Abstract
Background & Aims
Intestinal epithelial cells and the myenteric plexus of the mouse gastrointestinal tract contain a circadian clock–based intrinsic timekeeping system. Because disruption of the biological clock has been associated with increased susceptibility to colon cancer and gastrointestinal symptoms, we aimed to identify rhythmically expressed genes in the mouse distal colon.
Methods
Microarray analysis was used to identify genes that were rhythmically expressed over a 24-hour light/dark cycle. The transcripts were then classified according to expression pattern, function, and association with physiologic and pathophysiologic processes of the colon.
Results
A circadian gene expression pattern was detected in approximately 3.7% of distal colonic genes. A large percentage of these genes were involved in cell signaling, differentiation, and proliferation and cell death. Of all the rhythmically expressed genes in the mouse colon, approximately 7% (64/906) have been associated with colorectal cancer formation (eg, B-cell leukemia/lymphoma-2 [Bcl2]) and 1.8% (18/906) with various colonic functions such as motility and secretion (eg, vasoactive intestinal polypeptide, cystic fibrosis transmembrane conductance regulator).
Conclusions
A subset of genes in the murine colon follows a rhythmic expression pattern. These findings may have significant implications for colonic physiology and pathophysiology.
Circadian rhythms are cyclic changes in physiologic parameters that occur over approximately 24 hours. The gastrointestinal tract exhibits several circadian biological rhythms. For example, gastric acid secretion, gastric emptying, digestive enzyme expression and activity along the crypt-villus axis, and response to certain drugs used in the treatment of patients with colon cancer vary substantially with the time of day.1,2 Colonic motility follows a rhythm as well. Healthy people have bowel movements during the day, frequently following awakening or following a meal but rarely during the night.
A central pacemaker is located in the hypothalamic suprachiasmatic nucleus of the brain, which receives direct photic input via the retinohypothalamic tract. This central pacemaker or clock communicates with peripheral tissues via neuronal and humoral pathways by either driving rhythmic activity or, more likely, entraining peripheral oscillators, controlling the expression of a subset of tissue-specific processes and genes, thereby regulating organ-specific physiologic functions. It has been estimated that approximately 8%–10% of genes in peripheral organs are expressed in a rhythmic manner, suggesting that they are either directly or indirectly controlled by clock genes.
The molecular basis for biological rhythms is believed to be regulated by a set of so-called “clock genes” and their products within the suprachiasmatic nucleus as well as peripheral tissues. Clock genes are a group of genes that participate in an interlocked transcription-translation feedback loop, in which the “positive elements” (CLOCK and brain and muscle Arnt-like protein 1 [BMAL1]) form heterodimers and enter the nucleus. There, they bind to the promoter regions of the “negative elements” (Period genes [Per] and Cryptochrome genes [Cry]) and activate their transcription. The PER and CRY proteins slowly accumulate as heterodimers and feed back to inhibit CLOCK-BMAL1– dependent transcription, thereby closing the central loop.3 A second interlocking loop involving the transcription of retinoic acid orphan-like receptor a (Rora) and Reverbα, which are also activated by CLOCK-BMAL1, amplifies the central loop by regulation of Bmal1 transcription. The 2 interlocking loops are believed to generate the approximately 24-hour period of the molecular oscillator.4
In addition, these clock genes and their products can directly or indirectly regulate the transcription of a subset of tissue-specific genes,3 affecting the output path-way(s) of the molecular clock. Previous gene array studies have suggested that approximately 5%–15% of genes in peripheral organs follow a rhythmic expression profile.5 We have recently shown that clock genes are rhythmically expressed in the mouse gastrointestinal tract and are capable of shifting their phase of expression in response to changes in the feeding cycle.6 Within the gastrointestinal tract, clock genes are expressed predominantly in the epithelial cells and the myenteric plexus, suggesting a possible role for clock genes in the coordination of cell proliferation, differentiation, and motility.6 In addition, clock genes may drive the rhythmic expression of the sodium-hydrogen exchanger NHE3 in the rat colon, suggesting a possible role for rhythmic regulation of salt absorption as well.7
Several clinical observations suggest a role for clock genes in the pathogenesis of gastrointestinal symptoms and disease. First, disruption of our cyclic environment secondary to shift work, time zone traveling, or space flights is associated with gastrointestinal symptoms such as abdominal discomfort, constipation, or diarrhea.8 –12 Second, irritable bowel syndrome is more common in nurses participating in shift work.13 Finally, the gastrointestinal clock may play an important role in the pathogenesis of colon cancer. Data from the Nurses’ Health Study showed that women who worked at least 3 nights per month for 15 or more years had a significantly greater risk of developing colorectal cancer when compared with women who never worked rotating night shifts.14 On the basis of such studies, the International Agency for Research on Cancer, the cancer arm of the World Health Organization, considers overnight shift work a “probable carcinogen.”15
Based on these observations, we hypothesized that a subset of genes in the gastrointestinal tract is transcriptionally regulated by the products of clock genes. These genes are likely to be involved in the regulation of rhythmic gastrointestinal events such as motility, cell differentiation, and proliferation. This study therefore aimed to determine temporal patterns of gene expression in the mouse distal colon. This profiling of gene expression was used to (1) determine the extent to which the transcriptome was rhythmically expressed in the distal colon and (2) characterize the functional distribution of rhythmically expressed genes within metabolic and signaling pathways relevant to gastrointestinal physiology.
Materials and Methods
Animals
Mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Male C57BL/6J mice were used for all experiments. Mice were 8 –12 weeks of age at the onset of the experiment with body weights of 20 –25 g. Experimental protocols involving animals were approved by the institutional animal care and use committee in accordance with the guidelines provided by the National Institutes of Health.
