluorescence could be quenched by high concentrations of compounds absorbing in the UV-range such as those with aromatic rings and nitrogen heterocycles. Also, in some combinatorial libraries, numerous false positives could be generated since some heterocyclic compounds such as 2-aminoquinoline, 4-aminoquinaldine [13] and 2-[4-(dimethylamino)phenyl] benzothiazole [14] can be fluorescent. Once potent leads are found, these properties are unlikely to cause serious problems. However, initial library screens will be problematic. Therefore, in this study, our aim was to obtain fluorescent reporters with both the excitation and emission wavelengths red-shifted. We used these reporters for developing novel assays for aminopeptidases. Finally, we investigated the use of such assays for inhibitor screening, and measuring LAP activities from cancer cell extracts as a potential diagnostic tool.
Materials and methods
Chemicals
Porcine kidney microsomal leucine aminopeptidase, L-leucine-p-nitroanilide (I, Fig. 1), 7-amino-4-methylcoumarin, and N-(tert-butoxycarbonyl)-L-leucine monohydrate were purchased from Sigma-Aldrich (Saint Louis, MI, USA). L-Leucine-(4-methyl-7-coumarinyl amide (II, Fig. 1) was purchased from Acros Organics (New Jersey, USA). 3-Amino-6-methoxy-2-picoline, 5-amino-2-methoxypyridine, and 2-bromo-5-nitropyridine were purchased from Asychem (Durham, NC, USA). Pooled human liver S9 was purchased from BD Biosciences (Woburn, MA, USA).
Figure 1.
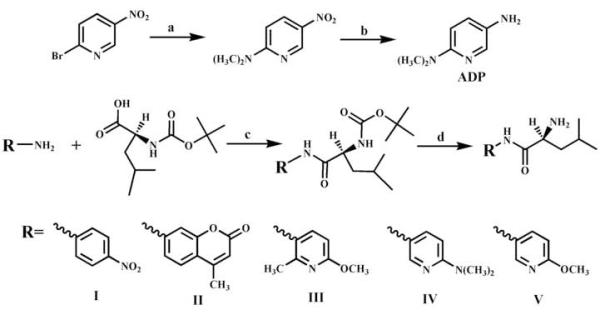
Schematic preparation of a novel aminopyridine-based fluorescent reporter (ADP) and LAP substrates. Reagents and conditions: (a) DMSO/150-160 °C, overnight, 60%; (b) Pt/H2 in CH3OH, 4 h, 90%; (c) (N-(tert-butoxycarbonyl)-L-leucine monohydrate, DBU, 1,3-diisopropylcarbodiimide/CH2Cl2, 2h, 96%; (d) TFA, 2 h, 55%.
Synthesis
The fluorescent reporter (5-amino-2-dimethylaminopyridine, ADP) was prepared as described in Fig. 1 steps a and b. This is similar to the procedure described by Heindel et al. [15]. The LAP substrates III, IV, and V were prepared as described in Fig. 1 steps c and d. Commercially available and optically pure L-leucine derivatives were used as starting material, and the reaction performed did not alter the chirality of the products, thus, substrates III, IV, and V are optically pure (S)-enantiomers. Structural identification of synthesized compounds was based on data from proton nuclear magnetic resonance (1H-NMR), which were acquired from a Mercury 300 spectrometer (Varian, Palo Alto, CA), and by mass spectrometry. Mass spectra were obtained on a Micromass liquid chromatograph orthogonal acceleration time-of-flight mass spectrometer (Waters-Micromass, Manchester, UK) by using electrospray ionization in the positive mode, and elution was performed isocratically with a solvent mixture of 89.9:10.0:0.1 acetonitrile: water:formic acid. The chemical purity of the final products was supported by the spectra described above, a single spot on TLC under 254 nm wavelength, and lack of fluorescence on TLC at 254 or 360 nm wavelengths. The NMR and mass spectral data suggested purities above 97% for all synthetic compounds, and contamination by the fluorescent reporter to be less than 0.01% for substrates III to V.
N-(6-Methoxy-2-methylpyridin-3-yl), (S)-2-amino-4-methylpentanamide III
Reddish oil (overall 66% yield). 1H NMR (CDCl3): 9.38 (br, 1H, NH), 8.12 (d, J =9.00, 1H, pyridine), 6.58 (d, J =9.00, 1H pyridine), 3.89 (s, 3H, OCH3), 3.56 (t, J =10.5, 1H, CHNH2), 2.41 (s, 3H, CH3), 1.75-1.86 (3H, CH2CH), 0.99 (d, J =6.30, 3H, CH3), 0.97 (d, J =6.30, 3H. CH3). Calculated mass: 251.16, found [M+H]+ = 252.16.
