Summary
Degradation by the proteasome typically requires substrate ubiquitination. Two ubiquitin receptors exist in the proteasome, S5a/Rpn10 and Rpn13. Whereas Rpn13 has only one ubiquitin-binding surface, S5a binds ubiquitin with two independent ubiquitin interacting motifs (UIMs). Here, we use NMR and analytical ultracentrifugation to define at atomic level resolution how S5a binds K48-linked diubiquitin, in which K48 of one ubiquitin subunit (the “proximal” one) is covalently bonded to G76 of the other (the “distal” subunit). We demonstrate that S5a’s UIMs bind the two subunits simultaneously with a preference for UIM2 binding to the proximal subunit while UIM1 binds to the distal one. In addition, NMR experiments reveal that Rpn13 and S5a bind K48-linked diubiquitin simultaneously with subunit specificity, and a model structure of S5a and Rpn13 bound to K48-linked polyubiquitin is provided. Altogether, our data demonstrate that S5a is highly adaptive and cooperative towards binding ubiquitin chains.
Keywords: proteasome, ubiquitin, protein degradation, S5a, Rpn13
Introduction
Ubiquitin conjugation plays a key regulatory role in a wide array of biological processes. In its most established role, ubiquitination targets proteins for degradation by the 26S proteasome (Ciechanover, 1994), a process important for controlling the lifespan of regulatory proteins, removing misfolded proteins, producing immunocompetent peptides (Rock and Goldberg, 1999), and regulating cell cycle progression (Ciechanover et al., 1984; Finley et al., 1984).
The 26S proteasome contains a 20S catalytic core particle (CP) capped at either or both ends by 19S regulatory particles (RPs), which prepare substrates for hydrolysis in the CP. RP ubiquitin receptors S5a/Rpn10 (Deveraux et al., 1994) and Rpn13 (Husnjak et al., 2008) capture substrates by recognizing their covalently attached ubiquitin chains, which are removed and disassembled by three deubiquitinating enzymes Rpn11 (Verma et al., 2002), Ubp6/Usp14 (Leggett et al., 2002) and Uch37/UCHL5 (Lam et al., 1997). These actions of the RP’s ubiquitin processing enzymes are complemented with that of six ATPases that unfold protein substrates to allow their passage into the CP for degradation.
Ubiquitin receptors play an integral role in substrate capture and apparently contribute to ubiquitin chain deconjugation, as Rpn13 binds and activates deubiquitinating enzyme Uch37 (Hamazaki et al., 2006; Qiu et al., 2006; Yao et al., 2006). S5a and Rpn13 may receive ubiquitinated proteins from extra-proteasomal receptors, including the hHR23 and hPLIC proteins, which bind to them with ubiquitin-like (UBL) domains (Hiyama et al., 1999; Husnjak et al., 2008; Walters et al., 2002) and to ubiquitin with ubiquitin-associated (UBA) domains (Bertolaet et al., 2001; Wang et al., 2003; Wilkinson et al., 2001). UBL-UBA proteins can also dock substrates into the proteasome by binding to Rpn1/S2 (Elsasser et al., 2002), a scaffolding protein that may also bind S5a (Wilkinson et al., 2000). S5a is also abundant free of the proteasome, where it has been demonstrated in budding yeast to prevent the hPLIC homolog Dsk2 from binding to proteasome and inhibiting substrate degradation (Matiuhin et al., 2008). Although not essential in budding yeast (van Nocker et al., 1996), S5a is essential in mice (Hamazaki et al., 2007) and Drosophila melanogaster (Szlanka et al., 2003).
Unique roles for the ubiquitin receptors associated with proteasome degradation are suggested by their diverse ubiquitin binding properties. hHR23a is sandwiched between the ubiquitin subunits of K48-linked diubiquitin (Varadan et al., 2005), effectively sequesters subunits of polyubiquitin up through octaubiquitin (Kang et al., 2007), and protects ubiquitin chains from deubiquitination (Raasi and Pickart, 2003; Verma et al., 2004). By contrast, Rpn13 binds preferentially to the proximal subunit of K48-linked diubiquitin leaving the distal one available for other interactions (Schreiner et al., 2008), presumably including that of Uch37, which may facilitate its distal end (Lam et al., 1997) deubiquitinating activity (Hamazaki et al., 2006; Qiu et al., 2006; Yao et al., 2006).
Although S5a was the first proteasome ubiquitin receptor discovered (Deveraux et al., 1994), its structure complexed with polyubiquitin has not yet been solved. Like Rpn13, the yeast Rpn10 protein is significantly truncated compared to its human S5a homologue (Figure 1A). Whereas mammalian Rpn13 gains a deubiquitinating enzyme binding domain (that of Uch37), the extension in mammalian S5a affords a second UIM that binds ubiquitin more strongly than the conserved one (Wang et al., 2005). In previous work, we solved the structure of S5a (196–306) alone and complexed with monoubiquitin to reveal that its two UIMs act independently in each of these structures (Wang et al., 2005).
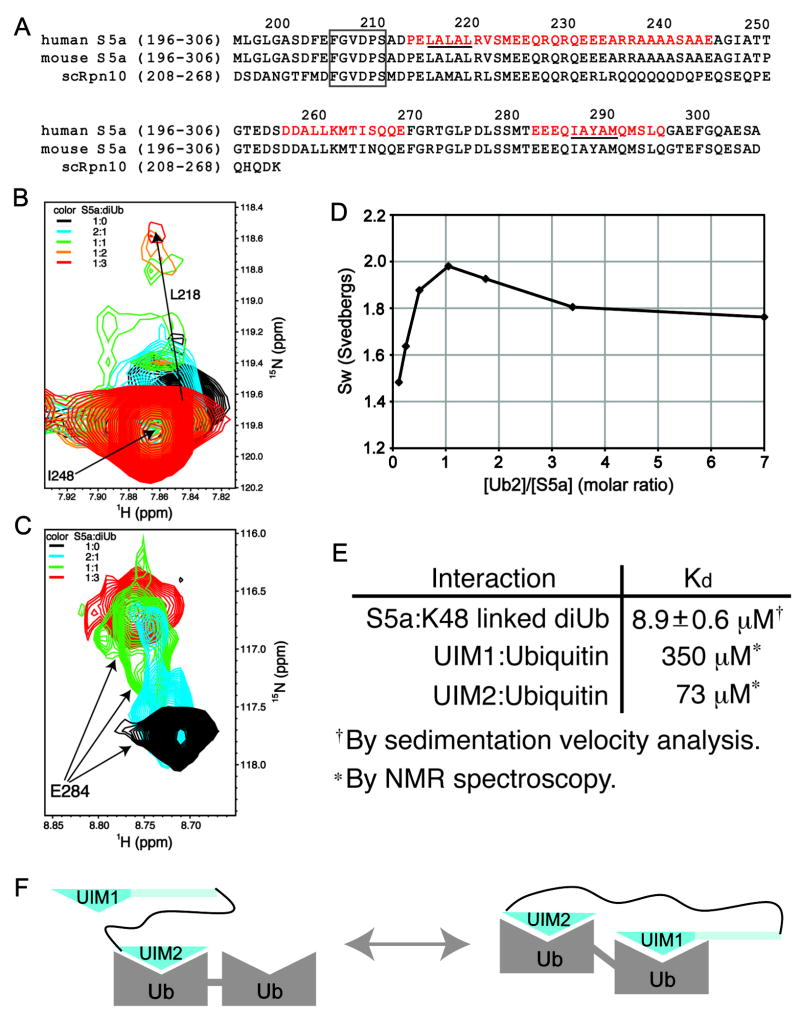
Figure 1. S5a’s affinity for diubiquitin is enhanced by its two UIMs binding to ubiquitin subunits simultaneously.