Time-of-Day Experiments
Mice were maintained for 2 weeks on a 12-hour light/dark cycle (lights on at 7 AM, lights off at 7 PM) before all experiments. Throughout this report, time is indicated using Zeitgeber time (ZT) as the indicator for the phase of the rhythm, whereby ZT0 refers to the time that lights went on (7 AM) and ZT12 refers to the time that lights went off (7 AM). Samples were collected every 4 hours starting at ZT1 with n = 3 mice per time point for all experiments. For the reference experiment, mice had ad libitum access to food and were kept under a strict 12-hour light/dark cycle. For a rhythm to be considered circadian, it has to persist in noncycling environmental conditions. Therefore, for the clock genes and a subset of colonic genes, we assessed rhythmicity in the absence of light or feeding. To remove the effect of light, mice were released into constant darkness for a total of 48 hours with ad libitum access to food. To remove the effect of feeding, mice were released into constant darkness for a total of 24 hours while fasting for solids but with ad libitum access to water. Under constant darkness, circadian time was referenced to the previous ZT such that the beginning of the subjective day was labeled ZT0 and the beginning of the subjective night was labeled ZT12.
Total RNA Extraction
The distal colon was identified following laparotomy and resected approximately 0.5 cm above the anus. Total RNA was isolated by modified guanidinium thio-cyanate/phenol/chloroform extraction method and was treated with deoxyribonuclease I (Promega, Madison, WI) at 37°C for 30 minutes. RNA samples were quantified using a NanoDrop Spectrophotometer (NanoDrop Technologies, Wilmington, DE) and qualified by analysis on an RNA Nano Chip using the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc, Santa Clara, CA).
Affymetrix GeneChip Analysis
Gene expression analysis was completed by the Molecular Genomics Core at the University of Texas Medical Branch using Affymetrix GeneChips mouse 430 2.0 (Affymetrix, Santa Clara, CA) according to protocols described in the Affymetrix GeneChip Expression Analysis Technical Manual. In brief, first-strand complementary DNA was synthesized from 15 μg of total RNA using an oligo(dT) primer encoding a bacteriophage T7 RNA polymerase promoter (SuperScript II Reverse Transcriptase; Invitrogen, Carlsbad, CA). Samples were prepared with varying concentrations of prokaryotic genes dap, thr, lys, phe (poly (A) controls that serve to address technical adequacy of the labeling and transcription process) and bio B, bio C, bio D, and cre (to address array sensitivity). Following second-strand synthesis, in vitro transcription was performed using T7 RNA polymerase in the presence of biotin-labeled nucleotides. Hybridization of the arrays was performed at 45°C for 16 hours in 0.1 mol/L 2-N-morpholinoethanesulfonic acid (0.1 mol/L; pH 6.6) sodium chloride (1 mol/L), EDTA (0.02 mol/L), and 0.01% Tween 20. Arrays were scanned using a Gene Array Scanner (Hewlett Packard, Houston, TX) and analyzed using the Affymetrix GeneChip Analysis Suite (GCOS v1.4) software.
Quantitative Reverse-Transcriptase Polymerase Chain Reaction
Validation of expression analysis of specific genes was performed using quantitative polymerase chain reaction (PCR). Synthesis of complementary DNA from distal colonic RNA was performed using the High Capacity cDNA Reverse Transcription Kit from Applied Biosystems (ABI, Foster City, CA) according to the manufacturer’s protocol.
PCR primers and probes for the individual clock genes were obtained from ABI. Data were normalized using TaqMan Ribosomal RNA Control Reagents from ABI. The TaqMan Universal PCR Master Mix with UNG from ABI was used to prepare the assays according to the manufacturer’s protocol. Real-time PCR was performed on an Eppendorf real-time PCR machine (Eppendorf North America, Westbury, NY). All reactions were performed in 96-well plates. Thermal cycling conditions were 50°C for 2 minutes, 95°C for 10 minutes, and 40 cycles at 95°C for 15 seconds and 60°C for 1 minute. Each gene-specific PCR was performed in duplicate.
Western Blotting
Western blotting was completed as previously described.6 Antibodies used for Western blotting were BCL2 (Chemicon, Billerica, MA) 1:1000, cystic fibrosis transmembrane conductance regulator (CFTR; Abcam, Cambridge, MA) 1:500, neuronal nitric oxide synthase (nNOS; Abcam) 1:1000, and actin (Santa Cruz Biotechnology Inc, Santa Cruz, CA) 1:1000.
Data Analysis
Array preprocessing
Raw probe-level expression data (CEL files) were normalized using the GC-Robust Multichip analysis method. GC-Robust Multichip analysis models intensity of probe-level data as a function of GC contents based on the theory that probes with high GC contents will have increased binding. The theory above with additional steps from the RMA background correction methods, quantiles normalizations. Medianpolish was used to summarize the probe sets. Probe sets absent across all the chips were filtered out before differential expression testings. S+ Array Analyzer 2.1, a module in S-PLUS 7.0 (Insightful Inc, Seattle, WA), was used for background correction, summarization, and normalization.
Array analysis
To identify genes with statistically significant rhythmic expression, preprocessed array data were analyzed by maSigPro (microarray Significant Profiles), a 2-step regression-based method for the analysis of single and multiple time series microarray experiments.16 The first step is a gene selection step that applies the least-square technique to estimate the parameters of regression models and calculate the analysis of variance table for each gene. The P value associated with F statistics for each gene is used to select significant genes. The P value is corrected for the multiple comparisons using Benjamini & Hochberg, a false discovery rate method. The second step is a variable selection step that applies the stepwise regression approach to identify statistically significant profiles based on the R2 value of the second regression model between the experimental groups.16 The maSigPro analysis identified 906 significant genes at a false discovery rate of 0.01 and an R2 threshold of 0.7.
Array expression clusters
Results were visualized using the clustering algorithm in maSigPro. The clusters were obtained using the estimated regression coefficients, which help discard the noise from the original data. With this approach, multiple genes were identified with statistically significant rhythmic expression over a 24-hour period. To identify those genes with similar expression patterns, a single output of genes was created using vars = “all” in get.siggenes function and then clustered by the values of their regression coefficients using the parameter cluster.data = 2 in the function see.genes. Clusters with homogeneous patterns were identified that respond to similar significant models, because clustering is not based on the original expression patterns but on the coefficients obtained by the modeling function. All genes with similar rhythmic expression patterns fit in 7 different clusters. When categorizing rhythmically expressed genes by function, most genes were found to be involved in cell signaling and cell cycle regulation.
