Abstract
ADAM10, a member of a disintegrin and metalloprotease family, is an α-secretase capable of anti-amyloidogenic proteolysis of the amyloid precursor protein. Here, we present evidence for genetic association of ADAM10 with Alzheimer's disease (AD) as well as two rare potentially disease-associated non-synonymous mutations, Q170H and R181G, in the ADAM10 prodomain. These mutations were found in 11 of 16 affected individuals (average onset age 69.5 years) from seven late-onset AD families. Each mutation was also found in one unaffected subject implying incomplete penetrance. Functionally, both mutations significantly attenuated α-secretase activity of ADAM10 (>70% decrease), and elevated Aβ levels (1.5–3.5-fold) in cell-based studies. In summary, we provide the first evidence of ADAM10 as a candidate AD susceptibility gene, and report two potentially pathogenic mutations with incomplete penetrance for late-onset familial AD.
INTRODUCTION
Alzheimer's disease (AD) is a progressive neurodegenerative disorder and the leading cause of dementia in the elderly. AD is pathologically characterized by abundant amyloid plaques and neurofibrillary tangles. The amyloid β-protein (Aβ), the primary component of the amyloid plaques, is generated via serial proteolytic cleavage of the amyloid precursor protein (APP) by β-secretase followed by γ-secretase, which employs the presenilins as the catalytic subunits. Over 200 missense mutations in the APP and the presenilin 1 and 2 (PSEN1; PSEN2) genes cause early-onset, autosomal dominant, familial AD. APP is usually cleaved toward the middle of the Aβ sequence by α-secretase, which precludes Aβ production. This anti-amyloidogenic pathway also generates a soluble N-terminal fragment (sAPPα), for which neurotrophic and neuroprotective mechanisms have been proposed (1–4). α-Secretase activity can be regulated by several signaling pathways involving protein kinase C (PKC), tyrosine kinases, the mitogen-activated protein kinases and extracellular signal-regulated kinases (5–7). Activation of the PKC signaling cascade by phorbol esters has been shown to increase sAPPα secretion and to significantly alleviate Aβ formation (8–13). Three members of the ADAM (a disintegrin and metalloprotease) family, ADAM9, ADAM10 and ADAM17, have been shown to possess α-secretase activity (14). Single knockout mutants of the ADAM proteases do not completely abolish α-secretase activity. For example, ADAM10-deficient mice are embryonic lethal due to defective Notch signaling; however, embryonic fibroblasts from these mice maintain α-secretase activity (15). In ADAM17 (tumor necrosis factor-α convertase, TACE)-knockout mice, phorbol ester-induced secretion of sAPPα was abolished while the constitutive release of sAPPα was preserved (16,17). Finally, in vivo deletion of ADAM9 did not lead to changes in α-secretase activity (18). Hence, these three ADAM proteases may exhibit considerable functional redundancy (14,15). Each ADAM member has unique features, i.e. ADAM10 has been shown to be responsible for both constitutive and regulated α-secretase activities (19,20), while TACE appears to be mainly involved in regulated activity (16,17,21,22). Although all three proposed α-secretase candidates contribute to APP cleavage, ADAM10 is unique due to its combined constitutive and regulated activity, high enzymatic stability under conditions for α-secretase cleavage (23) and coordinated mRNA expression with APP expression in mouse and human cortical neurons (24). Reduced levels of ADAM10 have been reported in sporadic AD patients along with lower sAPPα levels (25). Additionally, moderate neuronal overexpression of ADAM10 in mice has been shown to lead to elevated sAPPα release, reduction in Aβ formation and plaque deposition and improved cognition (26).
The domain structure of ADAMs consists of a prodomain, a metalloprotease domain, a disintegrin domain, a cystein-rich domain, an EGF-like domain, a transmembrane domain and a cytoplasmic tail (27). The primary function of the prodomain is to maintain the metalloprotease site in an inactive state via a cysteine switch (28). A zinc atom in the catalytic site is coordinated by a conserved cysteine residue in the prodomain and the metalloprotease domain remains in a latent conformation. Inactivation prevents ADAMs from auto-catalysis during biosynthesis. To generate the active protease, the prodomain must be cleaved from the rest of the protein by proprotein convertases in trans-Golgi network (TGN) (20,29). In ADAM10, proprotein convertase recognition sequence (RKKR) is essential for activation of the zymogen, and both furin and PC7 can act as proprotein convertases (29). Following maturation, the majority of the mature ADAM10 is transported to the plasma membrane, where it functions as an ectodomain sheddase for several cell surface proteins. In addition, the prodomain is suggested to act as an intramolecular chaperone during biosynthesis and secretion of the catalytic domain, facilitating proper folding and structure of the catalytic active site, as well as assisting transit throughout the secretory pathway of the protease (27).
Late-onset AD is a genetically complex and heterogeneous disease. The ε-4 allele of apolipoprotein E (APOE) is the only established genetic risk factor for late-onset AD. However, it has been suggested that APOE and the three early-onset familial AD genes, APP, PSEN1 and PSEN2 account for less than half of the genetic variance of AD (30). With the aim of assessing the potential role of ADAM10 as an AD susceptibility gene, we initially tested nine single nucleotide polymorphisms (SNPs) in this gene for genetic association with AD in over 400 AD families comprising the National Institute of Mental Health (NIMH) AD Genetics Initiative family sample. These association findings were later confirmed by data generated in a recently completed genome-wide association study (GWAS) in the same sample (31). Ultimately, these data led to the identification of two rare, non-synonymous mutations in the region of ADAM10 encoding the prodomain. We assessed the functional consequences of these two prodomain mutations by examining their effects on α-secretase activity, Aβ levels, as well as ADAM10 maturation and biogenesis in a series of Chinese hamster ovary (CHO) cell lines stably overexpressing APP and ADAM10 constructs.