(A) Sequence alignment of the ubiquitin-binding region of human and mouse S5a with their Saccharomyces cerevisiae homologue Rpn10. The helices of S5a are highlighted in red and a strictly conserved region N-terminal to UIM1 is boxed. The UIM’s hallmark LALAL/IAYAM motif is underlined. (B) UIM1 resonances shift and broaden upon binding to K48-linked diubiquitin. 1H,15N HSQC spectra expanded to view L218 are displayed for 15N labeled S5a (196–306) alone (black) and at 2:1 (blue), 1:1 (green), 1:2 (orange), and 1:3 (red) S5a:K48-linked diubiquitin molar ratio. The amide resonance of I248, which is not involved in diubiquitin-binding and does not shift with increasing quantities of diubiquitin, overlaps with the L218’s unbound resonance. (C) UIM2 resonances exhibit slow exchange dynamics upon binding to K48-linked diubiquitin. 1H, 15N HSQC spectra expanded to view E284 are displayed for S5a alone (black) and at 2:1 (blue), 1:1 (green), and 1:3 (red) S5a:K48-linked diubiquitin molar ratio. (D) Sedimentation velocity results reveal S5a (196–306) to bind K48-linked diubiquitin with 1:1 stoichiometry and with an affinity of 8.9±0.6 μM. Experimentally determined averaged sedimentation coefficients (diamonds) are plotted. (E) Binding affinities are displayed for S5a (196–306) binding to diubiquitin, as determined with the data of (D), and for each UIM binding to monoubiquitin (Ryu et al., 2003; Wang et al., 2005). (F) Model depicting the two S5a:diubiquitin states present at 1:1 molar ratio. The weaker binding UIM1 exchanges between an unbound and bound state. Ub, ubiquitin; UIM1, S5a’s ubiquitin interacting motif 1; UIM2, S5a’s ubiquitin interacting motif 2.
In this manuscript, we use NMR spectroscopy and analytical ultracentrifugation to solve the S5a (196–306):K48-linked diubiquitin structure. In contrast to S5a’s binding of monoubiquitin, its two UIMs bind a common diubiquitin molecule rather than recruit two diubiquitins simultaneously. Moreover, it exhibits subunit specificity and binding determinants that make its interaction with ubiquitin chains unique from that of Rpn13 or hHR23a. In addition, we demonstrate that S5a and Rpn13 bind K48-linked diubiquitin simultaneously with subunit specificity. This research provides fundamental information on how the proteasome recognizes polyubiquitinated substrates.
Results
S5a’s UIMs bind to ubiquitin subunits of K48-linked diubiquitin simultaneously and with different affinities
We used NMR spectroscopy to determine whether S5a’s two UIMs bind independently to K48-linked diubiquitin. We added unlabeled K48-linked diubiquitin to 15N labeled human S5a (196–306), which contains its two UIMs (Figure 1A), and used 1H,15N HSQC experiments to detect binding. NMR signals belonging to UIM1 and UIM2 residues were affected by diubiquitin addition, thus demonstrating diubiquitin’s interaction with both UIMs. Different binding behavior was observed for each of the UIMs however. Upon diubiquitin addition, UIM1 signals shift and broaden progressively to their fully saturated state (Figure 1B), indicating that these residues exchange rapidly on the NMR timescale (100 milliseconds) between their free and bound states. UIM2 signals by contrast exhibit two peaks prior to saturation, which are not broadened by exchange (Figure 1C, blue spectrum) and are representative of their free and diubiquitin-bound states, according to comparisons of spectra recorded on free (Figure 1C, blue spectrum) and fully saturated S5a (Figure 1C, red spectrum). Hence, UIM2 with its readily observed diubiquitin-bound state is the stronger binding partner.
Unexpectedly, diubiquitin causes signals from the internal helix of S5a (196–306), which spans residues D257-E269, to shift (Supplementary Figure 1A). Such perturbations were not observed in S5a with monoubiquitin (Supplementary Figure 1B). This region does not contain a known ubiquitin-binding element and shows no direct contact with monoubiquitin. We tested whether it contacts diubiquitin by using a 13C half-filtered NOESY spectrum in which S5a (196–306) was 13C labeled and diubiquitin was unlabeled. This experiment records intermolecular NOE interactions between S5a and diubiquitin hydrogen atoms that are within 5 Å while selecting against intramolecular NOEs. Whereas such interactions were observed between S5a’s UIMs and diubiquitin (as described below), no residues within the intervening helix exhibited such intermolecular NOE interactions (data not shown). Therefore, the spectral perturbation of residues in this region upon diubiquitin addition is not due to its direct contact with diubiquitin. Moreover, the helical structure of this region is preserved according to 15N and 13C dispersed NOESY spectra (Supplementary Figure 2). We therefore hypothesized that the spectral changes to this region are caused by physical constraints imposed by the simultaneous binding of both UIMs to diubiquitin.
To test directly whether the two UIMs of S5a bind to the same diubiquitin molecule, we performed sedimentation velocity experiments. Data were recorded at molar ratios of diubiquitin to S5a (196–306) spanning 1:7 to 8:1 and the weight average sedimentation coefficients (sw) were plotted against molar ratio (Figure 1D). This analysis revealed a maximum at a 1:1 mixing ratio, indicating 1:1 binding stoichiometry. The decrease in sw at lower and higher mixing ratios is due to the presence of free diubiquitin and S5a, respectively. Global analysis of the sedimentation velocity data using this 1:1 binding model gives a best-fit Kd of 8.9±0.6 μM. This value is substantially lower than that determined for UIM1 and UIM2 binding to monoubiquitin (Wang et al., 2005) (listed in Figure 1E). Altogether, these data indicate that the two UIMs bind ubiquitin subunits of diubiquitin simultaneously and that this coordinated binding affords a greater affinity.
The estimated Kd for S5a (196–306) binding to diubiquitin predicts that 86% of S5a is bound to diubiquitin when these two proteins are at equimolar ratio and 0.4 mM protein concentration. As mentioned above, however, S5a’s UIM2 exhibits stronger affinity compared to UIM1. The NMR titration experiments revealed that the binding between S5a’s UIM1 and diubiquitin is not saturated until diubiquitin is at 3-fold molar excess (Figure 1B). Two shifted peaks are observed for L278, S279, E284, A288 and S294 of S5a’s UIM2 at equimolar ratio (as demonstrated for E284 in Figure 1C), one of which remains when diubiquitin is at 3-fold molar excess. We propose that the peak present with 3-fold molar excess diubiquitin corresponds to a state in which UIM2 and UIM1 fully occupy diubiquitin whereas the other stems from diubiquitin-bound UIM2 when UIM1 is not bound (Figure 1F).
UIM2 binds the proximal subunit while UIM1 binds the distal one
To determine whether distinct binding preferences exist between K48-linked diubiquitin’s subunits and S5a’s UIMs, we acquired 3D 13C half-filtered NOESY experiments on two different samples (Figure 2A, top panel). In these, S5a (196–306) is unlabeled and mixed with diubiquitin with either its proximal (Figure 2B, black and Supplementary Figure 3A) or distal (Figure 2B, red) ubiquitin subunit 13C labeled. A hydrophobic patch formed by L8, I44, and V70 is used by ubiquitin to bind many of its receptors. These residues in the proximal, but not distal ubiquitin subunit, exhibit multiple intermolecular NOEs with S5a (196–306). Similarly, NOEs unique to the distal subunit were observed for the C-terminal β-strand of ubiquitin, especially L71 and L73. Importantly, intermolecular NOEs were observed to the residues in the diubiquitin linker region, namely K48 and Q49 of the proximal subunit and G75 or G76 of the distal subunit. Altogether, our data indicate that the two ubiquitin subunits form unique interactions with S5a and that contacts are made to the diubiquitin linker region of K48-linked chains.
