Abstract
A cluster of three nicotinic acetylcholine receptor genes on chromosome 15 (CHRNA5/CHRNA3/CHRNB4) has been shown to be associated with nicotine dependence and smoking quantity. The aim of this study was to clarify whether the variation at this locus regulates nicotine intake among smokers by using the level of a metabolite of nicotine, cotinine, as an outcome. The number of cigarettes smoked per day (CPD) and immune-reactive serum cotinine level were determined in 516 daily smokers (age 30–75 years, 303 males) from the population-based Health2000 study. Association of 21 SNPs from a 100 kb region of chromosome 15 with cotinine and CPD was examined. SNP rs1051730 showed the strongest association to both measures. However, this SNP accounted for nearly a five-fold larger proportion of variance in cotinine levels than in CPD (R2 4.3% versus 0.9%). The effect size of the SNP was 0.30 for cotinine level, whereas it was 0.13 for CPD. Variation at CHRNA5/CHRNA3/CHRNB4 cluster influences nicotine level, measured as cotinine, more strongly than smoking quantity, measured by CPD, and appears thus to be involved in regulation of nicotine levels among smokers.
INTRODUCTION
Genetic effects are known to influence smoking behavior and nicotine dependence, causing a major public health problem worldwide (1,2). Nicotine is the central addictive substance in tobacco. The role of genetic variants on nicotinic acetylcholine receptor (nAChR) genes has been a target of several recent studies as the physiological effects of nicotine are largely mediated through nAChRs (3). Thus, identification of functional variants in the receptor genes increases our understanding of individual differences in the effects of nicotine. Nine different nicotinic cholinergic receptor subunits (α2–α7, β2–β4) are expressed in the human brain. These subunits combine with each other in particular patterns to form a functional pentameric nAChR. The different receptor subtypes are distinguished by subunit composition and sensitivity to nicotine (4).
A cluster of three nAChR subunit genes, CHRNA5, CHRNA3 and CHRNAB4 (nicotinic cholinergic receptor alpha 5, alpha 3 and beta 4, respectively) residing on chromosome 15q25.1 has been identified to influence smoking behaviors. Recent studies have revealed that multiple genetic variants in this locus are associated with smoking quantity and nicotine dependence as well as many diseases influenced by smoking: lung cancer, peripheral arterial disease and chronic obstructive pulmonary disease (5–10). However, the biology underlying these genetic associations remains unclear (11).
Most studies use self-reported smoking status, nicotine dependence or smoking quantity as phenotype, often considered as proxy measures of nicotine intake. However, self-reported smoking behaviors obtained using questionnaires or interviews may be influenced by underreporting, particularly when there is a perceived social pressure not to smoke (12). The correlation between amount smoked and cotinine level is also affected by variation in the nicotine content of the cigarettes, use of other cigarette products and patterns of smoking. Cotinine levels are a more objective measure of the intake of nicotine (13). Approximately 80% of nicotine is metabolized to cotinine in the liver by CYP2A6 enzyme (14). Cotinine is a relatively stable compound with a half-life of ∼15–20 h (15) and its level reflects not only recent intake, but also a cumulative intake of ∼7 days, as well as nicotine intake from passive smoking (13).
The aim of this study was to clarify whether the cluster of three nAChR genes on chromosome 15 is more strongly associated with nicotine intake, measured by immune-reactive serum cotinine concentration, than the smoking quantity, and thus to clarify the biology underlying nicotine dependence.