RESULTS
Family-based association of ADAM10 SNPs in the NIMH AD family sample
To test the potential role of ADAM10 as an AD susceptibility gene, we initially tested a total of nine SNPs in the ADAM10 gene for genetic association with AD in the NIMH AD Genetics Initiative family sample (1439 DNAs from 436 multiplex AD families (32)). The nine SNPs were chosen from publicly available databases as proxies to tag the most common variants in the ADAM10 gene. One of these SNPs (rs2305421) showed evidence of genetic association in the NIMH families (P-value = 0.003; Table 1). Upon stratification of families by APOE-ε4, the association with rs2305421 became more pronounced (P-value = 0.0005 in the APOE4+ families), and two other SNPs of the nine tested (rs605928 and rs4775083) now showed marginal association (P-values = 0.02 and 0.06, respectively; Table 1). These associations were later confirmed on a larger number of SNPs genotyped as part of a GWAS on the same families from the NIMH collection (31). On the GWAS array, there were a total of 53 SNPs ± 100 kb from the ADAM10, and 11 of these showed nominal association (P-value ≤ 0.05) with AD risk, in good agreement with the SNPs genotyped in the initial phase of this study (Supplementary Material, Table S1). Despite the consistent evidence for association between AD risk and genetic variants in ADAM10 in the NIMH sample, none of the initially genotyped SNPs showed evidence of association in the independent Consortium on Alzheimer's Genetics (CAG) sample consisting of primarily of discordant sibpairs (489 DNAs, 217 families (33)), most likely due to lower power of this sample (see Supplementary Material, Table S2). Nonetheless, the combined NIMH and CAG samples still yielded overall significant evidence for association of AD with ADAM10 SNP rs2305421 (P-value = 0.008 in the unstratified NIMH and CAG samples and P-value = 0.0007 in the APOE ε4-pos families only).
Table 1.
Association results for nine SNPs in ADAM10 gene region and AD
| SNP | Mb | MAF | Fams | P-value (risk)* | P-value (HWE) |
|---|---|---|---|---|---|
| Unstratified NIMH sample | |||||
| rs605928 | 56833455 | 0.296 | 170 | −0.114 | 0.984 |
| rs593742 | 56833066 | 0.284 | 159 | −0.304 | 0.214 |
| rs514049 | 56829655 | 0.427 | 195 | −0.327 | 0.034 |
| rs442495 | 56809907 | 0.313 | 105 | −0.441 | 0.374 |
| rs7161889 | 56770717 | 0.309 | 105 | −0.737 | 0.588 |
| rs714696 | 56750279 | 0.274 | 170 | −0.271 | 0.898 |
| rs2305421a | 56690375 | 0.123 | 103 | −0.003 | 0.509 |
| rs4775083 | 56679967 | 0.344 | 185 | −0.180 | 0.852 |
| rs1869135 | 56669261 | 0.197 | 85 | −0.847 | 0.454 |
| APOE ε4-pos families only | |||||
| rs605928 | 56833455 | 0.295 | 139 | −0.023 | 0.789 |
| rs593742 | 56833066 | 0.280 | 131 | −0.128 | 0.210 |
| rs514049 | 56829655 | 0.425 | 161 | −0.264 | 0.069 |
| rs442495 | 56809907 | 0.318 | 91 | −0.370 | 0.251 |
| rs7161889 | 56770717 | 0.312 | 91 | −0.626 | 0.565 |
| rs714696 | 56750279 | 0.275 | 144 | −0.177 | 1.000 |
| rs2305421a | 56690375 | 0.122 | 86 | −0.0005 | 0.772 |
| rs4775083 | 56679967 | 0.344 | 152 | −0.057 | 0.942 |
| rs1869135 | 56669261 | 0.199 | 75 | 0.999 | 0.790 |
MAF, minor allele frequency as determined by PBAT. Fams, informative families.
aSNP was used to select families for sequencing.
*Negative P-values indicate undertransmission of minor allele to affecteds. All P-values were calculated by PBAT v.3.6.
We next sequenced 32 families of the NIMH sample that showed particularly strong evidence of association, i.e. those in which affected individuals were homozygous for the risk allele of the best-associated SNP (rs2305421). This led to the identification of two rare, non-synonymous changes in exon 5 of ADAM10, Q170H and R181G, located near the consensus sequence for the cysteine switch in the prodomain of ADAM10 in two families out of 32. Screening of the remaining NIMH samples for the Q170H and R181G mutations revealed further three families, resulting in a total of five families (three for Q170H and two for R181G) carrying these potential ADAM10 mutations (Table 2). Combining both mutations to one aggregate genotype revealed significant evidence for association with AD risk across the NIMH sample [P-value = 0.0043 (including all individuals), and a P-value = 0.06 (after exclusion of probands initially selected for sequencing)]. While none of the unaffected individuals of these families were found to be mutation carriers, for each mutation there was one family in which one of the affected siblings was not a carrier. The affected R181G non-carrier also carried an APOE-ε4 allele, while the affected Q170H non-carrier did not. Neither mutation was found in the total set of 678 unaffected subjects in the combined NIMH and CAG samples.
Table 2.
Seven AD families harboring either Q170H or R181G mutation in ADAM10 gene
| Cohort | Family | Subject | Mutation | Diagnosisa | Gender | Ageb | APOE | Genotypec |
|---|---|---|---|---|---|---|---|---|
| NIMH | 165d | I | Q170H | AD (def) | M | 69 | 34 | Het |
| NIMH | 165d | II | Q170H | AD (prob) | M | 67 | 23 | Het |
| NIMH | 165d | III | Q170H | AD (prob) | F | 65 | 34 | Het |
| NIMH | 165d | IV | Q170H | Unaffected | F | 73 | 23 | Wild-type |
| NIMH | 165d | V | Q170H | Unaffected | F | 63 | 23 | Wild-type |
| NIMH | 165d | VI | Q170H | Unaffected | F | 70 | 34 | Wild-type |
| NIMH | 254 | I | Q170H | AD (def) | F | 71 | 34 | Het |
| NIMH | 254 | II | Q170H | AD (def) | F | 72 | 33 | Wild-type |
| NIMH | 254 | III | Q170H | Unaffected | F | 80 | 33 | Wild-type |
| NIMH | 198 | I | Q170H | AD (prob) | M | 69 | 44 | Het |
| NIMH | 198 | II | Q170H | AD (def) | F | 68 | 44 | Het |
| NIMH | 261d | I | R181G | AD (def) | F | 67 | 34 | Het |
| NIMH | 261d | II | R181G | AD (def) | F | 66 | 34 | Wild-type |
| NIMH | 261d | III | R181G | Unaffected | F | 73 | 34 | Wild-type |
| NIMH | 388 | I | R181G | AD (prob) | F | 75 | 34 | Het |
| NIMH | 388 | II | R181G | AD (prob) | F | 70 | 34 | Het |
| NIMH | 388 | III | R181G | Unaffected | M | 79 | 34 | Wild-type |
| NIMH | 388 | IV | R181G | Unaffected | F | 74 | 34 | Wild-type |
| NIA | 25_6 | I | Q170H | AD (prob) | F | 63 | 34 | Het |
| NIA | 25_6 | II | Q170H | AD (prob) | F | 71 | 33 | Wild-type |
| NIA | 25_6 | III | Q170H | AD (prob) | F | 65 | 34 | Wild-type |
| NIA | 25_6 | IV | Q170H | Unaffected | F | 63 | 33 | Het |
| NIA | 15_3051 | I | R181G | AD (prob) | M | 80 | 34 | Het |
| NIA | 15_3051 | II | R181G | AD (prob) | F | 74 | 34 | Wild-type |
| NIA | 15_3051 | III | R181G | Unaffected | M | 83 | 34 | Het |
aAD (def), ‘definite’ AD (neuropathologically confirmed); AD (prob), ‘probable’ AD (based on clinical diagnosis).
bOnset age in AD patients; age at last examination in unaffected subjects.
cHet, heterozygous for amino-acid change.
dMutations identified upon sequencing of families showing strongest association with rs2305421.