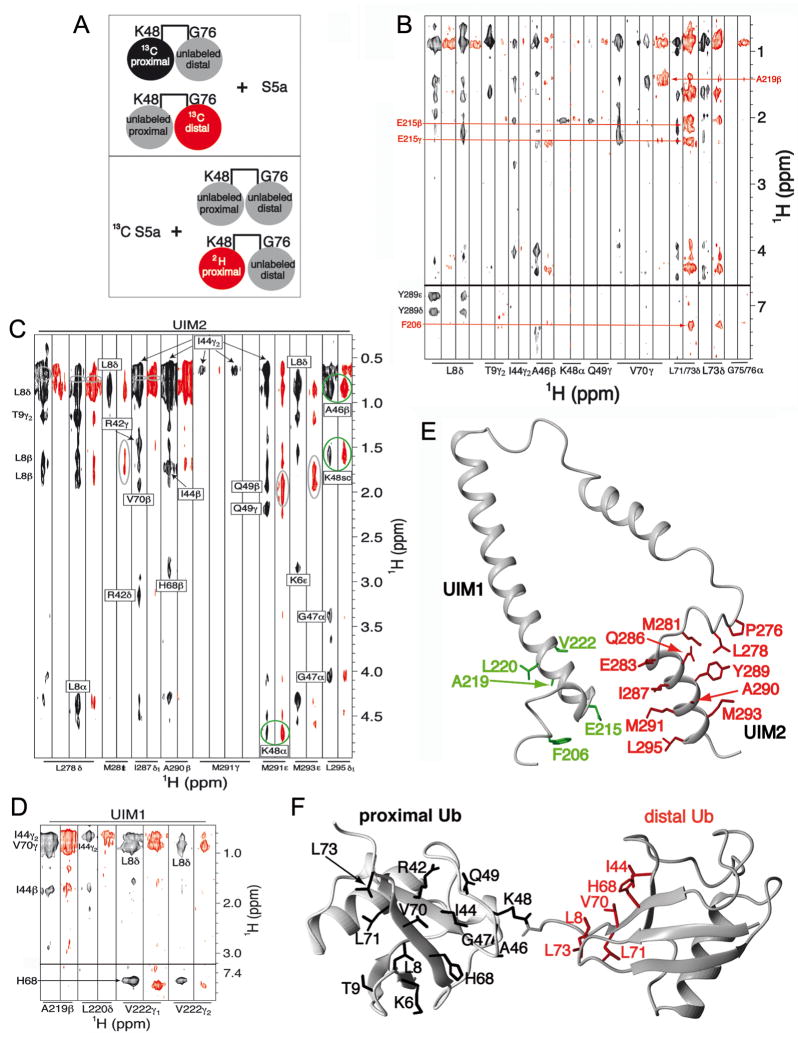
Figure 2. S5a prefers to bind K48-linked diubiquitin with its UIM1 bound to the distal subunit while its UIM2 binds the proximal one.
(A) Schematic illustrating the NMR samples used for the 13C half-filtered NOESY spectra displayed in (B; top panel) and (C and D; bottom panel). Coloring matches the associated spectrum, such that the labeled black and red subunits produced the spectra displayed in black and red, respectively. (B) 13C half-filtered NOESY experiments recorded on K48-linked diubiquitin with either its proximal (black) or distal (red) subunit 13C labeled and mixed with unlabeled S5a (196–306). Abundant NOE interactions are observed between UIM2 and the proximal subunit as well as between UIM1 and the distal subunit. Selected interactions are labeled and greater detail is provided in Supplementary Figure 3A. (C, D) 13C half-filtered NOESY spectra recorded with 13C-labeled S5a (196–306) and 3-fold molar excess unlabeled K48-linked diubiquitin (black) or K48-linked diubiquitin with its proximal subunit deuterated (red). Most interactions with UIM2 residues are mitigated by 2H labeling the proximal subunit (C), whereas those involving UIM1 resonances are not (D). Breakthrough diagonal peaks, which should be treated as noise, are circled in grey. NOE crosspeaks involving UIM2 that are only slightly affected by deuterating the proximal subunit are circled in green; these indicate the presence of a minor species in which UIM2 binds the distal subunit. The sidechain atoms of (E) S5a and (F) diubiquitin residues that form intermolecular NOE contacts in the major binding species are highlighted with UIM1 and UIM2 in green and red, respectively, and diubiquitin’s proximal and distal subunits in black and red, respectively.
We next sought to establish whether UIM1/2 binds to the proximal versus distal subunit of diubiquitin. We were able to assign all of the intermolecular NOEs involving diubiquitin’s proximal subunit to residues within UIM2 (Figure 2B and Supplementary Figure 3A). To confirm these assignments and to facilitate assigning the intermolecular NOEs involving the distal subunit, we acquired 3D 13C half-filtered NOESY experiments on samples containing 13C labeled S5a (196–306) and unlabeled diubiquitin (Figure 2C and 2D, black) or diubiquitin in which the proximal subunit is 100% 2H labeled (Figure 2C and 2D, red), illustrated in the bottom panel of Figure 2A. The latter spectrum contains NOE interactions between S5a and the distal ubiquitin subunit only as the 2H labeling silences the proximal subunit in this experiment.
Consistent with our analysis of the 13C half-filtered NOESY experiments described above (Supplementary Figure 3A), residues of UIM2 displayed abundant intermolecular NOEs with the L8-I44-V70 hydrophobic patch of diubiquitin (Figure 2C, black). Most of these were either eliminated or greatly reduced when the proximal subunit was silenced with 2H labeling (Figure 2C, red). A few intermolecular NOEs were only slightly affected by 2H labeling of the proximal ubiquitin subunit; these are between M291 and L295 in the UIM2 motif and A46 and K48 of diubiquitin (Figure 2C, circled in green). The comparable intensities of these intermolecular NOEs in the two spectra indicate that a population of S5a:diubiquitin is formed with UIM2 binding to the distal subunit. Moreover, the unique NOE pattern between UIM2 and diubiquitin’s distal versus proximal subunit suggests slightly different UIM2:ubiquitin binding motifs, as necessitated by the involvement of K48, which is chemically different in the two subunits.
M291’s and L295’s side chain methyl groups have slightly different chemical shift values when bound to diubiquitin’s proximal versus distal subunit (Supplementary Figure 4). By obtaining the volume of M291’s methyl group NOE to K48 Hα in the proximal versus distal subunit, we estimated that UIM2 binding to diubiquitin’s proximal subunit is at 3-fold molar excess compared to distal binding. Consistent with this analysis, NOE interactions between UIM1 and diubiquitin were only slightly affected by 2H labeling of the proximal subunit (Figure 2D, black versus red). Altogether, these data provide strong evidence that UIM2 is predominately bound to the proximal subunit of diubiquitin while UIM1 binds the distal subunit, and that a minor population exists with UIM2 bound to the distal subunit. In the next section, we present these two structures; however, it is worth noting here that the difference in the volume of the NOEs from M291’s methyl group and diubiquitin’s proximal versus distal K48 Hα is not due to differences in their distances from each other in the two structures as only a 0.2 Å difference is observed. We used standard thermodynamics equations relating population to binding constants and Gibbs free energy (see Supplement) to estimate that an energy difference of 2.7 kJ/mol is sufficient for a 3:1 binding preference.