RESULTS
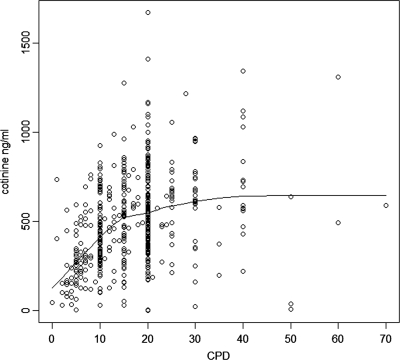
Among daily smokers, immune-reactive serum cotinine concentration (range 1–1671 ng/ml, mean 500 ng/ml, SD 255 ng/ml) and cigarettes smoked per day (CPD) (range 0–70, mean 16.9, SD 9.2) correlated significantly with a Spearman correlation coefficient of 0.41 (P < 0.0001). A lowess plot of the distribution of these variables is presented in Figure 1. Significant sex differences were observed in both variables [for cotinine t(515)=44, P < 0.0001 and for CPD t(505)=37, P < 0.0001]. Males smoked more (mean CPD 19.1) than females (mean CPD 13.7) and had higher cotinine levels (mean 529 ng/ml) than females (mean 458 ng/ml). Case–control status also influenced immune-reactive cotinine levels: controls had higher cotinine levels than cases (530 ng/ml versus 461 ng/ml). Four individuals reporting to be daily smokers had a cotinine level lower than the threshold for identifying a regular smoker suggested by Benowitz et al. (16). These cases were not removed from the data as there was no reason to assume that they would have over-reported their smoking quantity.
Figure 1.
Relationship between number of cigarettes smoked daily (CPD) and immune-reactive serum cotinine levels with a lowess curve within daily smokers (n = 516).
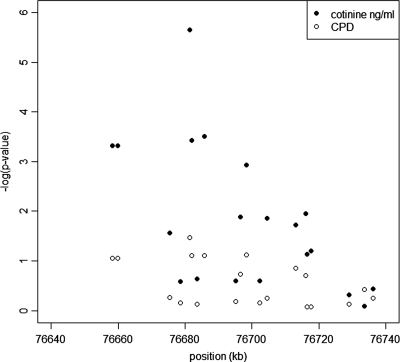
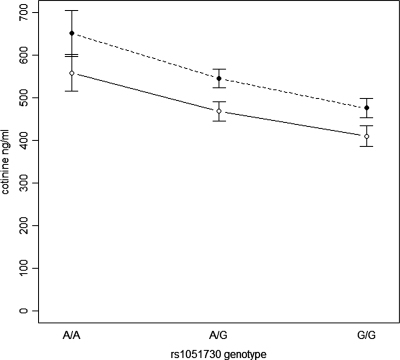
Nominal P-values (−log transformation) of association analysis for both outcome variables are presented in Figure 2 and the corresponding SNP genotype means and coefficients of determination (R2) in Table 1. The SNP rs1051730 is associated with both variables, but the effect is stronger on immune-reactive cotinine level (nominal P = 2.23×10−6, empirical P = 2×10−6) than on CPD (nominal P = 0.034, empirical P-value P = 0.043). This SNP accounted for nearly a five-fold larger proportion of variance in immune-reactive cotinine levels [R2 4.3%; 95% confidence interval (CI) 1.4–8.6] than in CPD (R2 0.9%; 95% CI 0.03–3.1). Regression analysis revealed that each A allele of this SNP increased immune-reactive cotinine level by 77 ng/ml (95% CI 45–108) and CPD by 1.2 cigarettes (95% CI 0.07–2.35), corresponding to effect sizes of 0.30 and 0.13 for immune-reactive cotinine level and CPD, respectively. The immune-reactive cotinine levels according to the rs1051730 genotype in males and females are presented in Figure 3.
Figure 2.
Results of the association analysis for immune-reactive cotinine level (ng/ml) and number of cigarettes smoked per day (CPD).
Table 1.