To search for additional AD families carrying these mutations, we further screened 1111 individuals from 351 AD pedigrees from the National Institute on Aging (NIA) Study Sample (Table 2). We found one additional AD family for each of the mutations. In both of these families, the mutations did not perfectly segregate with disease status (Table 2). For the Q170H mutation, one AD patient, with onset age of 63 years, carried the mutation while the other two (onset age of 65 and 71 years) did not. In addition, a 63-year-old unaffected family member carried the mutation. These results suggest that other genetic factors contribute to AD risk in this family and, potentially, incomplete penetrance of the Q170H mutation. It is worth noting that the patient with the earliest onset carries the Q170H mutation as well as an APOE-ε4 allele. For the R181G mutation, one of two affected individuals carried the mutant allele (onset 80 years) while the other (onset 74 years) did not. In addition, an unaffected family member (age 83 years) carried the mutant allele. All three family members were heterozygous for APOE-ε4. As was observed with Q170H, these findings suggest incomplete penetrance for the R181G mutation, and the probable involvement of additional genetic factors. Combining both mutations to one aggregate genotype revealed significant evidence for association with AD risk across the NIMH and NIA samples [P-value = 0.0186 (including all individuals), and a P-value = 0.212 (after exclusion of probands initially selected for sequencing)].
Novel prodomain mutations in ADAM10 attenuate constitutive α-secretase activity
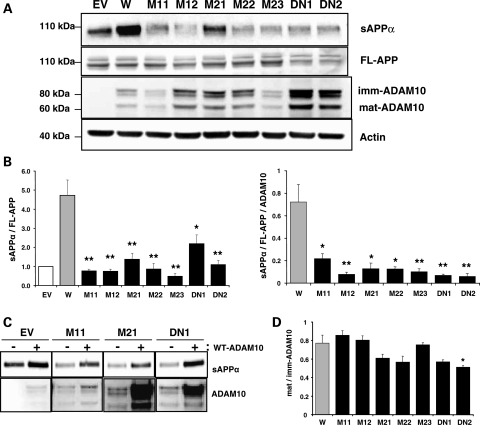
To investigate the potential functional effects of Q170H and R181G mutations in the ADAM10 prodomain, we generated CHO cell lines stably expressing wild-type or mutant ADAM10 together with APP. For this purpose, CHO-APP751 cells were transfected with a HA-tagged ADAM10 cDNA, with or without the prodomain mutations. CHO-APP751 cells transfected with empty vector or a dominant negative (E384A) mutant form of ADAM10 were also generated to serve as negative and positive controls, respectively. Among several ADAM10-expressing, single cell-originated clones, we selected multiple clones for the mutant constructs, two for Q170H, three for R181G and two for E384A, and then measured sAPPα levels. As shown in Figure 1A and B, relative levels of sAPPα normalized to full-length APP (FL-APP) levels were >4-fold higher in wild-type ADAM10 cells (W) when compared with empty vector-transfected cells (EV), indicating increased α-secretase activity resulting from overexpression of ADAM10. However, overexpression of ADAM10-Q170H and ADAM10-R181G mutant forms did not elevate sAPPα levels to the same extent as wild-type ADAM10. sAPPα levels, normalized to either FL-APP (sAPPα/FL-APP) or FL-APP and ADAM10 (sAPPα/FL-APP/ADAM10), were significantly decreased (by >72%) in cells expressing ADAM10 harboring the Q170H (M11, M12) and R181G mutations (M21, M22, M23) versus those in wild-type cells. sAPPα levels in the Q170H and R181G cells were comparable to those in cells expressing a known dominant negative (E384A) ADAM10 mutant (DN1 and DN2). The inhibitory effects of the dominant negative (E384A) ADAM10 mutation on the constitutive and regulated α-secretase activities of ADAM10 have been previously defined (20). Decreased sAPPα levels were observed in the multiple independent mutant clones and could be restored by transient overexpression of the wild-type ADAM10 (Fig. 1C), suggesting that the observed results were not a clone-specific effect of stable cells but due to genuine changes in ADAM10 catalytic activity.
Figure 1.
Decrease in constitutive α-secretase activity in CHO cells stably expressing APP and wild-type or prodomain mutant forms of ADAM10. CHO-APP751 cells were used to generate cell lines stably overexpressing different ADAM10 constructs, wild-type (W), Q170H mutant (M11, M12), R181G mutant (M21–M23), E384A dominant negative mutant (DN1, DN2) and empty vector (EV). Each stable cell line was grown to exponential phase and conditioned media and cell lysates were collected to measure levels of sAPPα, ADAM10 and APP. (A) Representative western blots. FL-APP, full length APP; imm-ADAM10, immature ADAM10; mat-ADAM10, mature ADAM10. (B) Graphs for relative sAPPα levels normalized with full-length APP levels (sAPPα/FL-APP) and further with ADAM10 levels (sAPPα/FL-APP/ADAM10). n = 4–8, mean ± SEM. (C) Restoration of the reduced sAPPα levels in ADAM10 mutant cells by overexpression of wild-type ADAM10. Each ADAM10 stable cell line was transiently transfected with wild-type ADAM10 construct and levels of sAPPα and ADAM10 were measured. (D) Ratios of mature and immature ADAM10. Mature and immature ADAM10 bands in the western blot in (A) were quantified and their ratio in each cell was calculated. n = 3–6, mean ± SEM. Significance of changes in the mutant cells was calculated when compared with wild-type. *P < 0.05, **P < 0.005 (two-tailed Student's t-test).