Structure of S5a (196–306):K48-linked diubiquitin
We assigned 59 and 31 NOE interactions between UIM2 and diubiquitin’s proximal and distal subunits, respectively. Unambiguous NOEs were also identified between F206, E215, A219, L220 and V222 and diubiquitin’s distal subunit (Figure 2B and 2D and Supplementary Figure 3B). We were surprised to find intermolecular NOEs between L73’s methyl groups of ubiquitin and F206, a conserved S5a residue that is nine amino acids N-terminal to the well-known LALAL ubiquitin-binding motif (Figure 1A). The S5a and diubiquitin residues that exhibited intermolecular NOEs are highlighted in Figures 2E and 2F.
By using our intermolecular NOEs, we calculated a well defined structure for the major S5a (196–306):K48-linked diubiquitin species with no NOE violation above 0.3 Å (Figure 3, Table 1). For the minor species, we were able to use intermolecular NOEs between UIM2 and distal ubiquitin (Table 1). However, there were no NOE interactions between UIM1 and the proximal subunit, most likely because the protein concentration of this species is low (estimated at 0.125 mM) and, in contrast to UIM2, UIM1’s binding to diubiquitin causes resonance broadening (Figure 1B). Therefore, we modeled the UIM1:proximal ubiquitin interaction based on our UIM1:distal ubiquitin NOEs. In the S5a (196–306):monoubiquitin complex, the relative position of UIM1 and UIM2 is not defined as the two UIMs are separated by two flexible linker regions in addition to the intervening α-helix (Wang et al., 2005). The relative orientation of the two UIMs is more restricted in the complexes with diubiquitin by the simultaneous binding of the two UIMs to the ubiquitin subunits (Figure 3A). This restriction provides an explanation for the resonance broadening and shifting observed for residues in the intervening helix (Supplementary Figure 1A). Diubiquitin’s flexible linker region does, however, enable some freedom in the location of UIM1 relative to UIM2 (Supplementary Figure 5).
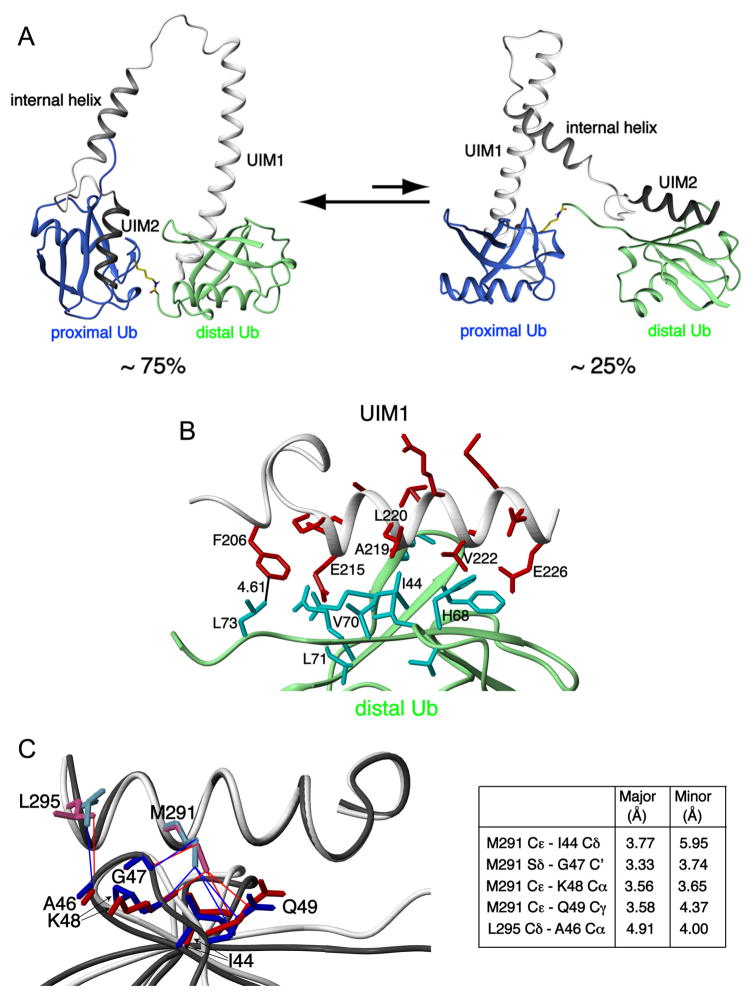
Figure 3. Structure of S5a:K48-linked diubiquitin complexes.
(A) Ribbon representation of the major and minor binding modes for S5a:K48-linked diubiquitin with S5a’s UIM1 and UIM2 motifs in white and black, respectively, and diubiquitin’s proximal and distal subunits in blue and green, respectively. Diubiquitin’s K48-G76 linkage is highlighted in yellow. (B) Expanded view of the UIM1:distal ubiquitin complex reveals critical interactions. Residues of UIM1 and distal ubiquitin are highlighted in red and cyan, respectively, whereas their ribbon structures are grey and green respectively. (C) Multiple contacts become more compact when UIM2 binds to the proximal subunit of K48-linked diubiquitin. UIM2’s M291 and L295 are displayed bound to proximal (pink and red) or distal (blue) ubiquitin. Selected distances are listed between M291 and L295 of UIM2 and proximal or distal ubiquitin atoms.
Table 1.
Structural statistics for the major and minor S5a (196–306):K48-linked diubiquitin complexes.
| Major | Minor | |
|---|---|---|
| Intermolecular NOE distance restraints (total) | 46 | 36 |
| UIM1:distal ubiquitin | 12 | N/A |
| UIM2:proximal ubiquitin | 34 | N/A |
| UIM1:proximal ubiquitin | N/A | 12* |
| UIM2:distal ubiquitin | N/A | 24 |
|
| ||
| Ramachandran plot | ||
| Most-favorable region (%) | 79.5 | 79.0 |
| Additionally allowed region (%) | 20.0 | 20.4 |
| Generously allowed region (%) | 0.5 | 0.6 |
| Disallowed region (%) | 0.0 | 0.0 |
|
| ||
| r.m.s.d of backbone atoms to average structure (Å) | ||
| UIM1: F206-M224; ubiquitin: M1-G76 | 1.15 ± 0.25 | 1.60 ± 0.85 |
| UIM2: P276-L295; ubiquitin: M1-G76 | 0.91 ± 0.28 | 0.76 ± 0.20 |
| r.m.s.d of all heavy atoms to average structure (Å) | ||
| UIM1: F206-M224; ubiquitin: M1-G76 | 1.47 ± 0.20 | 1.88 ± 0.73 |
| UIM2 P276-L295; ubiquitin: M1-G76 | 1.27 ± 0.27 | 1.13 ± 0.22 |
The distance restraints for UIM1:proximal ubiquitin used to calculate the structure of the minor S5a:K48-linked diubiquitin complex are derived from UIM1:distal ubiquitin intermolecular NOEs. See supplement for details.
The structure of S5a (196–306) complexed with K48-linked diubiquitin revealed interesting changes to the strictly conserved 206FGVDPS211 region N-terminal to UIM1 (Figure 1A). Contacts between F206 and distal ubiquitin’s L73, as demonstrated in Figure 2B (red) and Figure 3B, promote additional intramolecular interactions between 206FGVDPS211 and the LALAL motif to result in a more ordered structure. This consequence is apparent in our NMR data, as NOEs between P210 and A217 were strong in diubiquitin-bound S5a (196–306), but weak in spectra recorded on the unbound protein (data not shown).