5-Amino-2-dimethylaminopyridine ADP
Brownish oil (overall 55% yield). 1H NMR (CDCl3): δ 7.77 (d, J=3.00, 1H, pyridine), 6.99 (dd, J1=3.00, J2=9.00, 1H, pyridine), 6.46 (d, J=9.00, 1H, pyridine), 3.74 (br, 2H, NH2), 3.00 (s, 6H, 2CH3). Calculated mass: 137.10, found [M+H]+ = 138.12.
(S)-2-Amino-N-(6-(dimethylamino)pyridin-3-yl)-4-methylpentanamide IV
Yellowish oil (overall 42% yield). 1H NMR (CDCl3): δ 9.26 (br, 1H, NH), 8.16 (d, J=2.70, 1H, pyridine), 7.89 (d, J=9.00, 1H, pyridine), 6.49 (d, J=2.70, 1H, pyridine), 3.53 (m, 1H, CH), 3.04(s, 6H, 2CH3), 2.15 (br, 2H, NH2), 1.65-1.83 (m, 3H, CHCH2), 0.98 (dd, J1=3.90, J2=2.40, 6H, 2CH3). Calculated mass: 250.18, found [M+H]+ = 251.18.
N-(6-Methoxypyridin-3-yl), (S)-2-amino-4-methylpentanamide V
Reddish oil (overall 51% yield). 1H NMR (CDCl3): δ 9.47 (br, 1H, NH), 8.23 (d, J=2.40, 1H, pyridine), 8.01 (dd, J1=2.70, J2=8.70, 1H pyridine), 6.72(d, J=8.70, 1H pyridine), 3.91 (s, 3H, OCH3), 3.52 (t, J=10.5, 1H, CHNH2), 1.75-1.86 (3H, CH2CH), 0.99 (d, J=6.30, 3H, CH3), 0.97 (d, J=6.30, 3H. CH3). Calculated mass: 237.15, found [M+H]+ = 238.15.
Optical properties determination
Optical properties (molar extinction coefficient, excitation and emission wavelengths, and quantum yield) of 5-amino-2-dimethylaminopyridine (ADP) were determined using the procedures described by Huang et al. [12]. The molar extinction coefficient was determined at 290 nm for final concentrations of ADP between 10 and 50 μM. To ensure that the substrates containing an aminopyridine group as a reporter (III, IV & V) do not fluoresce significantly and do not interfere with the reporter fluorescent signal, their emission spectrum in 0.1 M pH 8.0 phosphate buffer with [S]final =50 μM was measured from 360 to 600 nm with the same excitation wavelength as their corresponding reporters (see Table 1).
Table 1.
Measurement parameters for the colorimetric and fluorescent substrates.
| Substrate | Reporter structure | λex. (nm) |
λem. (nm) |
|---|---|---|---|
| I | 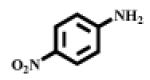 |
382* | - |
| II | 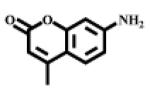 |
351 | 430 |
| III | 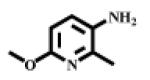 |
304 | 392 |
| IV | 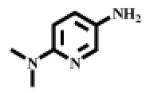 |
334 | 450 |
| V | 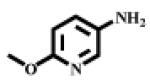 |
302 | 396 |
Absorbance wavelength.
Optimization of assay conditions
To optimize the pH of the assay buffer, the activity of porcine kidney microsomal LAP was measured in sodium phosphate buffer (0.1 M, pH 6.0 to 8.0) and Tris/HCl buffer (20 mM, pH 9.0). In a 96-well polystyrene flat-bottom microtiter plate, 10 μL of a diluted (0.2 μg/μL) LAP solution was added to 190 μL of buffer. The plate was then pre-incubated for 10 min at 27 °C. To each well was added 1 μL of a 10 mM solution of substrate I in ethanol ([S]final = 50 μM). Activity was immediately measured for 10 min at 27 °C by following the appearance of 4-nitroaniline at 382 nm every 30 seconds using a SpectraMax M2 spectrophotometer (Molecular Devices, Sunnyvale, CA). Assays in absence of protein were used as blank. Results were averaged and compared statistically. The optimal buffer was then used to examine the effect of temperature (25, 28, 31, 34, and 37 °C) on LAP activity with substrate I. Assays were performed in quadruplicate. The results are given as the average ± standard deviation.
Enzymatic assays
All assays were performed in 96-well polystyrene flat-bottom microtiter plates, clear for substrate I and black for substrates II-V. In each well, 10 μL of diluted protein solution (or buffer for the blank) was added to 190 μL of sodium phosphate buffer (0.1 M pH 8.0) to yield the appropriate protein concentration. The reaction was started by the addition of 1 μL of a 10 mM substrate solution in ethanol ([S]final = 50 μM). The reaction was followed for 10 min at 31 °C by measuring the appearance of the product every 30 seconds with a SpectraMax M2 spectrophotometer. The measurement parameters for the colorimetric I, and fluorescent II-V substrates are given in Table 1. Standard curves of the various reporters (from 0 to 5 nmol) were used to quantify the measured signal (r2 > 0.99 for all of them). Assays were performed in quadruplicates. The results are given as the average ± standard deviation of at least three separate measurements. The limit of detection (L.O.D.) was defined as the amount of enzyme needed to generate activity that was three times the value of the blank. The signal to background ratio (S/B) was calculated by dividing the reaction average velocity in presence of enzyme (Av.enz.) by the average background hydrolysis rate (Av.blank). The signal to noise ratio (S/N) was calculated by dividing the reaction velocity in presence of enzyme by the standard deviation of the background hydrolysis (SDblank). The Z’ factor was calculated following the method of Zhang et al. [16] using the formula: Z’ = (1 − {[3*(SDenz. + SDblank)]/(Av.enz. − Av.blank)}.