To identify those genes of interest to gastroenterological researchers, we searched the PubMed database for each rhythmically expressed gene against “colon.” Only those genes described in the literature in association with the main physiologic and pathophysiologic events of the colon (motility, colorectal cancer formation, and inflammation/inflammatory bowel disease) are listed in Table 1.
Table 1.
Rhythmically Expressed Genes Associated With Colon Cancer and Various Colonic Functions Including Motility and Secretion
| Category | Gene symbol | Gene title |
|---|---|---|
| DNA repair, cell differentiation and proliferation | Alkbh1 | alkB, alkylation repair homologue 1 (Escherichia coli) |
| E2F2 | E2F transcription factor 2 | |
| Klf9 | Kruppel-like factor 9 | |
| Rad17 | RAD17 homologue (Schizosaccharomyces pombe) | |
| Top1 | Topoisomerase (DNA) I | |
| Transcription regulators | HIF1a | hypoxia inducible factor 1, α subunit |
| Arnt | aryl hydrocarbon receptor nuclear translocator | |
| Atp2a2 | ATPase, Ca2+transporting, cardiac muscle, slow twitch 2 | |
| ATR | ataxia telangiectasia and Rad3 related | |
| Mbd1 | methyl-CpG binding domain protein 1 | |
| Apoptosis regulators | BCl2 | B-cell leukemia/lymphoma |
| Becn1 | beclin 1 (coiled-coil, myosin-like BCL2-interacting protein) | |
| Bfar | bifunctional apoptosis regulator | |
| Birc4 | baculoviral IAP repeat-containing 4 | |
| Casp4 | caspase 4, apoptosis-related cysteine peptidase | |
| Rnf7 | ring finger protein 7 | |
| Oncogenes | Braf | Braf transforming gene |
| Ccne1 | cyclin E1 | |
| Drctnnb1a | down-regulated by Ctnnb1, a | |
| k-ras | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homologue | |
| Nras | neuroblastoma ras oncogene | |
| Pim3 | proviral integration site 3 | |
| Wnt/β-catenin signaling | Bcr | breakpoint cluster region homologue |
| Ctnna1 | catenin (cadherin-associated protein), α1 | |
| Ereg | epiregulin | |
| Mycbp | c-myc binding protein | |
| TCF4 | transcription factor 4 | |
| Wnt6 | wingless-related MMTV integration site 6 | |
| Ccl28 | Chemokine (C-C motif) ligand 28 | |
| Genes associated with colorectal cancer | Cd1d1 | CD1d1 antigen |
| Cks2 | CDC28 protein kinase regulatory subunit 2 | |
| Cldn8 | claudin 8 | |
| Col4A5 | procollagen, type IV, α5 | |
| Col5a2 | procollagen, type V, α2 | |
| Col6a3 | procollagen, type VI, α3 | |
| Cxcl12 | chemokine (C-X-C motif) ligand 12 | |
| Cyld | cylindromatosis (turban tumor syndrome) | |
| Dek | DEK oncogene (DNA binding) | |
| GHR | growth hormone receptor | |
| Hes6 | hairy and enhancer of split 6 (Drosophila) | |
| Hhip | Hedgehog-interacting protein | |
| Hnrpa1 | heterogeneous nuclear ribonucleoprotein A1 | |
| IGF1R | insulin-like growth factor I receptor | |
| Incenp | inner centromere protein | |
| Ivd | isovaleryl coenzyme A dehydrogenase | |
| Kitl | kit ligand | |
| Mcc | mutated in colorectal cancers | |
| mki67 | antigen identified by monoclonal antibody Ki 67 | |
| Mobk1b | MOB1, Mps One Binder kinase activator-like 1B | |
| Mrpl12 | mitochondrial ribosomal protein L12 | |
| Nap1l1 | nucleosome assembly protein 1-like 1 | |
| Ndufc2 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2 | |
| Nfat5 | nuclear factor of activated T-cells 5 | |
| Nup88 | nucleoporin 88 | |
| Pde4b | phosphodiesterase 4B, cAMP specific | |
| Pfkfb3 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | |
| Phf14 | PHD finger protein 14 | |
| Sdccag1 | serologically defined colon cancer antigen 1 | |
| Snap25 | synaptosomal-associated protein 25 | |
| Thbs1 | thrombospondin 1 | |
| VEGFA | vascular endothelial growth factor A | |
| Inflammation/motility/secretion | Atg16l1 | autophagy-related 16-like 1 |
| Cftr | cystic fibrosis transmembrane conductance regulator homologue | |
| Chuk | conserved helix-loop-helix ubiquitous kinase | |
| HIF1a | hypoxia inducible factor 1, α subunit | |
| Hspb1 | heat shock protein 1 | |
| Ikbkb | inhibitor of κB kinase β | |
| Il18 | interleukin 18 | |
| Irak4 | interleukin-1 receptor-associated kinase 4 | |
| Mki67 | antigen identified by monoclonal antibody Ki 67 | |
| Nr5a2 | nuclear receptor subfamily 5, group A, member 2 | |
| Pde4b | phosphodiesterase 4B, cAMP specific | |
| Vdac1 | voltage-dependent anion channel 1 | |
| Htr7 | 5-hydroxytryptamine (serotonin) receptor 7 | |
| Itpr1 | inositol 1,4,5-triphosphate receptor 1 | |
| Nos1 | nitric oxide synthase 1, neuronal | |
| VIP | vasoactive intestinal polypeptide |
PCR
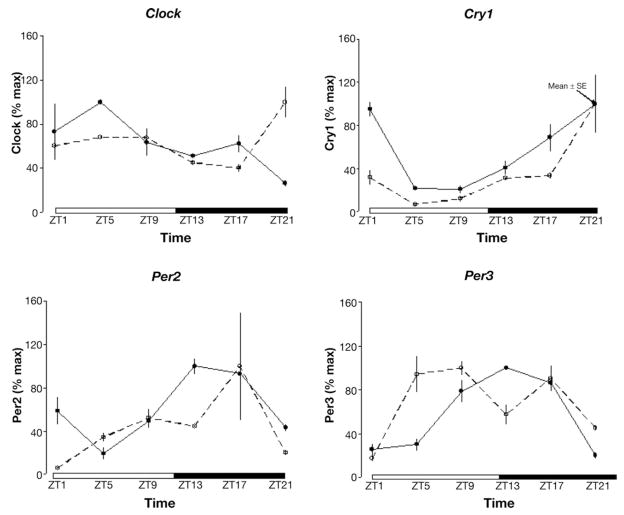
Gene expression was analyzed using relative PCR amplification analysis method (2−ΔΔCt). Changes in gene expression were referenced to the level of expression at the ZT1 time point. Rhythmicity was assessed by the zero amplitude test using the single cosinor method to fit a 24-hour cosine curve to the data and, in the case of vasoactive intestinal polypeptide (VIP) and nNOS, a 2-component model consisting of cosine curves with periods of 12 hours.17 Parameter tests were used to test the equality of circadian MESORs, amplitudes, and/or acrophases of gene expression assessed by quantitative PCR or maSigPro as previously described.6,17 Data were presented by expressing individual values for both array and quantitative PCR data as a percentage of maximum expression, which allowed for array and PCR data and standard error bars to be graphically presented in the same figure (Figure 2).