We sought to understand why the preferred diubiquitin binding mode was with UIM2 bound to the proximal subunit and UIM1 bound to the distal subunit and conversely, how the two UIMs could accommodate the minor binding species with such a small energy difference between the two. Comparison of the UIM2:distal ubiquitin and UIM2:proximal ubiquitin structures revealed M291 to form unique van der Waals interactions with I44 and more compact contacts with G47 and Q49 of the proximal compared to distal subunit (Figure 3C). These differences are derived from our experimental data. For example, the intermolecular NOEs between G47’s Hα protons and L295’s methyl group are greatly diminished in the spectrum recorded on the sample with the proximal subunit silenced by deuterium labeling (Figure 2C). Some distances however lengthen in the proximal subunit, the most significant of which is that between one of L295’s methyl groups and A46’s Cβ, as supported by the corresponding intermolecular NOEs displayed in Figure 2C. Therefore, UIM2 favors slightly different interactions with the two subunits.
UIM1 showed the strongest interactions with the C-terminal end of distal ubiquitin (Figure 2B, red). This region’s dynamic motions are more restricted in the distal subunit due to the presence of the isopeptide linker (Fushman et al., 2004), which apparently enhances its interaction with UIM1. These effects likely contribute to S5a’s preferred diubiquitin binding mode. UIM1’s importance in determining S5a’s diubiquitin binding mode is supported by previous studies on a UIM2 fragment, which exhibited little preference for the proximal versus distal subunit (Haririnia et al., 2007).
S5a binds the distal subunit of K48-linked diubiquitin while Rpn13 binds the proximal subunit
Rpn13’s ubiquitin binding domain binds to K48-linked diubiquitin with an affinity of 90 nM (Husnjak et al., 2008). Therefore, we tested whether it precludes S5a from binding to K48-linked diubiquitin. We added equimolar quantities of K48-linked diubiquitin and Rpn13 (1–150) to 13C labeled S5a (196–306). A 1H,13C HMQC experiment revealed S5a UIM1 and UIM2 resonances that shift upon addition of K48-linked diubiquitin (Figure 4A, green) to appear at similar positions when Rpn13 was added with diubiquitin (Figure 4A, blue). Hence, Rpn13 does not preclude S5a from binding to K48-linked diubiquitin. To test whether Rpn13 and S5a bind diubiquitin simultaneously, we added equimolar quantities of unlabeled K48-linked diubiquitin and S5a to 15N labeled Rpn13. Rpn13 amino acids that are close to the isopeptide bond linkage when bound to the proximal subunit shift in a different manner when bound to K48-linked polyubiquitin versus monoubiquitin (Schreiner et al., 2008). We found these Rpn13 resonances to shift to their proximal-bound state when S5a (196–306) is present (Figure 4B). Therefore, we hypothesized that Rpn13 binds to the proximal subunit while S5a binds to the distal subunit.
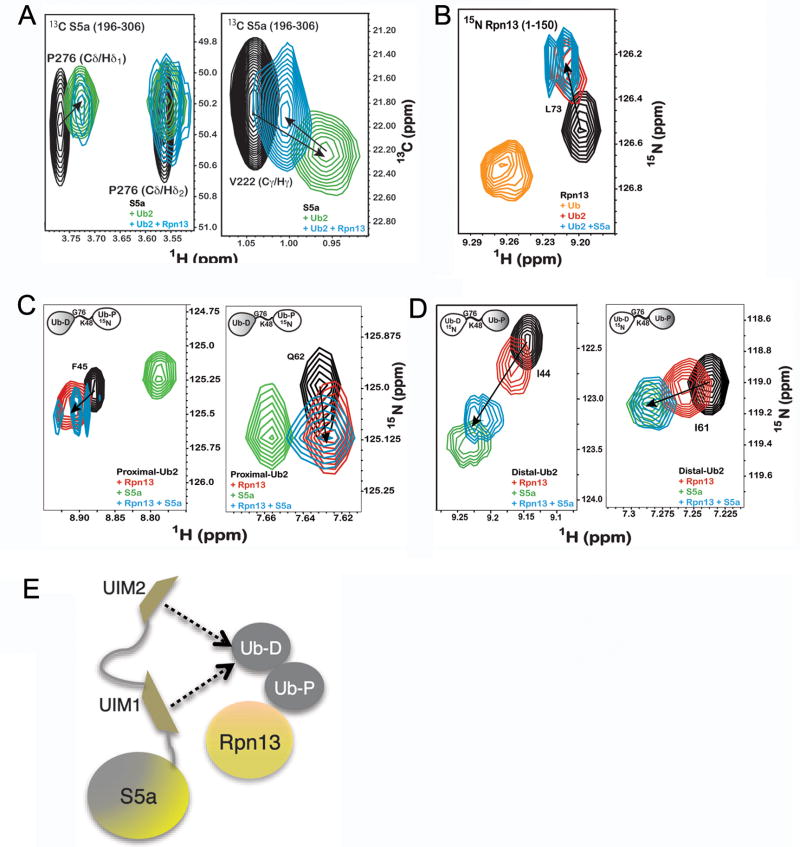
Figure 4. Ubiquitin receptors Rpn13 and S5a bind K48-linked diubiquitin simultaneously.
A) 1H, 13C HSQC experiments recorded on 13C labeled S5a (196–306) alone (black) or with equimolar K48-linked diubiquitin (green) or K48-linked diubiquitin and Rpn13 (1–150) (blue) reveal P276 (left panel) and V222 (right panel) to interact with diubiquitin when Rpn13 is present. B) 1H, 15N HSQC experiments on 15N labeled Rpn13 (1–150) alone (black) and with monoubiquitin (orange), K48-linked diubiquitin (red) or K48-linked diubiquitin and S5a (196–306) (blue) reveals L73 to shift to its diubiquitin-bound state when S5a is present. K48-linked diubiquitin with its (C) proximal or (D) distal subunit 15N labeled alone (black) or with Rpn13 (1–150) (red), S5a (196–306) (green), or both of these receptors (blue) indicates shifting that mimics Rpn13 binding to the proximal subunit, but S5a binding to the distal one, as shown for F45 and Q62 of the proximal subunit and I44 and I61 of the distal subunit. Additional data is provided in Supplementary Figure 6. (E) Model illustrating Rpn13 bound to K48-linked diubiquitin’s proximal subunit and S5a’s UIMs interacting dynamically with the distal one.
To test more directly which receptor binds K48-linked diubiquitin’s proximal subunit, we added S5a (196–306) and Rpn13 (1–150) or just one of these proteins to equimolar quantities of K48-linked diubiquitin with its proximal subunit 15N labeled. The observed chemical shift changes with both receptors present were similar to those caused by only Rpn13 addition, as demonstrated for F45 and Q62 (Figure 4C; see also Supplementary Figure 6). By contrast, the addition of equimolar quantities of S5a (196–306) and Rpn13 (1–150) to K48-linked diubiquitin with its distal subunit 15N labeled revealed the larger chemical shift changes characteristic of S5a (196–306) interaction, as illustrated for I44 and I61 in Figure 4D. Altogether, our data indicate that S5a binds predominately to diubiquitin’s distal subunit while the stronger binding Rpn13 maintains its position at the proximal subunit.