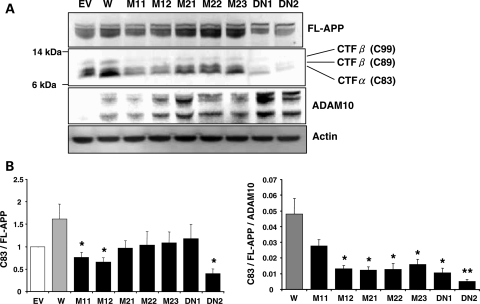
To confirm these results, levels of APP C-terminal fragment (CTF) of α-secretase cleavage were examined in the ADAM10 stable cells (Fig. 2). In order to accurately measure the production of CTFs, the γ-secretase inhibitor (DAPT) was used to inhibit cleavage of APP-CTFs. Consistent with the observed changes in sAPPα levels, the relative CTFα (C83) levels were significantly lower in the Q170H and R181G mutants when compared with cells expressing wild-type ADAM10. These data further support defective α-secretase activity in the cells expressing ADAM10 harboring the Q170H and R181G mutations. In contrast, we observed no detectable increase in levels of APP-CTFβs [C89 and C99; confirmed by treatment with a BACE1 inhibitor and by western blotting with Aβ1–16 specific antibody (data not shown)] in the mutant versus wild-type ADAM10 cells.
Figure 2.
Decrease in APP-CTFα levels in CHO cell lines stably expressing prodomain mutant forms of ADAM10. CHO-APP-ADAM10 stable cell lines were treated with 250 nM DAPT for 24 h to inhibit γ-secretase and APP-CTFs (C83, C89, C99) levels were examined in total cell lysates. (A) Representative western blots. (B) Graphs for relative CTFα levels normalized with full-length APP levels (C83/FL-APP) and further with ADAM10 levels (C83/FL-APP/ADAM10). n = 7, mean ± SEM. Significance of changes in the mutant cells was calculated when compared with wild-type. *P < 0.05, **P < 0.005 (two-tailed Student's t-test). EV, empty vector-transfected cells; W, wild-type ADAM10 cell; M11, M12, Q170H mutant cells; M21–M23, R181G mutant cells; DN1, DN2, E384A dominant negative mutant cells.
Since the Q170H and R181G mutations are located close to the potential consensus sequence for the cysteine switch in the ADAM10 prodomain and proprotein convertase recognition sequence (RKKR) in the metalloprotease domain, we next assessed the synthesis and maturation of ADAM10 in the mutant versus wild-type ADAM10 cells. Protein levels of total ADAM10, mature ADAM10 (64 kDa) and immature ADAM10 (90 kDa), were detectable but variable across the mutant clone cell lines (Fig. 1A). Variable levels were not likely to result from an enhanced protein degradation or auto-catalysis during ADAM10 biosynthesis because the ADAM10 levels were not consistent among cells expressing the same mutations nor did they correlate with α-secretase activity. The ratio of steady-state levels of mature to immature ADAM10 did not exhibit any significant or consistent changes in the mutant versus wild-type ADAM10 cell lines (Fig. 1D). Collectively, these data suggest that these two mutations do not alter ADAM10 maturation.
Elevated Aβ levels in the presence of the novel prodomain mutations in ADAM10
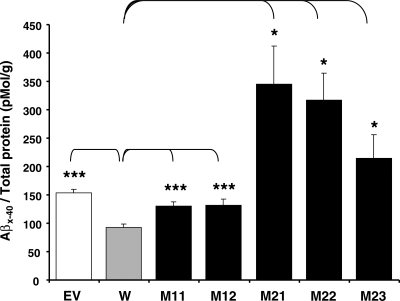
Next, we investigated potential effects of the ADAM10 prodomain mutations on Aβ levels in the CHO stable cell lines as measured by ELISA using Aβx-40-specific antibodies. The Aβ40 levels were significantly (P < 0.0005) decreased upon overexpression of wild-type ADAM10 (Fig. 3). In contrast, overexpression of the Q170H and R181G mutants did not lead to reduced Aβ40 levels, consistently showing Aβ levels equivalent to or higher than those of EV control. Aβ40 levels were significantly increased about 1.5-fold and 2∼3.5-fold in the Q170H mutants (P < 0.0005) and the R181G mutants (P < 0.05), respectively, when compared with those in wild-type cells. Collectively, these results suggest that the ADAM10 Q170H and R181G mutations attenuate constitutive α-secretase activity of ADAM10, concurrently increasing Aβ levels.
Figure 3.
Increase of Aβ40 levels in the prodomain mutant ADAM10 stable cell lines. CHO-APP-ADAM10 stable cell lines were grown to exponential phase. Twenty four hour after changing media, the conditioned media were collected and levels of Aβx-40 were measured by ELISA. Aβ40 levels were normalized with amount of total protein in each corresponding cell lysate. n = 6–12, mean ± SEM. Significance of differences of the mutant and empty-vector transfected cells versus wild-type was calculated. *P < 0.05, ***P < 0.0005 (two-tailed Student's t-test). EV, empty vector-transfected cells; W, wild-type ADAM10 cell; M11, M12, Q170H mutant cells; M21–M23, R181G mutant cells.
Attenuation of PKC-inducible α-secretase activity of ADAM10 by two novel prodomain mutations
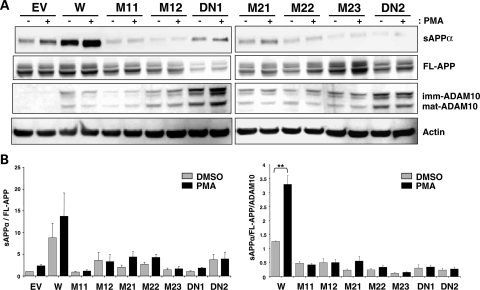
We next investigated whether phorbol ester-inducible α-secretase activity of ADAM10 is affected by the Q170H and R181G mutations. For this purpose, CHO-APP-ADAM10 stable cell lines were treated with PMA (Fig. 4). In the wild-type ADAM10 cells, sAPPα levels (normalized to FL-APP and ADAM10) were significantly increased (>2.5-fold) following PMA-treatment when compared with DMSO-treated controls. However, none of the mutant ADAM10 cells (Q170H, R181G and E384A) showed significant increases in sAPPα levels upon PMA treatment, leading to further decrease in sAPPα levels (>85%) in the mutant versus the wild-type ADAM10 cells under PKC-activation. These data indicate that the two novel prodomain mutations not only reduce constitutive activity of ADAM10, but also impair PKC-inducible α-secretase activity of ADAM10.