Figure 2.
Array data (solid line) and quantitative PCR (dotted line) for a subset of clock genes. Data represent mean fold change ± SE as percentage of maximum expression.
Results
Rhythmic gene expression profiles were detected in approximately 3.7% of distal colonic genes (1248 of ≈34,000 total mouse genes). These rhythmically expressed genes oscillate in a time-dependent manner as graphically presented via a gene tree (Figure 1A). The clustering analysis showed a diverse pattern of messenger RNA (mRNA) expression with 7 clusters of peaking transcript abundance over a 24-hour period (Figure 1B). Clusters 1 and 2 are indicative of transcripts that peak at the end of the dark cycle (ZT22), cluster 2 peaks toward the end of the light (ZT9) and the end of the dark cycle (ZT22), clusters 3 and 6 peak at the mid-dark cycle (ZT17), clusters 4 and 5 peak at the onset of the light cycle (ZT1), and cluster 7 peaks at the onset of the dark cycle (ZT13). Upon correction and removal of transcripts for redundancy and unknown/unclassified transcripts, the group of genes expressed in a rhythmic manner was reduced to 906 unique classified genes (Supplementary Table 1; see Supplementary material online at www.gastrojournal.org). Of these unique classified genes, 71.3% (646/906) were grouped under cluster 1, 8.0% (73/906) under cluster 2, 7.7% (70/906) under cluster 3, 3.2% (29/906) under cluster 4, 3.5% (32/906) under cluster 5, 6.0% (55/906) under cluster 6, and 0.1% (1/906) under cluster 7.
Figure 1.
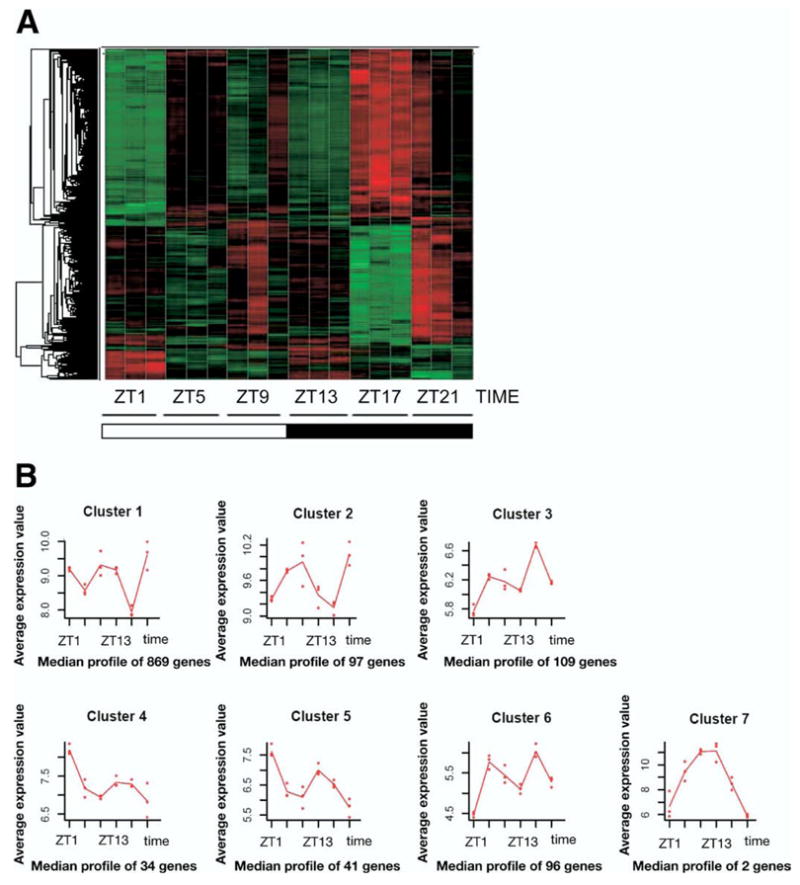
(A) Gene tree of colonic transcripts. Increased transcript abundance is presented in red and decreased abundance in green. (B) Cluster patterns. For each cluster, a trace is shown indicating the average profile of all oscillating transcripts in each respective cluster.
Clock Genes
The validity of the gene array analysis was confirmed by the identification of clock genes as genes with significant rhythmicity. Further confirmation was obtained through PCR analyses that were completed on the same samples. Both data were placed in the same figure (Figure 2). In addition, we used cosinor analysis on array as well as PCR data (cosinor curves not shown). When using parameter testing to compare circadian amplitudes for Clock and Cry1, no statistically significant difference was detected between the 2 methods. Similarly, when using parameter testing to compare circadian phase for Per2 and Per3, no statistically significant difference in phase was detected between the 2 methods.