Kinetic constant determination
Kinetic constants (Vmax and Km) were determined for substrates I and V, following the methods described above for enzyme activity. For I, an enzyme concentration of 10 μg/mL and [S]final between 0.78 and 50 μM were used. For V, an enzyme concentration of 0.12 μg/mL and [S]final between 1.6 and 100 μM were used. The assays were performed in quadruplicate. Km and Vmax were calculated by non-linear fitting of the Michaelis-Menten equation using the enzyme kinetic module of SigmaPlot 9.01 (Systat Software Inc.). The results are given as the average ± standard deviation of at least three separate measurements.
Inhibition assay
Inhibition potencies of bestatin for pig liver LAP were determined for substrates I and V, following the assay methods described above. Briefly, one microliter of the inhibitor in a DMSO solution was added to the enzyme diluted into the buffer (200 μL total; [E]final = 10 and 0.12 μg/mL for substrates I and V, respectively), to yield final concentrations of inhibitor between 0.78 and 50 μM. This mixture was incubated for 10 min at 31 °C, prior to the addition of the substrates. IC50 is defined as the concentration of an inhibitor that inhibited 50% of the enzyme activity. IC50s were calculated by non linear regression of at least five datum points using SigmaPlot. The results are given as the average ± standard deviation of at least three separate measurements.
Cell cultures
The human cancer cell lines used (293T, HepG2, HuH7, 22RV1, Du145, T24) were obtained from the American Type Culture Collection (Manassas, VA). These cell lines were cultured in RPMI1640 medium (Mediatech) supplemented with 10% fetal bovine serum (v/v) (BioWhittaker) under a 5% CO2 atmosphere at 37 °C. The cells were passaged at 90-100% confluence by washing with PBS and trypsinization (0.05% trypsin, 0.53mM EDTA) for 3-5 min at 37°C prior to subculture. Following trypsinization, the cell suspension was centrifuged at 400 g for 5 min at 5°C. The cell pellet was then resuspended in fresh RPM1640 medium.
Protein extracts preparation from cell lines
Protein extracts were released from 106 cells by sonication (Sonic Dismembrator Model 100, Fisher Scientific) for 3 sec in 500 μl of cell lysis buffer (20 mM sodium phosphate, pH 7.4; 1 mM PTU; 1 mM DTT; and 0.01% Tween-20). Cell debris was removed by centrifugation (10,000 g for 15 min at 4°C) and the supernatant was stored at -80°C until used. For each cell line, LAP activity was measured as described above using 40 μl of extract, corresponding to 8 × 104 cells. Protein concentrations were quantified by using the Pierce BCA assay using Fraction V bovine serum albumin (BSA) as the calibrating standard. LAP activity was measured on the extract as described above.
Results and Discussion
Characterization of a novel red-shifted amino-pyridine fluorescent reporter
Recently, we reported amino-pyridine derivatives as novel fluorescent reporters [12]. While, these small molecules have high quantum yields and large Stokes’ shift, they have near-UV excitation (280-300 nm) and emission (380-400 nm) wavelengths, which limit their usefulness. It has been shown that increasing the electron density on the electron withdrawing group of a fluorophore, yields red-shifted excitation and emission wavelengths [17]-[21]. Thus, to obtain a red shifted fluorescent signal, we replaced the methoxy group of 5-amino-2-methoxypyridine (AMP), the best reporter previously found [12], with a stronger electronic donor group (e.g., dimethylamine). This new amino-pyridine (ADP on Fig. 1) was synthesized following the method of Heindel et al. [15], with an overall yield of 55%. As expected, compared to AMP, this replacement resulted in a substantial red-shift in both excitation (∼30 nm) and emission (∼60 nm) wavelengths for ADP (Fig. 2). However, under the same conditions, the relative fluorescent intensity of 5-amino-2-dimethylaminopyridine was lower than that of AMP (Fig. 2). This was probably due to a broader emission peak, and slightly smaller quantum yield (0.89) compared to 5-amino-2-methoxypyridine (0.95) [12]. Interestingly, the molar extinction coefficient at 290 nm of 5-amino-2-dimethylaminopyridine (ADP) was found to be 5,600 ± 100 M-1cm-1, which is approximately 2-fold higher than the one for AMP, and even higher than any other aminopyridines examined. Overall, the novel amino-pyridine (ADP) seems to be a better fluorescent reporter than the ones described previously [12].