Figure 4.
Attenuated PMA-inducible α-secretase activity in CHO cells stably expressing the prodomain mutant forms of ADAM10. Each CHO-APP-ADAM10 stable cell line was treated with 1 µM PMA or equivalent amount of DMSO for 6 h and harvested to monitor sAPPα levels. (A) Representative western blots. (B) Graphs for relative sAPPα levels. n = 4–6, mean ± SEM. **P < 0.005 (two-tailed Student's t-test).
DISCUSSION
We have observed genetic association of ADAM10 with AD risk and identified two novel non-synonymous mutations in the ADAM10 prodomain in seven AD families; we also assessed their effects on ADAM10 function and APP processing. Using CHO-stable cell lines expressing ADAM10 and APP, we demonstrated that Q170H and R181G mutations significantly attenuate α-secretase activity of ADAM10, leading to decreased levels of sAPPα and C83, and increased levels of Aβ. Although the two novel mutations are located in the proximity of the conserved sequences for cysteine switch and proprotein convertase recognition, they did not affect either the maturation or biosynthesis of ADAM10.
The two novel ADAM10 prodomain mutations are rare, segregating in only seven AD families (three for one mutation and four for the other) out of 1004 AD families screened. For each mutation, there were two families in which one (or two) affected individuals were found to be non-carriers, suggesting that in these individuals disease onset was influenced by other genetic factors. Because multiple genetic and environmental factors likely contribute to risk for late-onset of the disorder, onset in one (or two) affected individuals in the absence of each ADAM10 mutation is not entirely unexpected. A similar situation has been observed for AD mutations observed in presenilin 2 (N141I (34)) and presenilin 1 (A79V (35)), most likely owing to high prevalence of AD in the elderly population. In addition, for each mutation, there was one family in which one unaffected subject carried each mutation, suggesting incomplete penetrance for late-onset AD.
Support for the potential pathogenicity of the ADAM10 Q170H and R181G mutations was derived from experiments assessing the functional impact of these missense mutations on ADAM10 activity. For this purpose, we tried to generate cell lines stably overexpressing both APP and ADAM10 using H4 neuroglioma cells, HEK293 cells and CHO cells. We were only able to select wild-type and mutant ADAM10-expressing clones in CHO cells, most likely due to cytotoxicity caused by overexpression of both ADAM10 (36,37) and APP in the two human cell lines. In the CHO-APP-ADAM10 stable cells, the Q170H and R181G mutants caused a dramatic reduction in the generation of sAPPα and APP-C83 when compared with wild-type ADAM10 (Figs 1 and 2). However, western blot analysis did not reveal increases in sAPPβ and APP-CTFβ (C99 and C89) levels in the mutant cell lines (data not shown and Fig. 2). This may be due to the relatively low sensitivity of our western blot analyses for sAPPβ and APP-CTFβ. Moreover, regulated α-secretase activity by TACE has previously been shown to directly compete with β-secretase activity for APP cleavage in TGN (38). However, catalytically active ADAM10 is mainly localized in plasma membrane (20); thus, concurrent changes in sAPPβ and APP-CTFβ levels may be difficult to detect upon ADAM10 overexpression using western blot analysis. Aβ levels, determined by a sensitive ELISA, were consistently increased in the presence of the two ADAM10 prodomain mutations. Collectively, these data indicate that both ADAM10 mutations lead to defective α-secretase activities and subsequently elevation in Aβ levels in vitro, suggesting potentially pathogenic roles for these mutations, pending confirmation in vivo.
The molecular mechanism underlying the effects of the two novel ADAM10 mutations likely involves the prodomain function. Both mutations are located near the proprotein convertase recognition sequence (RKKR), which is required for proteolytic activation of the zymogen (29). However, since steady-state levels of mature and immature ADAM10s were not altered significantly (Fig. 1D), the mutations most likely do not affect ADAM10 maturation. The mutations may instead affect the intramolecular chaperone function of the ADAM10 prodomain. The role of prodomain in proper folding of ADAM proteins, particularly the metalloprotease domain, is also supported by studies of other members of ADAM protein family. For example, the secreted soluble form of ADAM 12 (ADAM12-S) lacking the prodomain remains in the early endomembrane system and is not secreted from cells, suggesting that the prodomain might be required in folding of the metalloprotease domain to a secretion-competent conformation (39). Truncated forms of ADAM10 and TACE lacking the prodomain have been shown to be catalytically inactive (29,40). Intracellular degradation of TACE lacking the prodomain and its rescue by prodomain expression in trans strongly suggests a chaperone role for the prodomain. Moreover, the ability of the prodomain to hold the catalytic domain of TACE in a relatively open conformation that is inactive has also been shown (40,41). In the case of ADAM10, the absence of the prodomain does not lead to defective biogenesis and secretion/trafficking of the protease. ADAM10 lacking the prodomain and expressed at high levels has been detected on the plasma membrane; however, it was proteolytically inactive (29). Interestingly, as with TACE, the ADAM10 prodomain expressed in trans was able to restore the catalytic activity of ADAM10 lacking the prodomain. Prodomain expression has also been shown to inhibit proteolytic activity of endogenous and overexpressed wild-type ADAM10, implying its direct interaction with mature ADAM10 (29). Future studies will be necessary to test whether the Q170H and R181G mutations can affect the chaperone function of the ADAM 10 prodomain.
In summary, we have discovered two rare, partially penetrant, familial late-onset AD mutations in the ADAM10 gene that lead to defective α-secretase activity. The fact that these two mutations were both found in late- versus early-onset familial AD (average age of onset =69.5 years for both mutations) may reflect the relatively modest effects on Aβ accumulation relative to those of the early-onset familial mutations in APP, PSEN1 and PSEN2. Furthermore, since α-secretase activity is also exerted by TACE and ADAM9, one might expect a defect in ADAM10 activity to be compensated by molecular redundancy, possibly explaining the relatively late onset of AD in carriers of these two mutations and incomplete penetrance. The novel findings presented here provide the first genetic evidence in support of a possible role for the ADAM10 gene in the etiology and pathogenesis of late-onset AD. Moreover, given the location of these two mutations in ADAM 10, these data suggest that modulation of ADAM10 activity via the prodomain could represent a novel therapeutic target for the treatment and prevention of AD.