Furthermore, the transcripts for the PAR-domain basic leucine zipper transcription proteins albumin D-site-binding protein (DBP), the thyrotroph embryonic factor (TEF), and the hepatic leukemia factor (HLF), which are directly regulated by clock genes, thereby serving as a direct indicator of clock gene output, exhibited robust rhythmic expression (Supplementary Table 1).
Functional Categories of Rhythmically Expressed Transcripts
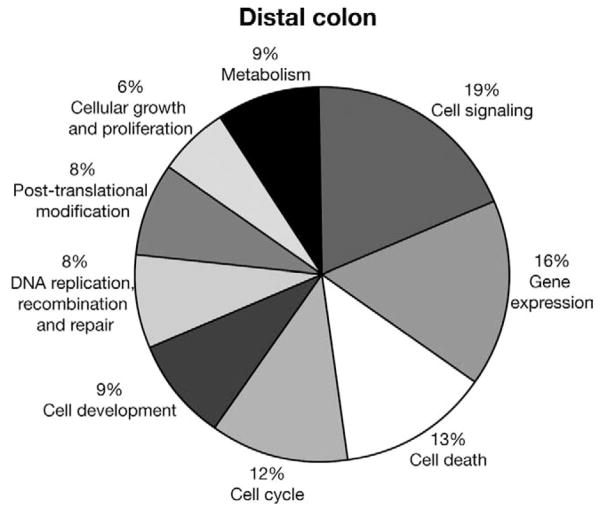
The majority of rhythmically expressed genes were involved in cell signaling, growth, and proliferation and cell death (Figure 3).
Figure 3.
Pie chart of functional categories of rhythmically expressed genes in mouse colon.
Because clock genes are predominantly expressed in epithelial cells and the myenteric plexus, rhythmically expressed genes were organized based on a potential role in the pathophysiology of colon cancer, inflammatory bowel disease, and gastrointestinal motility (Table 1). Of the total number of rhythmically expressed genes in the mouse colon, approximately 7% (62/906) have been associated with colorectal cancer formation and 1.8% (18/906) with various gastrointestinal functions such as motility and secretion (eg, VIP, CFTR). The majority of genes associated with colon cancer were expressed in cluster 1 (70.9% [44/62] and 50% [6/12], respectively). Examples of rhythmically expressed genes are shown in Figure 4 for 2 genes from each category. Other rhythmically expressed gene transcripts are shown in Supplementary Table 1.
Figure 4.
Subset of rhythmically expressed genes. (A) Genes involved in colon cancer formation, (B ) genes involved in inflammation, and (C) genes involved in other gastrointestinal functions such as motility and secretion. Data represent mean fold change ± SE as percentage of maximum expression.
Circadian Rhythmicity in the Mouse Colon
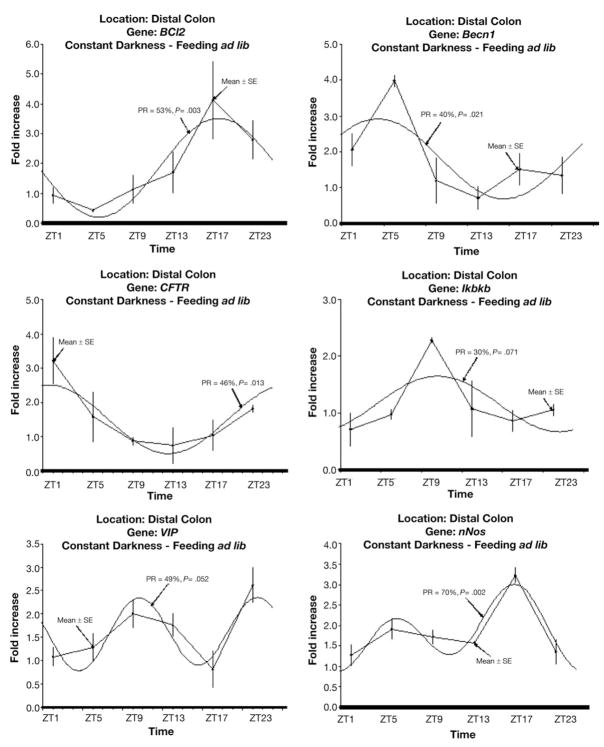
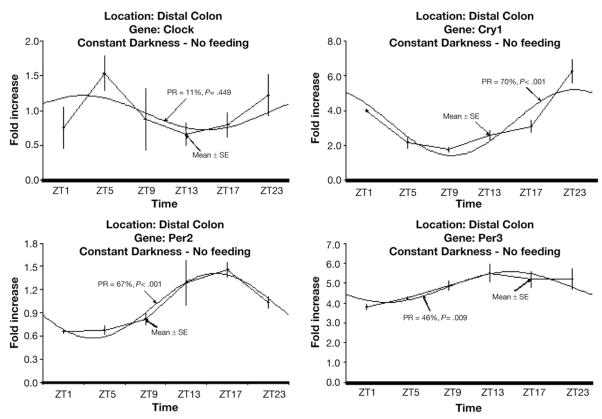
Rhythms that are circadian should persist in the absence of environmental stimuli such as the light/dark cycle and feeding. We previously showed that colonic clock gene expression persists in the absence of light. To determine whether our colonic genes of interest were potentially driven by clock genes, mice were placed in constant darkness to eliminate the effect of light as a synchronizer of gene expression. Under constant darkness, this subset of genes maintained rhythmic expression (Figure 5), supporting a role for clock genes in their transcriptional regulation.
Figure 5.
Daily profile of gene mRNA levels and corresponding fitted cosinor curve under constant darkness with ad libitum access to food. mRNA levels are expressed as means ± SE. PR, percentage rhythms; P value, probability from zero-amplitude (no rhythm) test.