Figure 2.
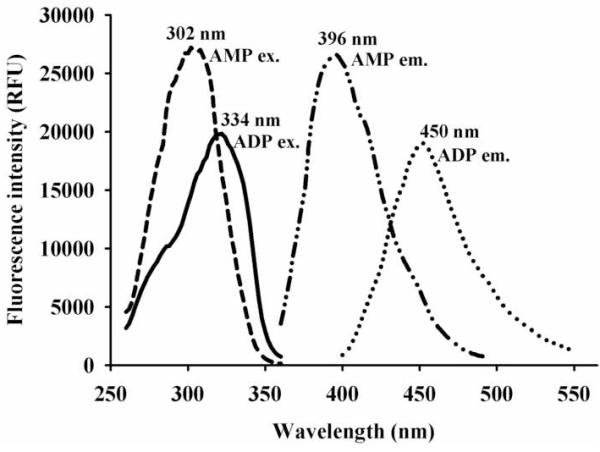
Comparison of excitation (ex.) and emission (em.) wavelengths of 5-amino-2-methoxypyridine (AMP) and 5-amino-2-dimethylaminopyridine (ADP).
Comparison and characterization of novel LAP substrates
The background hydrolysis in sodium phosphate buffer (0.1 M pH 8.0) of all of the LAP substrates tested in this study (compounds I to V Fig. 1) was determined at 31 °C. As shown in Table 2, dramatic differences in background hydrolysis were observed. Both commercial substrates (I and II) yielded the highest rates of background hydrolysis, with the absorbent substrate I being six times less stable than the fluorescent one II. For the amino-pyridine containing substrates, the presence of the dimethylamino group para to the amide function resulted in substrate IV that is roughly 10 fold less stable than the compounds (III and V) with a methoxy group at the same position. We did not observe striking differences in background hydrolysis rates over the range of pH (6.0 to 9.0) and temperature (25 to 37 °C) tested. There was a trend toward higher background hydrolysis rates with higher pH and temperature as one would expect. Overall, while a stronger electron donating group such as dimethylamino appeared to yield a red shifted fluorescent reporter, it also yielded a less stable amide. One could probably optimize the electron donor effect to balance the good optical properties and chemical stability.
Table 2.
The detection limit of porcine kidney leucine aminopeptidase (PKLAP) with colorimetric and fluorescent substrates.
| Substrate | Background hydrolysis (nmol.min-1) |
Specific activity (μmol.min-1.mg-1) |
L.O.D.* (μg/mL) |
S/B# | S/N# | Z’# |
|---|---|---|---|---|---|---|
| I | 2.5 ± 0.1 | 5.1 ± 0.1 | 5.0 | 3.0 | 56 | 0.89 |
| II | 0.42 ± 0.02 | 9.6 ± 0.7 | 1.0 | 4.5 | 69 | 0.69 |
| III | 0.029 ± 0.007 | 0.044 ± 0.003 | 10.0 | 3.0 | 83 | 0.80 |
| IV | 0.34 ± 0.01 | 8.4 ± 0.5 | 1.0 | 4.8 | 93 | 0.79 |
| V | 0.019 ± 0.001 | 6.5 ± 0.4 | 0.05 | 3.4 | 83 | 0.76 |
Limit of Detection: minimum amount of PKLAP yielding a significantly measurable activity.
Values calculated for [E] = L.O.D. The signal to background ratio (S/B) was calculated by dividing the reaction average velocity in presence of enzyme (Av.enz.) by the average background hydrolysis rate (Av.blank). The signal to noise ratio (S/N) was calculated by dividing the reaction velocity in presence of enzyme by the standard deviation of the background hydrolysis (SDblank). The Z’ factor was calculated following the method of Zhang et al. [16] using the formula: Z’ = (1 − {[3*(SDenz. + SDblank)]/(Av.enz. − Av.blank)}.
Using commercial porcine kidney leucine aminopeptidase (PKLAP), we investigated the enzymatic hydrolysis of the novel LAP substrates. We first optimized the assay conditions using substrate I. At 27 °C, PKLAP activity was the highest at pH 7.0 and 8.0 (0.1 M sodium phosphate buffer), but it was dramatically lower at lower and higher pHs. Over the time period of observation (10 min), PKLAP activity was the highest at 31 °C, and slowly decreased at higher temperatures. These results are consistent with previous observations [22]. Because the fluorescence of the amino-pyridines (III to V) is optimal around pH 8.0 [12], we decided to run the enzymatic activity at pH 8.0 (0.1 M sodium phosphate buffer) at 31 °C. Using these conditions, we determined the specific activities of PKLAP for the five substrates (Table 2). The commercially available substrates (I and II) and the pyridines (IV and V) that have para-substitution on the pyridine, gave similar specific activities. In contrast, substrate III that has a meta-substitution on the pyridine gave a specific activity that was roughly two order of magnitude lower. This meta substitution probably interferes sterically with the optimal placement of III in the active pocket of PKLAP. No such effect was observed previously for the hydrolysis of leucine-anilides by LAP from bovine lens [23].