MATERIALS AND METHODS
AD family samples
The National Institute of Mental Health (NIMH) AD Genetics Initiative Study Sample. Subjects were collected from January 1991 until September 1997 following a standardized protocol applying NINCDS/ADRDA criteria for the diagnosis of AD (42,43). Over the first 17 years that the participating families had been followed, a clinical diagnosis of AD was confirmed at autopsy in 94% of the cases (44). The full NIMH sample includes 1439 individuals (68.9% female) from 436 families with at least two affecteds, including 995 affected individuals [n = 995 affecteds (mean age of onset 72.4 ± 7.7 years, range 50–97 years), n = 411 unaffecteds, n = 34 with phenotype unknown; see Schjeide et al. for details (45)].
The Consortium on Alzheimer's Genetics (CAG) Study Sample. Subjects in this second, independently ascertained AD family sample were collected under the auspices of the ‘Consortium on Alzheimer's Genetics.’ NINCDS/ADRDA criteria were used for a clinical diagnosis of AD (42), and probands were included only if they had at least one unaffected living sibling willing to participate in this study. Unlike the NIMH sample, no affected individual beyond the proband was required, thus the majority of families include only one sample affected. Most sibships were consisted of just one discordant sibpair, while in 41 families, there were more than two siblings available. Data and specimen collection began in October 1999 (still ongoing), and is currently completed for 489 individuals (62.6% female) from 217 sibships in which all affected individuals displayed an onset age ≥50 years [n = 222 affecteds (mean age of onset 69.2 ± 9.0 years, range 50–89 years), n = 267 unaffecteds; see Schjeide et al. for details (45)].
The National Institute on Aging (NIA) Study Sample. Subjects of this AD family sample were obtained from the National Repository of Research on Alzheimer's Disease (NCRAD), and ascertainment and collection details can be found at the NCRAD website (http://www.ncrad.org). For this study, we used families of self-reported European ancestry with DNA available from at least two first-degree relatives (concordant or discordant) and in which all individuals affected with AD showed onset ages ≥50 years. It comprises 1111 subjects (62.1% female) from 351 pedigrees, including 803 affected individuals [n = 803 affecteds (mean age of onset 74.1 ± 7.1 years, range 52–98 years), n = 290 unaffecteds; see Schjeide et al. for details (45)].
Genotyping and association analyses
A total of nine SNPs (rs605928, rs593742, rs514049, rs442495, rs7161889, rs714696, rs2305421, rs4775083 and rs1869135) located in ADAM10 were initially identified from publicly available databases. SNP genotypes were generated by fluorescent polarization detected single-base extension (FP-SBE, on a ‘Criterion Analyst AD’, Molecular Devices, Inc.), using individually optimized PCR and single-base extension protocols as described previously (33). Approximately 10% of samples were run in duplicate to assess the genotyping efficiency (average across all nine SNPs >96.5%) and error rate (average across all nine SNPs <0.5%). Primer sequencing and genotyping conditions are available from the authors upon request. All nine SNPs were tested in the NIMH sample, and four of these (rs605928, rs714696, rs2305421 and rs4775083) were tested in the full CAG sample. Subsequently, NIMH families in which two risk alleles of the best-associated SNP (rs2305421) were transmitted to affected individuals were chosen for re-sequencing of the ADAM10 coding regions from genomic DNA, overall a total of 32 families.
GWAS SNPs were generated in a separate project on the Affymetrix' GeneChip Human Mapping 500K Array Set, using individually optimized genotyping and allele-calling procedures (31). SNPs selected for this project needed to be located within 100 kb of the start-/stop-codon of ADAM10, and show no deviation from Hardy–Weinberg equilibrium (P-value ≥ 0.01), and have an allele frequency of ≥0.001. Overall, this yielded 53 SNPs spanning a chromosomal interval of ∼340 kb that could be used in the statistical analyses (Supplementary Material, Table S1).
Statistical analyses
To test for association between SNPs in ADAM10 and AD risk, we used PBAT (v3.6) with an additive model and the same parameters as used in our GWAS (31). All analyses were first restricted to families of self-reported ‘Caucasian’ ancestry, and then repeated using families of all ancestries (with no change in results; data not shown). Hardy–Weinberg equilibrium was determined using ‘Haploview’ (v4.1; (46)). Association results were combined across the NIMH and CAG samples for overlapping SNPs P-values using the method described by Fisher (47) taking into account the direction of the transmissions in each individual sample.
DNA sequencing
We re-sequenced all exons and adjacent introns (∼200 bp) of ADAM10 in 96 individuals from 32 ADAM10-associated AD families (66 affected with AD and 30 unaffected) using standard capillary electrophoresis. In addition, we also re-sequenced ∼4 kb of the adjacent 5′ and ∼3 kb of the 3′ regions of ADAM10. The DNA sequence of exons 5 and 6 of ADAM10, encoding the cysteine switch and proprotein cleavage site, were further determined in all of the individuals making up the NIMH AD sample. Exons 9 and 10, encoding the active site of ADAM10, were sequenced in all 994 affected individuals from the NIMH AD sample. We used standard protocols for DNA sequencing, with minor modifications. PCR primers were designed to yield amplified DNA fragments between 500 and 900 bp in length. PCR products were amplified from ∼30 ng genomic DNA using HotStarTaq® (Invitrogen, Carlsbad, CA, USA) and individually optimized PCR conditions. The amplified DNA fragments were purified on QIAquick 96 PCR purification plates (QIAGEN, Valencia, CA, USA), and the yields were determined using PicoGreen (Molecular Probes, Eugene, OR, USA) according to the manufacturer's protocol. Sequencing reactions were performed using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA), according to the manufacturer's instructions, except that the volume of BigDye mix was reduced to 1 µl and supplemented with 2 µl 5x BigDye v3.1 sequencing buffer in each 20 µl reaction. The reactions were subjected to analysis on an ABI 3700 DNA sequencer (Applied Biosystems), and the data were evaluated using SequencherTM (Gene Codes Corporation, Ann Arbor, MI, USA) analysis software.
Plasmid construction
Mammalian expression peak12 plasmid containing human ADAM10 wild-type cDNA sequence with a C-terminal HA-tag and the empty vector were kindly provided by Dr Stefan Lichtenthaler at Ludwig-Maximilians-University, Germany (48). Q170H, R181G and E384A mutations were introduced to the wild-type ADAM10 sequence by site-directed mutagenesis using PCR with oligomeric primers containing the mutations and insert swapping with restriction enzyme digestion. The entire cDNA sequences of all ADAM10 constructs were verified.