SNPs, their locations, alleles, minor allele frequencies (MAF) and the genotype means and coefficients of determination (R2) for immune-reactive cotinine level and CPD
| SNP | Location | Alleles minor/major 1/2 | MAF | Cotinine level |
CPD |
||
|---|---|---|---|---|---|---|---|
| Genotype means (ng/ml); 11/12/22 | R2 (%) | Genotype means (ng/ml); 11/12/22 | R2 (%) | ||||
| rs6495306 | 76 652 948 | C/T | 0.37 | 480/463/553 | 2.4 | 16.6/16.3/17.6 | 0.6 |
| rs680244 | 76 658 343 | T/C | 0.37 | 480/463/553 | 2.4 | 16.6/16.3/17.6 | 0.6 |
| rs621849 | 76 659 916 | C/T | 0.37 | 480/463/553 | 2.4 | 16.6/16.3/17.6 | 0.6 |
| rs578776 | 76 675 455 | T/C | 0.32 | 431/482/524 | 0.9 | 17.5/16.1/17.4 | 0.1 |
| rs12910984 | 76 678 682 | C/T | 0.29 | 452/488/511 | 0.2 | 18.9/15.9/17.4 | <0.1 |
| rs1051730 | 76 681 394 | A/G | 0.34 | 612/511/451 | 4.3 | 17.6/17.5/15.9 | 0.9 |
| rs3743077 | 76 681 951 | A/G | 0.37 | 485/460/557 | 2.4 | 16.7/16.3/17.7 | 0.6 |
| rs938682 | 76 683 602 | C/T | 0.29 | 452/489/514 | 0.3 | 18.9/15.9/17.4 | <0.1 |
| rs12914385 | 76 685 778 | A/G | 0.36 | 578/506/460 | 2.5 | 17.4/17.4/16.0 | 0.6 |
| rs8042374 | 76 695 087 | C/T | 0.29 | 445/491/512 | 0.3 | 18.6/16.0/17.3 | <0.1 |
| rs3743075 | 76 696 507 | A/G | 0.35 | 500/464/538 | 1.2 | 16.6/16.6/17.3 | 0.4 |
| rs8192475 | 76 698 285 | T/C | 0.03 | NA/363/508 | 2.0 | NA/14.2/17.0 | 0.6 |
| rs6495309 | 76 702 300 | A/G | 0.27 | 444/489/511 | 0.3 | 18.4/16.2/17.2 | <0.1 |
| rs1948 | 76 704 454 | A/G | 0.32 | 490/468/534 | 1.2 | 17.7/16.4/17.2 | 0.1 |
| rs950776 | 76 713 073 | G/A | 0.32 | 487/474/528 | 1.1 | 15.8/16.8/17.2 | 0.4 |
| rs11636753 | 76 716 001 | T/G | 0.33 | 487/471/533 | 1.2 | 16.6/16.4/17.5 | 0.3 |
| rs12441998 | 76 716 427 | G/A | 0.29 | 421/486/517 | 0.6 | 17.7/16.6/17.0 | <0.1 |
| rs1316971 | 76 717 565 | A/G | 0.29 | 421/484/517 | 0.7 | 17.7/16.6/17.0 | <0.1 |
| rs3971872 | 76 729 090 | A/G | 0.13 | 595/482/503 | 0.1 | 16.7/16.8/16.9 | <0.1 |
| rs12594247 | 76 733 688 | A/G | 0.19 | 516/487/504 | <0.1 | 20.5/16.4/16.9 | 0.2 |
| rs12900519 | 76 736 182 | C/T | 0.13 | 575/484/503 | 0.2 | 16.0/16.5/17.0 | 0.1 |
The R2-values are presented for models adjusted for sex and case–control status. MAF, minor allele frequency.
Figure 3.
Mean immune-reactive cotinine levels (ng/ml) and their standard errors corresponding to different genotypes of rs1051730 within daily smokers successfully genotyped (n = 515).
Altogether 14 SNPs of the region were significantly associated (empirical P-value < 0.05) with immune-reactive cotinine level. After conditioning on rs1051730, association on two SNPs, rs12914385 (nominal P = 0.006, empirical P = 0.007) and rs8192475 (nominal P = 0.004, empirical P = 0.004) remained significant. Backwards elimination stepwise regression analysis implied that rs1051730 has the most prominent effect on immune-reactive cotinine level and additionally included SNPs rs12910984 and rs3743075 in the final model. Owing to these contradictory results, no clear conclusion on effects of one or more SNPs on immune-reactive cotinine level can be drawn from our data. Earlier studies have reported that rs578776 has an independent effect on nicotine dependence (5,8). To further study, whether rs1051730 and rs578776 have a genotype–genotype interaction effect, a cross-tabulation of genotype frequencies and cotinine levels based on combinations of the genotypes of these two SNPs was made (Table 2). It can be seen that the genotype AA of rs1051730 is only present with genotype CC of rs578776. This haplotype also corresponds to the highest immune-reactive cotinine level.