The limit of detection (L.O.D.) of PKLAP was defined as the concentration of protein for a particular substrate that yields an activity signal that is at least three times the one of the background hydrolysis rate (S/B ≥ 3) over a 10 min kinetic measurement. Because of its lower background hydrolysis and higher intensity of the reporter signal, substrate V gave the lowest L.O.D. (Table 2). In theory, using this substrate, one can detect 100 and 20 fold lower amounts of LAP activity in comparison to the commercial colorimetric I and fluorescent II substrates, respectively. Interestingly, because of its relatively high background hydrolysis, the substrate with the novel fluorescent reporter IV has a L.O.D. similar to the amino-coumarin based substrate II. For each substrate, we also calculated for [E] = L.O.D. S/N, and Z’ (Table 2). For all of the substrates, we found S/N above 50, reflecting a small variation in the reporter signal, and thus, robustness of the assays. We found Z’ values above 0.7, indicating a large separation band between the samples and blank signals. The Z’ value also indicated that these substrates could easily be used in high throughput screening assays [16]. To test this hypothesis, we followed the enzymatic hydrolysis of substrates I and V over 90 min. While substrate I was completely hydrolyzed after 20 min, only 5 % of substrate V was hydrolyzed after 60 min resulting in a linear response over time (r2 = 0.98). This was due to the higher sensitivity of V that allowed using a 100-fold lower amount of enzyme. Taken together, these data show that substrate V will be an excellent substrate for high throughput screening, either in a kinetic or end-point format.
To further compare substrate selectivity of porcine kidney microsomal LAP with other substrates published in the literature, we conducted a kinetic study with substrates I and V (Table 3). We found that PKLAP has a Vmax that was two fold higher for V than for I, and the Km for V was ∼60 fold lower for V than for I. Taken together, these data indicate that the PKLAP has a 92-fold preference for the novel fluorescent substrate V than for the commercial colorimetric substrate I. This is consistent with the lower L.O.D. obtained for substrate V (Table 2). Overall, our results clearly show that substrate V is an excellent substrate for LAP, and far better than the commercially available substrates.
Table 3.
Kinetic constants and substrate selectivity of porcine kidney LAP.
| Substrate |
Vmax (μmol.min-1.mg-1)* |
Km (μM) |
Vmax/Km (μmol.min-1.mg-1.μM-1) |
Selectivity ratio |
|---|---|---|---|---|
| I# | 3.9 ± 0.2 | 3000± 200 | 0.0013 ± 0.0003 | 1 |
| V# | 7.7 ± 0.2 | 65± 4 | 0.12 ± 0.01 | 92 |
Vmax data are per mg of protein.
Assays were carried out in sodium phosphate buffer (01.M pH 8.0), with 0.0236 μg/well of PKLAP, with final substrate concentrations between 1.56 and 200 μM. The reactions were followed for 10 min at 31°C.
Potential application of novel fluorescent reporters
Substituted aminopyridines used herein display high fluorescence and represent a novel class of fluorescent reporter that are structurally different from amino-coumarins, such as in substrate II. Substituted aminopyridines possess many advantages over aminocoumarins such as higher fluorescence, better aqueous solubility, and smaller size. To illustrate their potential applications and advantages over commercially available substrates, we used substrate V to determine inhibitor potency and detect LAP activities in cells. As shown on Fig. 3, we first looked at the inhibition of PKLAP by bestatin, a slow-binding competitive inhibitor of LAP [24]-[26]. The inhibition potency (IC50) obtained with substrate V was in the same order of magnitude as that obtained with substrate I. Because the enzyme concentrations used were different (one hundred fold lower for V), one can not directly compare the IC50s obtained with both substrates. Nevertheless, these data suggested that V could be easily used to accurately determine LAP inhibition, with the advantage of using much less enzyme, thus yielding a much more sensitive assay. This is especially useful for high throughput assay screening for potent inhibitors; because the ability of an assay to distinguish among potent inhibitors is largely dependent upon a low enzyme concentration.
Figure 3.
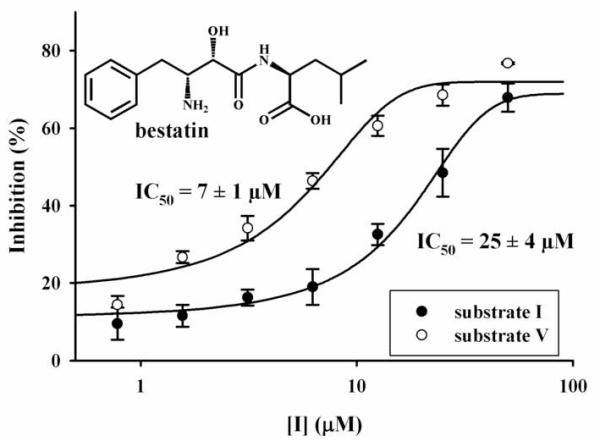
Inhibition of porcine kidney LAP by bestatin, using a colorimetric (substrate I) and a fluorescent (substrate V) assays.