Cell culture, transfection and generation of stable cells
The CHO cell line was grown in Dulbecco's modified Eagle's medium (BioWhittaker) supplemented with 10% fetal bovine serum (Sigma), 2 mm l-glutamine (Sigma), 100 units/ml penicillin (Sigma) and 100 µg/ml streptomycin (Sigma). CHO cells overexpressing APP751 were grown in the same medium with 200 µg/ml G418 (Sigma). To generate stable CHO cell lines expressing ADAM10, each cDNA construct harboring either wild-type, Q170H, R181G or E384A ADAM10 was transfected into the CHO-APP751 cells using the Amaxa nucleofector system (Lonza) as described in the manufacturer's instructions. Clonal ADAM10-expressing CHO-APP751 cells were selected in 250 µg/ml puromycin and 200 µg/ml G418. ADAM10 expression was confirmed by western blot analysis. Among several single cell-originated ADAM10-expressing clones, cell lines with similar levels of APP were identified by western blot analysis. The selected CHO-APP-ADAM10 stable cells were plated at the constant number and grown to exponential phase, followed by replacing the medium with the constant volume of fresh medium. After 24 h of incubation, the conditioned medium was collected and the cell lysate was prepared in M-PER mammalian protein extraction reagent (Pierce) for measuring sAPPα and ADAM10 levels, respectively. To monitor levels of APP-CTFs, the γ-secretase specific inhibitor DAPT (N-[N-(3,5-Difluorophenacetyl-L-alanyl)]-S-phenylglycine t-butyl ester, Sigma) was added to the cells (250 nM) and 24 h later cell lysate was collected and analyzed by western blot analysis. The phorbol ester effect was characterized 6 h after 1 µM of PMA (phorbol 12-myristate 13-acetate, Sigma), which was added to the cells.
For rescue of decreased sAPPα levels in ADAM10 mutant cells by wild-type ADAM10, the wild-type ADAM10 cDNA construct was transiently transfected to the CHO-APP-ADAM10 stable cells containing the mutants. Transfection efficiency was measured to be ∼90% based on counting of enhanced green fluorescent protein-positive cells. The medium was replaced with a constant volume of fresh medium 24 h after transfection. After 24 h of incubation, the conditioned medium and the cell lysate were collected.
Western blot analysis
Total protein lysates or conditioned media (the equal volume) were separated on 4–12% or 12% Bis-Tris polyacrylamide gel (Invitrogen), and transferred to immunoblot polyvinylidene difluoride membrane (Bio-Rad). For primary antibodies, anti-Aβ monoclonal antibody (6E10, Covance) was used to detect sAPPα levels in conditioned medium. Anti-APP C-terminal antibody (A8717, Sigma) was utilized for full-length APP and CTFs in cell lysates and anti-HA antibody (6E2, Cell Signaling) for HA-tagged ADAM10. Anti-actin monoclonal antibody (pan Ab-5, Neo-Markers) was used as control signal for equal loading. Protein signal was visualized using SuperSignal West Femto Maximum Sensitivity Substrate (Pierce) and the chemiluminescence signal was quantified by VersaDoc imaging system and Quantity One quantification software (Bio-Rad).
Aβ ELISA
Levels of Aβx-40 in cell culture media were determined using a human/rat Aβ-specific chemiluminescence ELISA kit (Wako Chemicals USA, Inc., VA, USA). Briefly, samples and Aβ standards were aliquoted to 96 well plates coated with monoclonal antibody against Aβ11–28 region. After overnight incubation, anti-Aβ40 specific antibody conjugated with horseradish peroxidase (HRP) was added to the captured Aβ in each well. Plates were then developed with a chemluminescent HRP substrate and the absorbance at 450 nm was read with microplate reader.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by the Cure Alzheimer's Fund and the National Institute of Mental Health (R37 MH60009).
ACKNOWLEDGEMENTS
We thank Dr Lars Bertram for planning and supervising the manual genotyping, being responsible for all aspects of the genetic association and segregation analyses. We thank Dr Stefan Lichtenthaler at Ludwig-Maximilians-University, Germany for providing a human ADAM10 wild-type cDNA construct and for his generous advice.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Saitoh T., Sundsmo M., Roch J.M., Kimura N., Cole G., Schubert D., Oltersdorf T., Schenk D.B. Secreted form of amyloid beta protein precursor is involved in the growth regulation of fibroblasts. Cell. 1989;58:615–622. doi: 10.1016/0092-8674(89)90096-2. [DOI] [PubMed] [Google Scholar]
- 2.Furukawa K., Sopher B.L., Rydel R.E., Begley J.G., Pham D.G., Martin G.M., Fox M., Mattson M.P. Increased activity-regulating and neuroprotective efficacy of alpha-secretase-derived secreted amyloid precursor protein conferred by a C-terminal heparin-binding domain. J. Neurochem. 1996;67:1882–1896. doi: 10.1046/j.1471-4159.1996.67051882.x. [DOI] [PubMed] [Google Scholar]
- 3.Meziane H., Dodart J.C., Mathis C., Little S., Clemens J., Paul S.M., Ungerer A. Memory-enhancing effects of secreted forms of the beta-amyloid precursor protein in normal and amnestic mice. Proc. Natl Acad. Sci. USA. 1998;95:12683–12688. doi: 10.1073/pnas.95.21.12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattson M.P., Guo Z.H., Geiger J.D. Secreted form of amyloid precursor protein enhances basal glucose and glutamate transport and protects against oxidative impairment of glucose and glutamate transport in synaptosomes by a cyclic GMP-mediated mechanism. J. Neurochem. 1999;73:532–537. doi: 10.1046/j.1471-4159.1999.0730532.x. [DOI] [PubMed] [Google Scholar]