Table 2.
Cross-tabulation of mean (±standard error) immune-reactive cotinine levels (ng/ml) of different haplotypes of rs1051730 and rs578776 among daily smokers
| rs578776 |
||||
|---|---|---|---|---|
| CC | CT | TT | ||
| rs1051730 | AA | 611.7 ± 32.2 (66) | N.A. (0) | N.A. (0) |
| AG | 491.0 ± 21.7 (133) | 537.4 ± 22.7 (108) | N.A. (0) | |
| GG | 500.0 ± 35.7 (64) | 428.2 ± 21.0 (107) | 430.6 ± 44.2 (38) | |
Corresponding haplotype frequencies presented in brackets.
The effect of exclusion of the covariates on the coefficient of determination (R2) of the most significant SNP on cotinine level was studied in order to determine whether the covariates, especially case–control status, actually explain part of the genetic variation on cotinine levels. The original R2 for rs1051730 was 4.3% with sex and case–control status as covariates. Removal of case–control status from the model resulted in decrease of R2 to 4.2% and further removal of sex to 3.9%. As the removal of covariates did not increase the R2 of the SNP, it appears that the covariates do not account for the genetic variance of cotinine level.
DISCUSSION
The cluster of three nicotinic cholinergic receptors on chromosome 15 is associated much more strongly with immune-reactive cotinine levels than with number of cigarettes smoked daily as reported by the study subjects. Thorgeirsson et al. (9) found that the same SNP giving the strongest association signal for both variables, rs1051730, was significantly associated to smoking quantity in a genome-wide association. Our results regarding CPD have a very similar magnitude; in the study of Thorgeirsson et al. (9), a copy of the A allele was estimated to increase smoking quantity by around one cigarette per day, whereas the estimate was 1.2 cigarettes per day in our study. The effect of the allele was 77 ng/ml on immune-reactive cotinine level. Thus the effect size of rs1051730 is 2.3 times greater on immune-reactive cotinine level (effect size 0.30) compared with CPD (0.13).
These results imply that the choice of the phenotype is of crucial importance to detect the association signals. If a genetic variant influences directly nicotine metabolism (in the brain or peripherally) rather than behavior (e.g. vulnerability to addiction), then a phenotype reflecting individual differences in metabolism, such as cotinine level, certainly results in more easily detectable signal than self-reported behavior, such as nicotine dependence or smoking quantity. However, all of these variables are correlated. With phenotypes not directly associated with the gene function, the signal might be detectable, but large sample sizes are needed [for example, more than 13 000 smokers were required in the study of Thorgeirsson et al. (9)] and the variance accounted by the genetic variant remains low.
Genetic association studies do not reveal which aspect of the genetic variant causing individual differences in immune-reactive cotinine level is related to the gene function. There are several possibilities. For example, the gene product may be directly related to metabolic rate of nicotine to cotinine, physiological effects of nicotine or regulation of nicotine level by receptor activity in the central nervous system. It is known that smokers are able to regulate their nicotine level by adjusting the smoking quantity and rate of inhalation. For example, the amount of nicotine needed to cause withdrawal symptoms, when starting to smoke, can be as low as one to two cigarettes per week, but soon the tolerance will develop and a larger nicotine intake, i.e. more frequent smoking, is needed (3,17). CPD is often used as a measure of smoking quantity, i.e. nicotine intake. However, cotinine levels usually reflect recent nicotine intake more closely than self-reported long-term averaged smoking quantity.