Although UIM2 has the stronger affinity for K48-linked diubiquitin, 13C resonances from both UIM motifs shift upon addition of K48-linked diubiquitin when Rpn13 is present (Figure 4A, blue). Rpn13 however either mitigates the chemical shift perturbations, as shown in Figure 4A for V222, or causes attenuation, as shown in Figure 4A for P276. These differences suggest that UIM1 and UIM2 compete for K48-linked diubiquitin’s distal subunit when Rpn13 occupies the proximal one (Figure 4E). Dynamic binding behavior was also evident in spectra recorded on the proximal subunit, although to a lesser extent. The observed spectral changes with both receptors present largely mimic those caused by only Rpn13; however, additional attenuations are observed for A46 and H68 (Supplementary Figure 6).
We tested the model presented in Figure 4E directly by labeling either S5a’s UIM1 or UIM2 region with the paramagnetic spin label S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL). MTSL causes distance-dependent attenuation with effects apparent up to distances of ~20 Å. It forms a disulfide bond with cysteine residues and since S5a (196–306) contains no cysteines, it was straightforward to produce proteins samples with either UIM1 or UIM2 labeled. UIM1 and UIM2 labeling was performed by substituting Q227 and A298 with cysteine, respectively. These two residues were chosen based on our NMR structure. They do not contact diubiquitin directly and their mutation is therefore not expected to disrupt S5a:diubiquitin interaction; however, they are close enough for MTSL-induced attenuation of diubiquitin atoms (model structures are provided for the major conformation in Figure 5A).
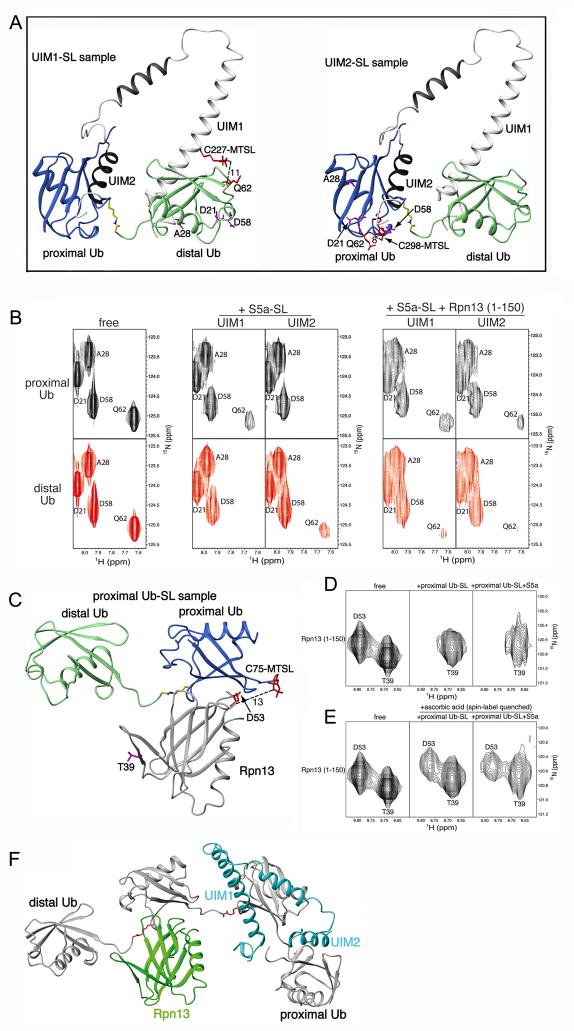
Figure 5. MTSL-labeling demonstrates hRpn13’s preference for K48-linked diubiquitin’s proximal subunit as S5a’s two UIMs bind the distal subunit.
(A) Ribbon representation of the major S5a (196–306):K48-linked diubiquitin structure displaying MTSL covalently attached to C227 (left) or C298 (right). Each of these cysteines was introduced for UIM1 or UIM2 spin labeling by MTSL. Q62 is also displayed as well as approximate distances in Å between its backbone amide proton atom and MTSL’s single-electron center (oxygen atom). It is worth noting that MTSL and the cysteine sidechain atoms are flexible such that the distances shown are only approximations. D21, A28 and D58 are also displayed and highlighted in purple, as they are used for comparison. (B) Expanded view of 1H, 15N HSQC spectra for the proximal (top panels) or distal (bottom panels) subunit of K48-linked diubiquitin alone (left panel) with added MTSL labeled C227 (UIM1) or C298 (UIM2) only (middle panel), or with hRpn13 (1–150) in addition (right panel), as labeled. S5a-SL refers to MTSL labeled S5a (196–306). (C) Ribbon representation of Rpn13:K48-linked diubiquitin structure displaying MTSL covalently attached to C75 of the proximal ubiquitin. D53 of hRpn13 is displayed as well as the approximate distance in Å between its backbone amide proton atom and MTSL’s single-electron center (oxygen atom). T39 is displayed and highlighted in purple, as it is used for comparison. (D) Expanded view of 1H, 15N HSQC spectra for the hRpn13 (1–150) alone (left panel) with MTSL labeled K48-linked diubiquitin only (middle panel), or with S5a (196–306) in addition (right panel), as labeled. Proximal Ub-SL refers to K48 diubiquitin with its proximal subunit MTSL labeled. (E) Expanded view of 1H, 15N HSQC spectra for the hRpn13 (1–150) alone (left panel) and with either spin label quenched K48-linked diubiquitin G75C only (middle panel) or with unlabeled S5a (196–306) in addition (right panel). All spectra of protein mixtures were acquired with all components at 0.3 mM. (F) Model of one possible binding configuration for Rpn13 (1–150) (green) and S5a (196–306) (blue) simultaneously bound to K48-linked tetraubiquitin (grey with K48-G76 linker region in red). In a tetraubiquitin chain, a ubiquitin subunit is available to each ubiquitin-binding module and K48 and G76 of the interior subunits are both engaged in isopeptide bonds with neighboring units. It is worth noting that the highly dynamic nature of S5a binding to polyubiquitin ensures that other configurations are possible.
We produced samples in which either diubiquitin’s proximal or distal subunit is 15N labeled and mixed these at 1:1 molar ratio with S5a (196–306) samples with either Q227C or A298C MTSL labeled. The amide resonance of Q62 from diubiquitin’s proximal subunit was partially attenuated by UIM1 spin labeling, but obliterated by UIM2 spin labeling (Figure 5B, middle panel compared to one on left). Q62 is not attenuated upon S5a (196–306) addition when S5a is not spin labeled (Supplementary Figure 7). Moreover, D21 and A28, which are over 20 Å away from each of S5a’s MTSL labels, are not greatly attenuated, whereas D58, which is ~15 Å away is slightly attenuated (Figure 5B). This finding is consistent with the presence of two S5a:diubiquitin conformations, with the predominant species having UIM2 bound to the proximal subunit and the minor species with it bound to the distal one. Consistent with this model, UIM1 spin labeling resulted in partial attenuation of Q62 from the proximal subunit, but obliterated this residue’s amide resonance from the distal subunit (Figure 5B, middle panel compared to one on left). Similarly, D58 is slightly affected by the induced paramagnetic relaxation effects from S5a MTSL labeling whereas the more remote D21 and A28 are not affected. Altogether, these data are consistent with the NOE-derived S5a:diubiquitin structures of Figure 3A. Confident that our approach is effective, we next tested the effect of having hRpn13 (1–150) present.
hRpn13 (1–150) and S5a (196–306) with either its UIM1 or UIM2 spin labeled was added to diubiquitin with either its proximal or distal subunits 15N labeled. All three proteins were at 0.3 mM concentration. In this case, general resonance broadening is observed due to the larger size of the ternary complex. However, Q62 of the proximal subunit is only slightly more broadened than D21, A28, and D58 when either UIM1 or UIM2 is MTSL labeled (Figure 5B, right panel compared to left one). By contrast, the NMR signal of this residue in the distal subunit is almost obliterated by UIM1 and UIM2 MTSL labeling. The greatly reduced attenuation of the proximal subunit’s Q62 upon Rpn13 addition indicates that S5a’s UIM2 is largely displaced from this subunit by Rpn13, as modeled in Figure 4E. That UIM2 spin labeling causes increased attenuation of Q62 from the distal subunit indicates that it now interacts with this ubiquitin moiety. The UIM2-distal ubiquitin interaction causes partial restoration of Q62 in the sample with UIM1 labeled and hRpn13 present (Figure 5B, right panel compared to one on left). Hence, UIM1 and UIM2 appear to compete for diubiquitin’s distal subunit when Rpn13 is present.