There are many reports on the relationship between the LAPs and carcinoma [3][27]-[30]. These reports show that LAPs not only play regulatory roles in tumor cell proliferation, invasion and/or angiogenesis via degradation/inactivation of target peptides (e.g., oxytocin, angiotensins, and endothelin-1) which act on cancer cells as stimulatory or inhibitory factors. These reports also suggest that LAPs possibly function to inhibit oxytocin-dependent signaling. Thus, it is important to measure LAP activity in cancer cells. However, it is difficult to measure LAP activity with commercial substrates such as I and II. As shown on Fig. 4, we were able to measure LAP activities in homogenates from 6 cancer cell lines with the substrate V. We were not able to measure LAP activity with substrate I on the same extracts. Like I, substrate V was designed to be a general LAP activity reporter rather than to be specific for a single LAP; thus, the LAP activities observed in the cell extract could result from more than one enzyme, and caution should be taken about the interpretation of the results. Furthermore, the use of a general synthetic substrate like V, does not allow monitoring of the biological functions of an enzyme, but rather it is an indication of its expression/repression. We found that there was a significant increase of LAP activities (p≤0.01) for T24, HepG2 and Huh7 cancer cell lines, but a significant decrease (p≤0.01) for 22 RV1, compared with that of human liver S9. There was no significant difference of LAP activity for 293T and Du145 cancer cell lines compared with that of human liver S9. These results also indicated that LAP activity can be easily measured using a small amount of sample (equivalent of 8 × 104 cells were used in each assay only) by using this highly sensitive fluorescent substrate. Caution is needed when using these new substrates directly on live cell cultures. While the substrate described herein are probably not too toxic to cells, the amino-pyridine products could be [31]. Thus, their possible toxicity could influence the results. However, their high fluorescence permits using low concentrations of substrate (< 50 μM) reducing possible toxic effects.
Figure 4.
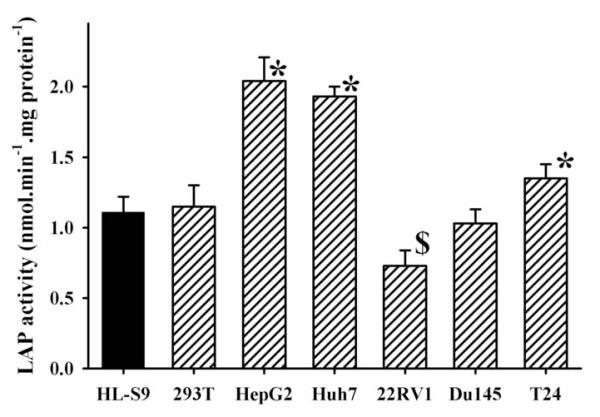
LAP activities of different cancer cell lines measured with the novel fluorescent substrate V. Compared to the human liver S-9 (HLS-9), LAP activity was: * significantly increased (p < 0.01), or $ significantly decreased (p < 0.01).
Rapid and efficient monitoring of LAP activity is important to understand the biological functions of this enzyme as well as following the development of associated diseases [3]. The sensitivity of such assays is especially important when the sample to be tested is in limited supply. Herein, we described novel fluorescent substrates for LAPs based on amino-pyridines. With these new substrates, we obtained up to a one hundred fold increase in sensitivity compared to currently available assays. Our assay will be highly useful for monitoring LAP activities in extracts from cancer cell lines, as well as for the high throughput screening of LAP inhibitors, which could potentially lead to new cancer treatments. Finally, the technology developed herein for LAPs could be easily adapted to other aminopeptidases and metalloproteases by simply changing the leucine on the substrate to a more appropriate amino acid or peptide. Furthermore, more specific substrates could be obtained by having specific amino acid at P1′ position and the reporter group in P1 [32].