Because of the possible effect of feeding on rhythmic gene expression, we then determined the effect of fasting on clock gene expression. Fasting under constant darkness did not substantially alter clock gene expression (Figure 6), suggesting that colonic clock gene oscillations are independent of intraluminal contents. Fasting under constant darkness did not change the rhythmic expression of other colonic genes such as VIP and nNOS either (Supplementary Figure 1; see Supplementary material online at www.gastrojournal.org).
Figure 6.
Daily profile of colonic clock gene mRNA levels and corresponding fitted cosinor curve under constant darkness and fasting conditions. mRNA levels are expressed as means ± SE. PR, percentage rhythms; P value, probability from zero-amplitude (no rhythm) test.
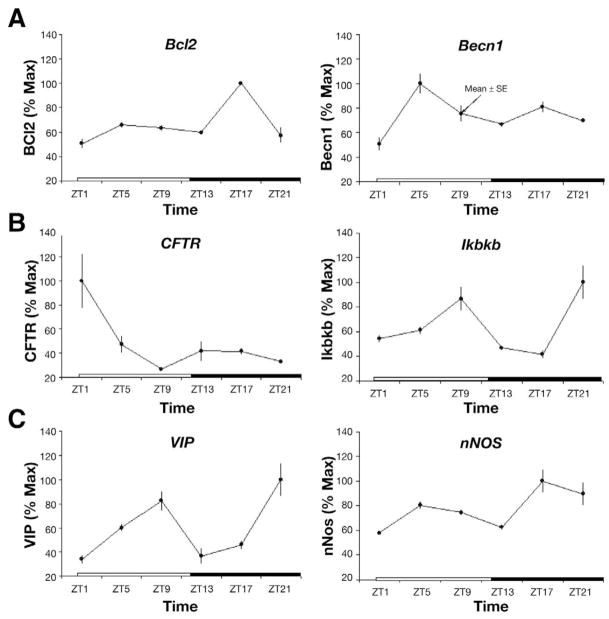
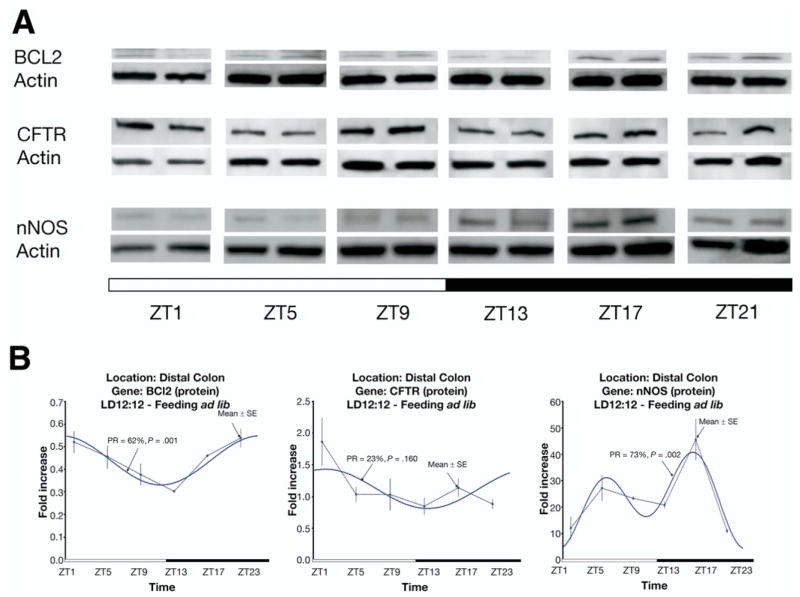
Protein Expression Patterns of Rhythmically Expressed Transcripts
To determine whether a subset of rhythmically expressed genes demonstrated rhythmic expression at the protein level as well, Western analyses were completed for BCL2, CFTR, and nNOS. Rhythmic protein expression was detected for BCL2 and nNOS but not for CFTR (Figure 7).
Figure 7.
(A) Western blotting for proteins (BCL2, CFTR, and nNOS) in mice with ad libitum access of food under a regular light/dark cycle. (B) Quantitative analysis of protein expression and corresponding fitted cosinor curve. Protein levels are expressed as means ± SE. PR, percentage rhythms; P value, probability from zero-amplitude (no rhythm) test.
Discussion
This study shows that a subset of genes in the distal colon is expressed in a rhythmic manner. The circadian expression of clock genes such as Clock, Per2, Per3, and Cry1 was consistent with previously characterized circadian clock gene expression in a variety of peripheral tissues such as the heart, liver, and adipose tissue18,19 as well as the gastrointestinal tract.6 In addition, the oscillation of Clock occurred in antiphase to those of Per and Cry, consistent with the autoregulatory mechanisms of the circadian clock. These observations suggest that our technical approach to the gene array studies and the statistical analyses represent a valid method for the identification of rhythmically expressed genes.
Several expression clusters demonstrated biphasic peaks in transcript expression. Biphasic patterns in rhythmic expression and physiology have been previously observed in other tissues such as the pineal gland.20 It raises the question of dual regulation of genes by endogenous and exogenous mechanisms. For example, a particular gene may be directly or indirectly controlled by the molecular clock mechanism (endogenous mechanism), thereby allowing the organ to be optimally prepared for its anticipated function for a particular time of the day. However, the same gene must be capable of responding to an unanticipated event at other times of the day as well (exogenous mechanism). This may explain why most array studies do not find clock-controlled genes to follow a perfect bell-shaped expression pattern.
The functional categorization of rhythmically expressed genes in this study identified a substantial number of genes in the mouse distal colon associated with cell replication. This is in line with previous thymidine incorporation studies that demonstrated rhythmic changes in DNA replication in mice as well as in the rectal mucosa of humans.21–23
The majority of colon cancer–associated genes was part of cluster 1, which had its peak expression at the end of the dark cycle. Thus, these genes are in antiphase with the Per genes, which peak at the beginning of the dark cycle, thereby suggesting that their transcription is more likely to be under indirect clock gene control and/or suppressed by PER activity. In contrast, the Bcl2 gene appeared cotranscribed (in phase) with Per2, suggesting potential direct control of transcriptional regulation by the CLOCK/BMAL1 dimer. Indeed, Bcl2 has an E-box in its promoter region, which could allow for direct clock-controlled transcriptional regulation.24 This observation has an important pharmacologic implication as well. Bcl2 enhances oncogenesis by preventing programmed cell death (apoptosis). Therapeutic strategies aiming at the promotion of apoptosis through modulation of Bcl2 RNA levels should therefore take their temporal pattern of expression into account.