We tested directly whether hRpn13 binds to K48-linked diubiquitin’s proximal subunit when S5a (196–306) is present by substituting G75 of the proximal subunit with cysteine and then labeling it with MTSL. Proximal G75 is not in the immediate Rpn13 binding surface and therefore its mutation is not expected to disrupt binding; however, it is close enough to some residues to cause paramagnetic attenuation when labeled with MTSL (Figure 5C). Addition of proximal G75C MTSL diubiquitin to 15N labeled hRpn13 (1–150) obliterates NMR signals from Rpn13 residues that are close to the proximal subunit in the Rpn13 (1–150):K48-linked diubiquitin complex (Husnjak et al., 2008), as demonstrated for D53 in Figure 5D, middle panel. When this experiment is repeated with unlabeled S5a (196–306) present, the obliteration due to hRpn13 binding to the proximal subunit is preserved (Figure 5D, right panel). These experiments were performed with all three proteins at 0.3 mM. Residues with distances to the MTSL label that are too far for paramagnetic relaxation enhancement effects do not exhibit such dramatic signal attenuation, as demonstrated for T39 in Figure 5D. Moreover, the D53 resonance reappears in the binary and ternary complexes after spin label quenching (Figure 5E). The quenching reaction was done by reducing the MTSL moiety with 5-fold molar excess ascorbic acid (Figure 5E). Therefore, the attenuation observed in Figure 5D is due to the distance-dependent paramagnetic relaxation enhancement induced by MTSL. This data indicates that Rpn13 predominately binds to the proximal subunit when S5a is present, as suggested by the chemical shift perturbation data and the results of spin labeling S5a.
Altogether, our data indicate that Rpn13 and S5a can bind to a common K48-linked polyubiquitin chain, even for diubiquitin, which has only two ubiquitin subunits. In longer chains, each ubiquitin-binding element can readily bind a ubiquitin subunit. The internal subunits of longer chains harbor the advantages of both the proximal and distal subunits of a diubiquitin chain, as K48 and G76 of these subunits are both engaged in isopeptide bonds with neighboring ubiquitin subunits. Internal ubiquitins therefore provide the additional contact of the linker region preferred by S5a’s UIM2 and Rpn13 as well as the increased rigidity of the C-terminal stretch of residues (Fushman et al., 2004), which is preferred by UIM1. A model of Rpn13 and S5a’s UIMs bound to internal and proximal subunits is provided in Figure 5F.
Discussion
We demonstrate here that the coordinated binding of S5a’s two UIMs greatly enhances its affinity for K48-linked diubiquitin over monoubiquitin and that although the UIMs can be accommodated on either subunit, a preference exists for UIM1 on the distal subunit while UIM2 occupies the proximal one. We find that S5a and Rpn13 are able to bind a common diubiquitin chain and that as Rpn13 retains its preferred position at the proximal subunit, S5a binds the distal one. In a diubiquitin chain, the two UIMs compete for the distal subunit when Rpn13 is present; however, in a longer chain these two ubiquitin binding elements would no doubt occupy separate subunits, as they did in Rpn13’s absence (Figure 3). In the absence of the proteasome, our data suggest that multiple binding configurations are available for Rpn13 and S5a binding to K48-linked tetraubiquitin, one of which is shown in Figure 5F.
It is not yet clear whether such coordinated binding occurs in the context of the proteasome; however, it is possible that Rpn13 and S5a bind chains simultaneously to orient substrates for optimal capture and deubiquitination. The distance across the proteasome’s regulatory particle is similar to that spanned by an opened tetraubiquitin structure and therefore, even if S5a and Rpn13 are not docked next to each other it is still conceivable for them to bind a common ubiquitin chain, as modeled in Supplementary Figure 8. The proteasome appears to require most substrates to be conjugated to a chain of four or more ubiquitins (Thrower et al., 2000). This minimal chain length may be preferred to allow for the coordinated binding of ubiquitin receptors, which may in turn, orient the chain in a manner conducive for the effective activity of deubiquitinating enzymes. For example, S5a and Rpn13 coordinated binding may lead to the substrate end of the ubiquitin chain being proximal to Rpn11, which performs degradation-coupled deubiquitination (Maytal-Kivity et al., 2002; Verma et al., 2002; Yao and Cohen, 2002) and the distal end of the chain near Uch37, which performs distal end deubiquitination (Lam et al., 1997). Future experiments are required to test this hypothetical model.
We focused here on K48-linked chains because of their established importance in triggering degradation by the proteasome. S5a can also bind to K63-linkages with no apparent reduction in affinity compared to K48-linked chains (Wang et al., 2005). Rpn13’s ubiquitin binding domain has high affinity for even monoubiquitin (Schreiner et al., 2008), suggesting that it too can bind chains of varying linkage. Although K63-linked chains mediate non-proteasomal events, they can support degradation by the proteasome (Hofmann and Pickart, 2001). We propose that the outcome of K63-linked ubiquitination is not determined by the proteasome’s ubiquitin receptors. Rather, as demonstrated here, S5a and Rpn13 are adaptive and collaborative in their binding to ubiquitin chains, which supports their primary job of enabling proteasome to capture ubiquitinated proteins.
Although contacts were revealed between the UIMs and diubiquitin that were unique to the conjugated subunits, their overall binding mode is similar to that used to bind monoubiquitin. Moreover the two UIMs share the same basic ubiquitin-binding scaffold. The key residues involved in binding to ubiquitin units are 216LALAL220 for UIM1 and 287IAYAM291 for UIM2 and, in both cases residues N-terminal to these helical structures participate in binding ubiquitin (Figure 3). UIM1 is conserved in Saccharomyces cerevisiae, and we propose that UIM2 evolved in higher eukaryotes by UIM1 duplication and optimization for ubiquitin binding, as UIM2 is the stronger binding partner for diubiquitin (Figure 1B and 1C) and monoubiquitin (Wang et al., 2005). We determined S5a (196–306)’s affinity for diubiquitin to be significantly higher than either UIM’s affinity for monoubiquitin (Figure 1E) (Ryu et al., 2003; Wang et al., 2005) and showed that S5a binds diubiquitin with 1:1 stoichiometry even when diubiquitin is 8-fold in excess. Thus, UIM2 is apparently not used to recruit in multiple substrates simultaneously, but rather to enhance affinity for each substrate.
Although not essential in budding yeast (van Nocker et al., 1996), S5a is required for murine embryonic development (Hamazaki et al., 2007). It is also essential in Drosophila melanogaster, in which its deletion results in larval-pupal polyphasic lethality, multiple mitotic defects, as well as accumulation of ubiquitinated proteins and defective 26S proteasome (Szlanka et al., 2003). Therefore, in higher eukaryotes, Rpn13 and the UBL-UBA proteins cannot substitute for S5a.