Approximately 80% of nicotine is metabolized to cotinine by cytochrome P450 2A6. Individual differences in the rate of nicotine and cotinine clearance are apparent and factors such as age (rate decreased with age), sex (rate higher in females than males), race/ethnicity (greater intake of nicotine and slower metabolism of cotinine in blacks than in whites) and diet (meals increase hepatic flow and thus nicotine clearance increases) have been shown to influence the metabolism rate (14,18). In our study, females showed overall lower cotinine levels but higher cotinine levels per cigarette, which is consistent with existing knowledge of females metabolizing nicotine to cotinine at a faster rate than males. There are two commonly used methods for cotinine level measurements: radioimmunoassay and chromatographic methods. It has to be noted that though the results of these methods are significantly correlated (r = 0.85), the radioimmunoassay provides higher results (28% higher, on average) and is less accurate than the chromatographic method (19). Thus, the cotinine levels in our study using radioimmunoassay method are not directly comparable to chromatographic measurements.
Variation in metabolism of nicotine via cotinine pathways is largely influenced by genetic effects (heritability, h2>60%). However, Swan et al. (20) reported that the heritability estimate decreases only slightly (nine percentage point drop from h2 = 60.2–51.8%) after controlling for genetic variants in CYP2A6. Thus, it appears that a large proportion of genetic variance in nicotine metabolism via cotinine pathways cannot be explained by variation in CYP2A6 gene.
The central role of the most widely expressed nAChR subtype, α4β2 receptors, in nicotine metabolism is evident as the function and number of these receptors is changed by nicotine exposure (4) and that varenicline, a medication for smoking cessation, is a partial antagonist of the α4β2 receptor (21). Additionally, a functional α4β2 receptor can contain an α5 subunit on one of its five positions. The role of the α5 subunit appears to be important, but not indispensable in modulating dopaminergic function: whereas deletion of α4 and β2 genes results in total elimination of acetylcholine-stimulated dopamine release in striatal synaptosomes, the deletion of α5 gene only significantly decreased it (4). Though our association signal was on the SNP rs1051730, which resides in CHRNA3 gene, the SNPS of CHRNA3 are in highly correlated with SNPs of CHRNA5 and CHRNB4 (for the LD plots, see Supplementary Material) and it is thus impossible to determine, via which gene the effect of the SNPs at this area are mediated. Thus, our results may reflect the differences in the physiological effects of nicotine caused by variation in the α5 subunit.
Comparing our results with those of Bierut et al. (5) and Saccone et al. (8), suggests that the cluster of three nAChRs on chromosome 15 harbors two separate loci influencing nicotine dependence, but such effect could not be clearly detected for immune-reactive serum cotinine level. It is very interesting to hypothesize that the stronger effect of this area on nicotine dependence, detected via rs1051730 [or rs16969968 used in Bierut et al. (5) and Saccone et al. (8) or other correlated SNP] is mediated via regulation of nicotine intake, and the other effect, detected via rs578776, has some other influence. It has to be noted that the inability to detect the second effect in our study may be due to smaller sample size.
In this study, we focused on a very strong candidate gene region which has been repeatedly implicated in studies of smoking and nicotine dependence with a high degree of biological plausibility and used a nationally representative sample. With only 21 markers, the sample of 516 individuals proved to be sufficient to detect the signal with relatively large effect size. Immuno-reactive cotinine levels were much more strongly associated with a variant at the region with three nAChR genes on chromosome 15 than the number of cigarettes smoked per day. The proportion of variance accounted for was nearly five-fold for cotinine (4.3%) compared with CPD (0.9%). Though number of cigarettes smoked reflects ones need for nicotine, it may include error variance due to misreporting. In addition, the nicotine levels among individuals smoking the same quantity may vary considerably due to metabolic differences, nicotine content of cigarettes, smoking patterns (e.g. rate of inhalation, proportion of a cigarette smoked) and exposure to other tobacco products, passive smoking as well as simultaneous use of nicotine replacement therapy. It appears that due to these differences, the studies concerning the effects of nicotine should strive to use cotinine level from serum or saliva (13) rather than self-reported smoking quantity as a measure of nicotine intake or its regulation. Our results imply that the chromosome 15 nAChR gene complex plays a major role on regulation nicotine level of smokers.