A subset of rhythmically expressed genes in the distal colon has been associated with inflammation. Of those, the more intriguing genes are CFTR, which have a crucial role in the regulation of intestinal electrolyte homeostasis and whose expression is modulated by colonic inflammation,25 and the inhibitor of κB kinase β. CFTR expression peaks at the beginning of the light cycle, in anti-phase with Per gene expression, again suggesting indirect clock-controlled transcriptional regulation and/or PER activity. Inhibitor of κB kinase β phosphorylates serine residues on the IκB proteins, marking them for destruction via the ubiquitination pathway, thereby allowing activation of the nuclear factor κB complex.26 Little is known about the role of rhythmic gene expression in colonic inflammation. However, it has been suggested that chronic inflammation can alter clock gene function and that clock genes may directly regulate immune function.27–29 Because clock gene expression has been identified in colonic epithelial cells,6 it is conceivable that clock genes play a significant role in the regulation of the host immune defense.
This study identified VIP and nNOS as being rhythmically expressed. The rhythmic expression of these genes persisted in the absence of external stimuli such as light and feeding and is therefore likely to be transcriptionally regulated by the molecular clock. VIP and nNOS are expressed in the myenteric plexus.30,31 Because previous data from our laboratory have shown that clock genes are expressed in the myenteric plexus, it is conceivable that clock genes may play a role in the modulation of colonic motility.
To date, few studies have determined to what extent the proteome is rhythmically expressed. We found that BCL2 and nNOS but not CFTR oscillate at the protein level. However, it has been well documented that the presence of rhythmically expressed mRNA levels does not necessarily translate into rhythmically expressed protein levels. For example, rhythmic regulation of protein function may actually occur at the posttranscriptional level through protein phosphorylation.32 These findings highlight the possible importance of rhythmically controlled posttranslational modifications for the regulation of colonic physiology.
In a recent editorial by Scheving and Russell, the authors recognized the importance of circadian clocks and biological rhythms in the interpretation of experimental results.33 In fact, the authors recommended that the “time of experiments should routinely be included in the methods section in a similar fashion to the current reportage of animal gender, strain, and age.” The findings of this study reemphasize this point. First, quantification of RNA or protein expression is routinely used in the assessment of gene function. However, because these findings may differ depending on time of day, misleading interpretations or contradicting findings are bound to occur. Second, the majority of rodent studies are still completed during the day (the time of their natural sleep cycle), which may impact on physiologic findings as well.
This study has several limitations. First, a subset of clock genes such as Per1 and Bmal1 was not identified as following a rhythmic expression pattern on our gene array studies. However, subsequent analyses using PCR on the same samples indicated both Per1 and Bmal1 to be rhythmic in the mouse distal colon (data not shown). This inconsistent result may represent an underestimation in the amount of PCR product identified by gene array as previously suggested by others and/or inter-mouse variation in gene expression levels rendering a rhythmic pattern as statistically insignificant.20 Second, our study aimed to identify a cohort of rhythmically expressed genes in the mouse distal colon under normal light/dark conditions. The colonic transcriptome in this study therefore includes genes that follow circadian (ie, endogenously controlled) as well as diurnal (ie, light-dependent) rhythms. We are ultimately interested in the relevance of rhythmically expressed genes in human disease. Thus, we chose the light/dark cycle, an environment in which humans normally exist.
In summary, the present study confirms the rhythmic expression of a subset of genes in the mouse distal colon. These findings should initiate further studies into the role of clock genes in gastrointestinal physiology and pathophysiology.
Supplementary Material
Acknowledgments
The authors thank Chung Owyang, MD, for his critical review of the manuscript and Brenda Vibbart for her administrative assistance and acknowledge the National Institute of Bioinformatics (www.inab.org), a platform of Genoma España. The accession number of repository for expression microarray data is series record GSE10644. The following link has been created to allow review of GSE10644: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=jhefhwuksymyuro&acc=GSE10644.
The authors disclose the following: Supported by grant R21 DK074477-01A1 (to W.A.H.).
Abbreviations used in this paper
- CFTR
cystic fibrosis transmembrane conductance regulator
- nNOS
neuronal nitric oxide synthase
- PCR
polymerase chain reaction
- ZT
Zeitgeber time
Footnotes
Supplementary Data
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at doi: 10.1053/j.gastro.2008.08.048.