In a prior study, we demonstrated S5a-bound K48-linked tetraubiquitin can also bind to hHR23a, even though hHR23a precludes additional UBL-UBA family members from binding a common ubiquitin chain (Kang et al., 2007). In this ternary complex, S5a’s UIM1 physically contacts the ubiquitin chain, as UIM2 binds to hHR23a’s UBL domain. S5a therefore appears to be able to share ubiquitinated substrates with other ubiquitin receptors. Altogether, our data support a model in which S5a has evolved to bind polyubiquitinated substrates in a manner that takes advantage of other ubiquitin receptors.
Experimental Procedures
Preparation of NMR samples and NMR spectroscopy
S5a (196–306), K48-linked diubiquitin and hRpn13 (1–150) were produced as described in the Supplement. Methods used for the NMR experiments and their analyses as well as for solving the S5a (196–306):K48-linked diubiquitin complexes and for modeling the S5a (196–306):Rpn13 (1–150):K48-linked tetraubiquitin complex are described in the Supplement.
Analytical ultracentrifugation
Sedimentation velocity experiments were performed as previously described (Kang et al., 2007). Seven samples were prepared in which the molar ratio of K48-linked diubiquitin to S5a (196–306) was varied from 1:7 to 8:1. The sw for each mixture was calculated as previously described (Kang et al., 2007). Dissociation constants were obtained by global analysis using SEDPHAT (Schuck, 2003).
Spin labeling experiments
Based on the S5a (196–306):K48-linked diubiquitin complex structure of the major species (Figure 3A), two S5a (196–306) mutants were designed for MTSL labelling, namely Q227C and A298C. These two residues are not directly involved in binding diubiquitin, so their mutation was not expected to disrupt binding. They were also chosen because they are close enough to K48-linked diubiquitin amide atoms to cause distance-dependent attenuations (Figure 5A). After being treated with the spin labeling reagent (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methane thiosulfonate (MTSL), a single electron (carried by oxygen atom) containing moiety will be attached to the sulfur atom of cysteine residue in S5a mutants. The strong magnetic field of the NMR instrument causes the single electron carried by oxygen atom to accelerate the relaxation of residues within ~20 Å. Sample preparation and NMR spectra recorded for spin labeling experiments are described in Supplementary Material.
Data Bank accession numbers
The atomic coordinates of S5a (196–306) in complex with K48-linked diubiquitin have been deposited in the Protein Data Bank with the accession codes of 2KDE and 2KDF.
Supplementary Material
Acknowledgments
NMR data were acquired in the NMR facility of the University of Minnesota as well as that at the University of Wisconsin at Madison (NMRFAM) and Biomolecular NMR facility at the University of Maryland (UMD). We are grateful to Drs. Klaas Hallenga (NMRFAM) and Dr. Marco Tonelli (NMRFAM) for their technical assistance as well as Dr. Daoning Zhang (UMD) for preparing some diubiquitin samples. Data processing and visualization occurred in the Minnesota Supercomputing Institute Basic Sciences Computing Lab. Analytical ultracentrifugation data was collected at the National Analytical Ultracentrifugation Facility at the University of Connecticut in Storrs. This work was funded by grants from the National Institutes of Health CA097004 (KJW) and GM065334 (DF), the UMN Grant-in-Aid of Research, Artistry, and Scholarship (KJW), and the Minnesota Medical Foundation (KJW, NMR facility).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, Reed SI. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat Struct Biol. 2001;8:417–422. doi: 10.1038/87575. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Finley D, Varshavsky A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell. 1984;37:57–66. doi: 10.1016/0092-8674(84)90300-3. [DOI] [PubMed] [Google Scholar]
- Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GA, Finley D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- Finley D, Ciechanover A, Varshavsky A. Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts85. Cell. 1984;37:43–55. doi: 10.1016/0092-8674(84)90299-x. [DOI] [PubMed] [Google Scholar]
- Fushman D, Varadan R, Assfalg M, Walter O. Determining domain orientation in macromolecules by using spin-relaxation and residual dipolar coupling measurements. Progress NMR Spectroscopy. 2004;44:189–214. [Google Scholar]
- Hamazaki J, Iemura S, Natsume T, Yashiroda H, Tanaka K, Murata S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. Embo J. 2006;25:4524–4536. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazaki J, Sasaki K, Kawahara H, Hisanaga S, Tanaka K, Murata S. Rpn10-mediated degradation of ubiquitinated proteins is essential for mouse development. Mol Cell Biol. 2007;27:6629–6638. doi: 10.1128/MCB.00509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haririnia A, D’Onofrio M, Fushman D. Mapping the interactions between Lys48 and Lys63-linked di-ubiquitins and a ubiquitin-interacting motif of S5a. Journal of molecular biology. 2007;368:753–766. doi: 10.1016/j.jmb.2007.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, Hoeijmakers JH, Hanaoka F. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J Biol Chem. 1999;274:28019–28025. doi: 10.1074/jbc.274.39.28019. [DOI] [PubMed] [Google Scholar]
- Hofmann RM, Pickart CM. In vitro assembly and recognition of Lys-63 polyubiquitin chains. J Biol Chem. 2001;276:27936–27943. doi: 10.1074/jbc.M103378200. [DOI] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Chen X, Lary JW, Cole JL, Walters KJ. Defining how Ubiquitin Receptors hHR23a and S5a Bind Polyubiquitin. J Mol Biol. 2007;369:168–176. doi: 10.1016/j.jmb.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Matiuhin Y, Kirkpatrick DS, Ziv I, Kim W, Dakshinamurthy A, Kleifeld O, Gygi SP, Reis N, Glickman MH. Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol Cell. 2008;32:415–425. doi: 10.1016/j.molcel.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. Embo J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raasi S, Pickart CM. Rad23 UBA domains inhibit 26S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J Biol Chem. 2003;278:8951–8959. doi: 10.1074/jbc.m212841200. [DOI] [PubMed] [Google Scholar]
- Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- Ryu KS, Lee KJ, Bae SH, Kim BK, Kim KA, Choi BS. Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a. The Journal of biological chemistry. 2003;278:36621–36627. doi: 10.1074/jbc.M304628200. [DOI] [PubMed] [Google Scholar]
- Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- Szlanka T, Haracska L, Kiss I, Deak P, Kurucz E, Ando I, Viragh E, Udvardy A. Deletion of proteasomal subunit S5a/Rpn10/p54 causes lethality, multiple mitotic defects and overexpression of proteasomal genes in Drosophila melanogaster. J Cell Sci. 2003;116:1023–1033. doi: 10.1242/jcs.00332. [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker S, Sadis S, Rubin DM, Glickman M, Fu H, Coux O, Wefes I, Finley D, Vierstra RD. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol Cell Biol. 1996;16:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Walters KJ, Kleijnen MF, Goh AM, Wagner G, Howley PM. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry. 2002;41:1767–1777. doi: 10.1021/bi011892y. [DOI] [PubMed] [Google Scholar]
- Wang Q, Goh AM, Howley PM, Walters KJ. Ubiquitin recognition by the DNA repair protein hHR23a. Biochemistry. 2003;42:13529–13535. doi: 10.1021/bi035391j. [DOI] [PubMed] [Google Scholar]
- Wang Q, Young P, Walters KJ. Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol. 2005;348:727–739. doi: 10.1016/j.jmb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Wilkinson CR, Ferrell K, Penney M, Wallace M, Dubiel W, Gordon C. Analysis of a gene encoding Rpn10 of the fission yeast proteasome reveals that the polyubiquitin-binding site of this subunit is essential when Rpn12/Mts3 activity is compromised. J Biol Chem. 2000;275:15182–15192. doi: 10.1074/jbc.275.20.15182. [DOI] [PubMed] [Google Scholar]
- Wilkinson CR, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.