Acknowledgements
This work was supported in part by NIEHS Grant R37 ES02710, NIH/NIEHS R01 ES013933, and NIH/NIEHS Superfund Basic Research Program P42 ES04699.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Tsujimoto M, Goto Y, Maruyama M, Hattori A. Biochemical and enzymatic properties of the M1 family of aminopeptidases involved in the regulation of blood pressure. Heart Fail Rev. 2008;13:285–291. doi: 10.1007/s10741-007-9064-8. [DOI] [PubMed] [Google Scholar]
- [2].Matsui M, Fowler JH, Walling LL. Leucine aminopeptidases: diversity in structure and function. Biol. Chem. 2006;387:1535–1544. doi: 10.1515/BC.2006.191. [DOI] [PubMed] [Google Scholar]
- [3].Mizutani S, Shibata K, Kikkawa F, Hattori A, Tsujimoto M, Ishii M, Kobayashi H. Essential role of placental leucine aminopeptidase in gynecologic malignancy. Expert Opin. Ther. Targets. 2007;11:453–461. doi: 10.1517/14728222.11.4.453. [DOI] [PubMed] [Google Scholar]
- [4].Yamazaki T, Akada T, Niizeki O, Suzuki T, Miyashita H, Sato Y. Puromycin-insensitive leucyl-specific aminopeptidase (PILSAP) binds and catalyzes PDK1, allowing VEGF-stimulated activation of S6K for endothelial cell proliferation and angiogenesis. Blood. 2004;104:2345–2352. doi: 10.1182/blood-2003-12-4260. [DOI] [PubMed] [Google Scholar]
- [5].Pilar CM, Ramírez-Expósito MJ, Dueñas B, Dolores MM, Jesús MG, De la Chica S, Cortés P, Ruíz-Sanjuan M, Martínez-Martos JM. Insulin-regulated aminopeptidase/placental leucil Aminopeptidase (IRAP/P-lAP) and angiotensin IV-forming activities are modified in serum of rats with breast cancer induced by N-methylnitrosourea. Anticancer Res. 2006;26:1011–1014. [PubMed] [Google Scholar]
- [6].Keller SR, Scott HM, Mastick CC, Aebersold R, Lienhard GE. Cloning and characterization of a novel insulin-regulated membrane aminopeptidase from Glut4 vesicles. J. Biol. Chem. 1995;270:23612–23618. doi: 10.1074/jbc.270.40.23612. [DOI] [PubMed] [Google Scholar]
- [7].Keller SR. Role of the insulin-regulated aminopeptidase IRAP in insulin action and diabetes. Biol. Pharm. Bull. 2004;27:761–764. doi: 10.1248/bpb.27.761. [DOI] [PubMed] [Google Scholar]
- [8].Shibata K, Kikkawa F, Mizokami Y, Kajiyama H, Ino K, Nomura S, Mizutani S. Possible involvement of adipocyte-derived leucine aminopeptidase via angiotensin II in endometrial carcinoma. Tumour Biol. 2005;26:9–16. doi: 10.1159/000084181. [DOI] [PubMed] [Google Scholar]
- [9].Saifuku K, Sekine T, Namihisa T, Takahashi T, Kanaoka Y. A novel fluorometric ultramicro determination of serum leucine aminopeptidase using a coumarine derivative. Clin. Chim. Acta. 1978;84:85–91. doi: 10.1016/0009-8981(78)90479-5. [DOI] [PubMed] [Google Scholar]
- [10].Roth M. A fluorimetric ultramicromethod for determination of leucine aminopeptidase in biological fluids. Clin. Chim. Acta. 1964;9:448–453. doi: 10.1016/0009-8981(64)90082-8. [DOI] [PubMed] [Google Scholar]
- [11].Grembecka J, Mucha A, Cierpicki T, Kafarski P. The most potent organophosphorus inhibitors of leucine aminopeptidase. Structure-based design, chemistry, and activity. J. Med. Chem. 2003;46:2641–2655. doi: 10.1021/jm030795v. [DOI] [PubMed] [Google Scholar]
- [12].Huang H, Nishi K, Tsai HJ, Hammock BD. Development of highly sensitive fluorescent assays for fatty acid amide hydrolase. Anal Biochem. 2007;363:12–21. doi: 10.1016/j.ab.2006.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kovi PJ, Capomacchia AC, Schulman SG. Electronic spectra of 2-aminoquinoline and 4-aminoquinaldine. Evidence for the cyclic amidine structures of the singly protonated cations. Anal. Chem. 1972;44:1611–1615. [Google Scholar]
- [14].Dey J, Dogra SK. Dual Fluorescence of 2-[4-(Dimethylamino)phenyl]benzothiazole and Its Benzimidazole Analog: Effect of Solvent and pH on Electronic Spectra. J. Phys. Chem. 1994;98:3638–3644. [Google Scholar]
- [15].Heindel ND, Kennewell PD. Dialkylamino-aminations with dialkylformamides. J. Chem. Soc. Chem. Commun. 1969;2:38. [Google Scholar]
- [16].Zhang J, Chung TDY, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- [17].Thiagarajan V, Ramamurthy P, Thirumalai D, Ramakrishnan VT. A novel colorimetric and fluorescent chemosensor for anions involving PET and ICT pathways. Org. Lett. 2005;7:657–660. doi: 10.1021/ol047463k. [DOI] [PubMed] [Google Scholar]