References
- 1.Hoogerwerf WA. Biologic clocks and the gut. Curr Gastroenterol Rep. 2006;8:353–359. doi: 10.1007/s11894-006-0019-3. [DOI] [PubMed] [Google Scholar]
- 2.Scheving LA. Biological clocks and the digestive system. Gastroenterology. 2000;119:536–549. doi: 10.1053/gast.2000.9305. [DOI] [PubMed] [Google Scholar]
- 3.Bell-Pedersen D, Cassone VM, Earnest DJ, et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 5.Albrecht U, Eichele G. The mammalian circadian clock. Curr Opin Genet Dev. 2003;13:271–277. doi: 10.1016/s0959-437x(03)00055-8. [DOI] [PubMed] [Google Scholar]
- 6.Hoogerwerf WA, Hellmich HL, Cornelissen G, et al. Clock gene expression in the murine gastrointestinal tract: endogenous rhythmicity and effects of a feeding regimen. Gastroenterology. 2007;133:1250–1260. doi: 10.1053/j.gastro.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Sladek M, Rybova M, Jindrakova Z, et al. Insight into the circadian clock within rat colonic epithelial cells. Gastroenterology. 2007;133:1240–1249. doi: 10.1053/j.gastro.2007.05.053. [DOI] [PubMed] [Google Scholar]
- 8.Caruso CC, Lusk SL, Gillespie BW. Relationship of work schedules to gastrointestinal diagnoses, symptoms, and medication use in auto factory workers. Am J Ind Med. 2004;46:586–598. doi: 10.1002/ajim.20099. [DOI] [PubMed] [Google Scholar]
- 9.Cassone VM, Stephan FK. Central and peripheral regulation of feeding and nutrition by the mammalian circadian clock: implications for nutrition during manned space flight. Nutrition. 2002;18:814–819. doi: 10.1016/s0899-9007(02)00937-1. [DOI] [PubMed] [Google Scholar]
- 10.Costa G. The impact of shift and night work on health. Appl Ergon. 1996;27:9–16. doi: 10.1016/0003-6870(95)00047-x. [DOI] [PubMed] [Google Scholar]
- 11.Knutsson A. Health disorders of shift workers. Occup Med (Lond) 2003;53:103–108. doi: 10.1093/occmed/kqg048. [DOI] [PubMed] [Google Scholar]
- 12.Vener KJ, Szabo S, Moore JG. The effect of shift work on gastrointestinal (GI) function: a review. Chronobiologia. 1989;16:421–439. [PubMed] [Google Scholar]
- 13.Lu W, Gwee KA, Ho KY. Functional bowel disorders in rotating shift nurses may be related to sleep disturbances. Eur J Gastroenterol Hepatol. 2006;18:623–627. doi: 10.1097/00042737-200606000-00008. [DOI] [PubMed] [Google Scholar]
- 14.Schernhammer ES, Laden F, Speizer FE, et al. Night-shift work and risk of colorectal cancer in the Nurses’ Health Study. J Natl Cancer Inst. 2003;95:825–828. doi: 10.1093/jnci/95.11.825. [DOI] [PubMed] [Google Scholar]
- 15.Straif K, Baan R, Grosse Y, et al. Carcinogenicity of shift-work, painting, and fire-fighting. Lancet Oncol. 2007;8:1065–1066. doi: 10.1016/S1470-2045(07)70373-X. [DOI] [PubMed] [Google Scholar]
- 16.Conesa A, Nueda MJ, Ferrer A, et al. maSigPro: a method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics. 2006;22:1096–1102. doi: 10.1093/bioinformatics/btl056. [DOI] [PubMed] [Google Scholar]
- 17.Bingham C, Arbogast B, Guillaume GC, et al. Inferential statistical methods for estimating and comparing cosinor parameters. Chronobiologia. 1982;9:397–439. [PubMed] [Google Scholar]
- 18.Storch KF, Lipan O, Leykin I, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 19.Zvonic S, Ptitsyn AA, Conrad SA, et al. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- 20.Bailey MJ, Beremand PD, Hammer R, et al. Transcriptional profiling of the chick pineal gland, a photoreceptive circadian oscillator and pacemaker. Mol Endocrinol. 2003;17:2084–2095. doi: 10.1210/me.2003-0121. [DOI] [PubMed] [Google Scholar]
- 21.Buchi KN, Moore JG, Hrushesky WJ, et al. Circadian rhythm of cellular proliferation in the human rectal mucosa. Gastroenterology. 1991;101:410–415. doi: 10.1016/0016-5085(91)90019-h. [DOI] [PubMed] [Google Scholar]
- 22.Marra G, Anti M, Percesepe A, et al. Circadian variations of epithelial cell proliferation in human rectal crypts. Gastroenterology. 1994;106:982–987. doi: 10.1016/0016-5085(94)90757-9. [DOI] [PubMed] [Google Scholar]
- 23.Scheving LE, Burns ER, Pauly JE, et al. Circadian variation in cell division of the mouse alimentary tract, bone marrow and corneal epithelium. Anat Rec. 1978;191:479–486. doi: 10.1002/ar.1091910407. [DOI] [PubMed] [Google Scholar]
- 24.McGill GG, Horstmann M, Widlund HR, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 25.McCole DF, Barrett KE. Epithelial transport and gut barrier function in colitis. Curr Opin Gastroenterol. 2003;19:578–582. doi: 10.1097/00001574-200311000-00011. [DOI] [PubMed] [Google Scholar]
- 26.Cheon JH, Kim JS, Kim JM, et al. Plant sterol guggulsterone inhibits nuclear factor-kappaB signaling in intestinal epithelial cells by blocking IkappaB kinase and ameliorates acute murine colitis. Inflamm Bowel Dis. 2006;12:1152–1161. doi: 10.1097/01.mib.0000235830.94057.c6. [DOI] [PubMed] [Google Scholar]
- 27.Arjona A, Sarkar DK. Circadian oscillations of clock genes, cytolytic factors, and cytokines in rat NK cells. J Immunol. 2005;174:7618–7624. doi: 10.4049/jimmunol.174.12.7618. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Malkani G, Shi X, et al. The circadian clock Period 2 gene regulates gamma interferon production of NK cells in host response to lipopolysaccharide-induced endotoxic shock. Infect Immun. 2006;74:4750–4756. doi: 10.1128/IAI.00287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palomba M, Bentivoglio M. Chronic inflammation affects the photic response of the suprachiasmatic nucleus. J Neuroimmunol. 2008;193:24–27. doi: 10.1016/j.jneuroim.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Mizuta Y, Takahashi T, Owyang C. Nitrergic regulation of colonic transit in rats. Am J Physiol. 1999;277:G275–G279. doi: 10.1152/ajpgi.1999.277.2.G275. [DOI] [PubMed] [Google Scholar]
- 31.Shi XZ, Choudhury BK, Pasricha PJ, et al. A novel role of VIP in colonic motility function: induction of excitation-transcription coupling in smooth muscle cells. Gastroenterology. 2007;132:1388–1400. doi: 10.1053/j.gastro.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 32.Reddy AB, Karp NA, Maywood ES, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 33.Scheving LA, Russell WE. It’s about time: clock genes unveiled in the gut. Gastroenterology. 2007;133:1373–1376. doi: 10.1053/j.gastro.2007.08.068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.