MATERIALS AND METHODS
Subjects
Subjects were drawn from a Health2000 study which includes a total of 8028 subjects aged 30 or over and is a nationally representative sample of adult Finnish population (22). The proportion of daily smokers was 29% among males and 18% among females and the proportion of daily smokers decreased by age. Here, we studied a subcohort (n = 2212, aged 30–75 years) selected for a case–control genome-wide association study on metabolic syndrome.
Phenotype data
A total of 6986 Health2000 subjects (87%) participated in a health interview conducted by Statistics Finland's interview staff at the home of the participants. The questionnaire extensively examined factors influencing the health, including a one-page questionnaire on smoking behavior. This questionnaire examined the frequency and quantity of smoking with seven items. The number of cigarettes smoked daily was inquired by one question ‘How much do you daily smoke currently or did prior to quitting? (a) Factory-made cigarettes, (b) self-rolled cigarettes, (c) pipefuls of pipe tobacco, (d) cigarillos/cigars’. The respondent was requested to indicate the number of each of the tobacco products as an open-ended question. These were summed to create the CPD (cigarettes per day) variable used in the analyses.
During the home interview, an appointment for the health examination was made (6770 subjects participated). This examination included a venous blood sample. The time of the day of blood sampling ranged from 8.15 a.m. to 10.25 p.m. (mean 1.37 p.m., SD 2 h 49 min) and was not correlated with the immuno-reactive cotinine level (r = 0.11) among daily smokers. The cotinine concentration (ng/ml) was determined from the serum using liquid-phase radioimmunoassay methodology (Nicotine Metabolite DOUBLE ANTIBODY kit, Diagnostic Products Corporation, Los Angeles, CA, USA).
Genotyped subsample can be regarded as presentative of the whole Health2000 Survey as there were no substantial differences in educational level or prevalence of smoking, neither in CPD or serum cotinine levels of daily smokers between the whole of Health2000 Survey and the genotyped subsample.
Genotypes
The individuals to be genotyped were selected according to the International Diabetes Federation Worldwide Definition of the Metabolic Syndrome (http://www.idf.org/home/index.cfm?node=1429) and controls were selected for not carrying the trait. Venous blood samples were drawn from the antecubital vein after an overnight fast. The samples were genotyped by Illumina 610 Quad V1 BeadChip (Illumina, Inc., San Diego, CA, USA) at the Sanger Wellcome Trust Institute. This chip provides whole-genome SNP genotyping information with 598203 SNP markers per individual genotyped with mean spacing of 4.7 kb. After initial data quality control for unspecified sex (one person removed), relatedness (13 individuals removed), contamination (three individuals removed), MDS plot outliers (14 individuals removed) and excess inbreeding (four individuals removed), and genotyping success of less than 95% (seven individuals removed), genotype information from 2131 individuals (1038 males and 1093 females) were available for analysis.
SNPs residing on a 100 kb area on chromosome 15 (76 640 000–76 740 000 bp) harboring a cluster of three nAChR subunit genes, CHRNA5, CHRNA3 and CHRNB4 were extracted for analysis from the whole-genome data. The area included 22 SNPs and after screening for Hardy–Weinberg equilibrium test P-value of more than 0.0001 (one SNP removed), minor allele frequency of more than 2% (the same SNP) and genotyping success rate of more than 95% (0 SNPs), 21 SNPs remained for analysis. These SNPs, their locations, alleles and allele frequencies are presented in Table 1.