- [18].Wang W, Alexander D, Li Q. Design and Synthesis of Efficient Fluorescent Dyes for Incorporation into DNA Backbone and Biomolecule Detection. Bioconjugate Chem. 2007;18:1036–1052. doi: 10.1021/bc060151c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhang R, Kang KD, Shan G, Hammock BD. Design, synthesis and evaluation of novel P450 fluorescent probes bearing α-cyanoether. Tetrahedron Lett. 2003;44:4331–4334. [Google Scholar]
- [20].Kang KD, Jones PD, Huang H, Zhang R, Mostovich LA, Wheelock CE, Watanabe T, Gulyaeva LF, Hammock BD. Evaluation of alpha-cyano ethers as fluorescent substrates for assay of cytochrome P450 enzyme activity. Anal. Biochem. 2005;344:183–192. doi: 10.1016/j.ab.2005.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huang H, Nishi K, Gee SJ, Hammock BD. Evaluation of chiral alpha-cyanoesters as general fluorescent substrates for screening enantioselective esterases. J. Agr. Food Chem. 2006;54:694–699. doi: 10.1021/jf0526083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Itoh C, Nagamatsu A. An aminopeptidase activity from porcine kidney that hydrolyzes oxytocin and vasopressin: purification and partial characterization. Biochim. Biophys. Acta. 1995;1243:203–208. doi: 10.1016/0304-4165(94)00151-m. [DOI] [PubMed] [Google Scholar]
- [23].Taylor A, Tisdell FE, Carpenter FH. Leucine aminopeptidase (bovine lens): synthesis and kinetic properties of ortho-, meta-, and para-substituted leucyl-anilides. Arch. Biochem. Biophys. 1981;210:90–97. doi: 10.1016/0003-9861(81)90167-3. [DOI] [PubMed] [Google Scholar]
- [24].Rich DH, Moon BJ, Harbeson S. Inhibition of aminopeptidases by amastatin and bestatin derivatives. Effect of inhibitor structure on slow-binding processes. J. Med. Chem. 1984;27:417–422. doi: 10.1021/jm00370a001. [DOI] [PubMed] [Google Scholar]
- [25].Stöckel-Maschek A, Stiebitz B, Koelsch R, Neubert K. Novel 3-amino-2-hydroxy acids containing protease inhibitors. Part 1: Synthesis and kinetic characterization as aminopeptidase P inhibitors. Bioorg. Med. Chem. 2005;13:4806–4820. doi: 10.1016/j.bmc.2005.05.040. [DOI] [PubMed] [Google Scholar]
- [26].Stamper CC, Bienvenue DL, Bennett B, Ringe D, Petsko GA, Holz RC. Spectroscopic and X-ray crystallographic characterization of bestatin bound to the aminopeptidase from Aeromonas (Vibrio) proteolytica. Biochemistry. 2004;43:9620–9628. doi: 10.1021/bi049126p. [DOI] [PubMed] [Google Scholar]
- [27].Kondo C, Shibata K, Terauchi M, Kajiyama H, Ino K, Nomura S, Nawa A, Mizutani S, Kikkawa F. A novel role for placental leucine aminopeptidase (P-LAP) as a determinant of chemoresistance in endometrial carcinoma cells. Int. J. Cancer. 2006;118:1390–1394. doi: 10.1002/ijc.21509. [DOI] [PubMed] [Google Scholar]
- [28].Yamashita M, Kajiyama H, Terauchi M, Shibata K, Ino K, Nawa A, Mizutani S, Kikkawa F. Involvement of aminopeptidase N in enhanced chemosensitivity to paclitaxel in ovarian carcinoma in vitro and in vivo. Int. J. Cancer. 2007;120:2243–2250. doi: 10.1002/ijc.22528. [DOI] [PubMed] [Google Scholar]
- [29].Tsukamoto H, Shibata K, Kajiyama H, Terauchi M, Nawa A, Kikkawa F. Aminopeptidase N (APN)/CD13 inhibitor, Ubenimex, enhances radiation sensitivity in human cervical cancer. BMC Cancer. 2008;8:74. doi: 10.1186/1471-2407-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Suzuki Y, Shibata K, Kikkawa F, Kajiyama H, Ino K, Nomura S, Tsujimoto M, Mizutani S. Possible role of placental leucine aminopeptidase in the antiproliferative effect of oxytocin in human endometrial adenocarcinoma. Clin. Cancer Res. 2003;9:1528–1534. [PubMed] [Google Scholar]
- [31].Lechat P, Deysson G, Lemeignan M, Adolphe M. Comparison of the acute toxicity of some aminopyridines in vivo (in mice) and in vitro (on cell cultures) Ann. Pharm. Fr. 1968;26:345–349. [PubMed] [Google Scholar]
- [32].Gu YQ, Walling LL. Specificity of the wound-induced leucine aminopeptidase (LAPA) of tomato activity on dipeptide and tripeptide substrates. Eur. J. Biochem. 2000;267:1178–1187. doi: 10.1046/j.1432-1327.2000.01116.x. [DOI] [PubMed] [Google Scholar]