Data analysis and statistical methods
Among the genotyped participants, we included in the analyses only current daily smokers. These comprised 24% of the dataset [n = 516, 29% of males (n = 303) and 20% of females (n = 213) were daily smokers]. The proportion of daily smokers was identical among cases and controls. The selection of daily smokers only was done because the half-life of cotinine in serum is known to be 15–20 h (15) and thus the immune-reactive serum cotinine concentration of occasional smokers or non-smokers was not considered to be a reliable measure of nicotine intake/exposure. Smokers were aged from 30 to 75 years (mean of 47.5 and SD 9.7 years), and 224 subjects had metabolic syndrome diagnosis and 292 were healthy controls. Of these, 11 subjects had missing data for CPD and one individual had low genotyping success (less than 95% of SNPs successfully genotyped).
First, we fitted a linear regression analysis to account for possible confounders. The analysis showed that sex and case–control status but not age significantly influenced the cotinine level and CPD. The effect of BMI on the phenotypes was also studied. When examined alone, it had a significant effect on immune-reactive cotinine levels (linear regression β = −13.1, P = 5.0×10−7). However, as the case–control status already explained most of its variation, BMI did not remain significant in the full model and inclusion of BMI in the analyses did not influence the results. Next, standardized residuals of immune-reactive serum cotinine concentration and CPD from a linear model including sex and case–control status were tested for association with the 21 SNPs using program package PLINK version 1.05 (23). The association of these quantitative traits was tested by Wald test. The significance of the results was further evaluated by calculating empirical P-values by permutation procedure. The thresholds for significance were set at an empirical P-value being lower than 0.05. The effect of covariates (sex and case–control status) was further studied by examining the effect of their inclusion on the coefficient of determination (R2) of the SNP(s) giving the most significant result(s). To assess whether this 100 kb area has only one or multiple distinct loci influencing immune-reactive cotinine levels, an association analysis conditional on the SNP giving the most significant results and stepwise linear regression analyses were performed.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
The study is funded by Center of Excellence in Disease Genetics, Academy of Finland to L.P. and J.K. and the NIH grant DA12854.
ACKNOWLEDGEMENTS
The authors would like to thank Dr Anu Loukola and the personnel of the Health2000 study and the interviewers of Statistics Finland.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Li M.D., Burmeister M.B. New insights into the genetics of addiction. Nat. Rev. Genet. 2009;10:225–231. doi: 10.1038/nrg2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. WHO Report on the Global Tobacco Epidemic, 2008: the MPOWER Package. Geneva, Switzerland: World Health Organization; 2008. [Google Scholar]
- 3.Benowitz N. Pharmacology of nicotine: addiction and therapeutics. Ann. Rev. Pharmacol. Toxicol. 1996;36:597–613. doi: 10.1146/annurev.pa.36.040196.003121. [DOI] [PubMed] [Google Scholar]
- 4.Collins A.C., Salminen O., Marks M.J., Whiteaker P., Grady S.R. The road to discovery of neuronal nicotinic cholinergic receptor subtypes. In: Henningfield J.E., London E.D., Pogun S., editors. Nicotine Psychopharmacology. Heidelberg, Germany: Spriger-Verlag; 2009. pp. 85–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bierut L.J., Stitzel J.A., Wang J.C., Hinrichs A.L., Grucza R.A., Xuei X., Saccone N.L., Saccone S.F., Bertelsen S., Fox L., et al. Variants in nicotinic receptors and risk for nicotine dependence. Am. J. Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caporaso N., Gu F., Chatterjee N., Sheng-Chih J., Yu K., Yeager M., Chen C., Jacobs K., Wheeler W., Landi M.T., et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS ONE. 2009;4:e4653. doi: 10.1371/journal.pone.0004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pillai S.G., Ge D., Zhy G., Kong X., Shianna K.V., Need A.C. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saccone N.L., Saccone S.F., Hinrichs A.L., Stitzel J.A., Duan W., Pergadia M.L., Agrawal A., Breslau N., Gruzca R.A., Hatsukami D., et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am. J. Med. Genet. B Neuropscyhiatr. Genet. 2009;150B:453–466. doi: 10.1002/ajmg.b.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorgeirsson T.E., Geller F., Sulem P., Rafnar T., Wiste A., Magnusson K.P., Manolescu A., Thorleifsson G., Stefansson H., Ingason A. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss R.B., Baker T.B., Cannon D.S., von Niederhausern A., Dunn D.M., Matsunami N. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4:e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorgeirsson T.E., Stefansson K. Genetics of smoking behavior and its consequences: the role of nicotinic acetylcholine receptors. Biol. Psychiatry. 2008;64:919–921. doi: 10.1016/j.biopsych.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarvis M., West R., Tunstall-Pedoe H., Vesey C. An evaluation of the intervention against smoking in the multiple risk factor intervention trial. Prev. Med. 1984;13:501–509. doi: 10.1016/0091-7435(84)90018-5. [DOI] [PubMed] [Google Scholar]
- 13.Gorber S.C., Schofield-Hurwitz S., Hardt J., Levasseur G., Tremblay M. The accuracy of self-reported smoking: a systematic review of the relationship between self-reported and cotinine-assessed smoking status. Nicotine Tob. Res. 2009;11:3–11. doi: 10.1093/ntr/ntn010. [DOI] [PubMed] [Google Scholar]
- 14.Hukkanen J., Jacob P., III, Benowitz N.L. Metabolism and disposition kinetics of nicotine. Pharmacol. Rev. 2005;57:79–115. doi: 10.1124/pr.57.1.3. [DOI] [PubMed] [Google Scholar]
- 15.Benowitz N.L., Kuyt F., Jacob P., III, Jones R.T., Osman A.L. Cotinine disposition effects. Clin. Pharmacol. Ther. 1083;34:604–611. doi: 10.1038/clpt.1983.222. [DOI] [PubMed] [Google Scholar]
- 16.Benowitz N.L., Bernert J.T., Caraballo R.S., Holiday D.B., Wang J. Optimal serum votinine levels for distinguishing cigarette smokers and nonsmokers within different racial/ethnic groups in the United States between 1999 and 2005. Am. J. Epidemiol. 2009;169:236–248. doi: 10.1093/aje/kwn301. [DOI] [PubMed] [Google Scholar]
- 17.DiFranza J.R. Hooked from the first cigarette. J. Fam. Pract. 2007;56:1017–1022. [PubMed] [Google Scholar]
- 18.Wagenknecht L.E., Cutter G.R., Haley N.J., Sidney S., Manolio T.A., Hughes G.L., Jacobs D.R. Racial differences in serum cotinine levels among smokers in the coronary artery risk development in (young) adults study. Am. J. Health Promot. 1990;80:1053–1056. doi: 10.2105/ajph.80.9.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byrd G.D., Davis R.A., Ogden M.W. A rapid LC-MS-MS method for determination of nicotine and cotinine in serum and saliva samples from smokers: validation and comparison with a radioimmunoassay method. J. Chromatogr. Sci. 2005;43:133–140. doi: 10.1093/chromsci/43.3.133. [DOI] [PubMed] [Google Scholar]
- 20.Swan G.E., Benowitz N.L., Lessov C.N., Jacob P., III, Tyndale R.F., Wilhemsen K. Nicotine metabolism: the impact of CYP2A6 on estimates of additive genetic influence. Pharmacogenet. Genomics. 2005;15:115–125. doi: 10.1097/01213011-200502000-00007. [DOI] [PubMed] [Google Scholar]
- 21.Oncken C.A., Gonzales F., Nides M., Rennard S., Watsky E.J., Coe J.W. Varenicline: an α4β2 nicotinic acetylcholine receptor partial agonist as an to smoking cessation. In: George T.P., editor. Medication Treatments for Nicotine Dependence. Boca Raton, FL, USA: Taylor & Francis; 2007. pp. 213–224. [Google Scholar]
- 22.Aromaa A., Koskinen S. Health and Functional Capacity in Finland. Helsinki, Finland: Publications of the National Public Health Institute, KTL B12; 2004. http://www.terveys2000.fi/julkaisut/baseline.pdf . [Google Scholar]
- 23.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.