- 5.Buxbaum J.D., Oishi M., Chen H.I., Pinkas-Kramarski R., Jaffe E.A., Gandy S.E., Greengard P. Cholinergic agonists and interleukin 1 regulate processing and secretion of the Alzheimer beta/A4 amyloid protein precursor. Proc. Natl Acad. Sci. USA. 1992;89:10075–10078. doi: 10.1073/pnas.89.21.10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Checler F. Processing of the beta-amyloid precursor protein and its regulation in Alzheimer's disease. J. Neurochem. 1995;65:1431–1444. doi: 10.1046/j.1471-4159.1995.65041431.x. [DOI] [PubMed] [Google Scholar]
- 7.Mills J., Reiner P.B. Mitogen-activated protein kinase is involved in N-methyl-D-aspartate receptor regulation of amyloid precursor protein cleavage. Neuroscience. 1999;94:1333–1338. doi: 10.1016/s0306-4522(99)00381-4. [DOI] [PubMed] [Google Scholar]
- 8.Caporaso G.L., Gandy S.E., Buxbaum J.D., Ramabhadran T.V., Greengard P. Protein phosphorylation regulates secretion of Alzheimer beta/A4 amyloid precursor protein. Proc. Natl Acad. Sci. USA. 1992;89:3055–3059. doi: 10.1073/pnas.89.7.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buxbaum J.D., Koo E.H., Greengard P. Protein phosphorylation inhibits production of Alzheimer amyloid beta/A4 peptide. Proc. Natl Acad. Sci. USA. 1993;90:9195–9198. doi: 10.1073/pnas.90.19.9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabuzda D., Busciglio J., Yankner B.A. Inhibition of beta-amyloid production by activation of protein kinase C. J. Neurochem. 1993;61:2326–2329. doi: 10.1111/j.1471-4159.1993.tb07479.x. [DOI] [PubMed] [Google Scholar]
- 11.Hung A.Y., Haass C., Nitsch R.M., Qiu W.Q., Citron M., Wurtman R.J., Growdon J.H., Selkoe D.J. Activation of protein kinase C inhibits cellular production of the amyloid beta-protein. J. Biol. Chem. 1993;268:22959–22962. [PubMed] [Google Scholar]
- 12.Jacobsen J.S., Spruyt M.A., Brown A.M., Sahasrabudhe S.R., Blume A.J., Vitek M.P., Muenkel H.A., Sonnenberg-Reines J. The release of Alzheimer's disease beta amyloid peptide is reduced by phorbol treatment. J. Biol. Chem. 1994;269:8376–8382. [PubMed] [Google Scholar]
- 13.Gandy S., Greengard P. Regulated cleavage of the Alzheimer amyloid precursor protein: molecular and cellular basis. Biochimie. 1994;76:300–303. doi: 10.1016/0300-9084(94)90162-7. [DOI] [PubMed] [Google Scholar]
- 14.Allinson T.M., Parkin E.T., Turner A.J., Hooper N.M. ADAMs family members as amyloid precursor protein alpha-secretases. J. Neurosci. Res. 2003;74:342–352. doi: 10.1002/jnr.10737. [DOI] [PubMed] [Google Scholar]
- 15.Hartmann D., de Strooper B., Serneels L., Craessaerts K., Herreman A., Annaert W., Umans L., Lubke T., Lena Illert A., von Figura K., et al. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum. Mol. Genet. 2002;11:2615–2624. doi: 10.1093/hmg/11.21.2615. [DOI] [PubMed] [Google Scholar]
- 16.Buxbaum J.D., Liu K.N., Luo Y., Slack J.L., Stocking K.L., Peschon J.J., Johnson R.S., Castner B.J., Cerretti D.P., Black R.A. Evidence that tumor necrosis factor alpha converting enzyme is involved in regulated alpha-secretase cleavage of the Alzheimer amyloid protein precursor. J. Biol. Chem. 1998;273:27765–27767. doi: 10.1074/jbc.273.43.27765. [DOI] [PubMed] [Google Scholar]
- 17.Merlos-Suarez A., Fernandez-Larrea J., Reddy P., Baselga J., Arribas J. Pro-tumor necrosis factor-alpha processing activity is tightly controlled by a component that does not affect notch processing. J. Biol. Chem. 1998;273:24955–24962. doi: 10.1074/jbc.273.38.24955. [DOI] [PubMed] [Google Scholar]
- 18.Weskamp G., Cai H., Brodie T.A., Higashyama S., Manova K., Ludwig T., Blobel C.P. Mice lacking the metalloprotease-disintegrin MDC9 (ADAM9) have no evident major abnormalities during development or adult life. Mol. Cell. Biol. 2002;22:1537–1544. doi: 10.1128/mcb.22.5.1537-1544.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez-Perez E., Zhang Y., Frank S.J., Creemers J., Seidah N., Checler F. Constitutive alpha-secretase cleavage of the beta-amyloid precursor protein in the furin-deficient LoVo cell line: involvement of the pro-hormone convertase 7 and the disintegrin metalloprotease ADAM10. J. Neurochem. 2001;76:1532–1539. doi: 10.1046/j.1471-4159.2001.00180.x. [DOI] [PubMed] [Google Scholar]
- 20.Lammich S., Kojro E., Postina R., Gilbert S., Pfeiffer R., Jasionowski M., Haass C., Fahrenholz F. Constitutive and regulated alpha-secretase cleavage of Alzheimer's amyloid precursor protein by a disintegrin metalloprotease. Proc. Natl Acad. Sci. USA. 1999;96:3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blacker M., Noe M.C., Carty T.J., Goodyer C.G., LeBlanc A.C. Effect of tumor necrosis factor-alpha converting enzyme (TACE) and metalloprotease inhibitor on amyloid precursor protein metabolism in human neurons. J. Neurochem. 2002;83:1349–1357. doi: 10.1046/j.1471-4159.2002.01228.x. [DOI] [PubMed] [Google Scholar]
- 22.Merlos-Suarez A., Ruiz-Paz S., Baselga J., Arribas J. Metalloprotease-dependent protransforming growth factor-alpha ectodomain shedding in the absence of tumor necrosis factor-alpha-converting enzyme. J. Biol. Chem. 2001;276:48510–48517. doi: 10.1074/jbc.M103488200. [DOI] [PubMed] [Google Scholar]
- 23.Fahrenholz F., Postina R. Alpha-secretase activation—an approach to Alzheimer's disease therapy. Neurodegener. Dis. 2006;3:255–261. doi: 10.1159/000095264. [DOI] [PubMed] [Google Scholar]
- 24.Marcinkiewicz M., Seidah N.G. Coordinated expression of beta-amyloid precursor protein and the putative beta-secretase BACE and alpha-secretase ADAM10 in mouse and human brain. J. Neurochem. 2000;75:2133–2143. doi: 10.1046/j.1471-4159.2000.0752133.x. [DOI] [PubMed] [Google Scholar]
- 25.Colciaghi F., Borroni B., Pastorino L., Marcello E., Zimmermann M., Cattabeni F., Padovani A., Di Luca M. α-secretase ADAM10 as well as αAPPs is reduced in platelets and CSF of Alzheimer disease patients. Mol. Med. 2002;8:67–74. [PMC free article] [PubMed] [Google Scholar]
- 26.Postina R., Schroeder A., Dewachter I., Bohl J., Schmitt U., Kojro E., Prinzen C., Endres K., Hiemke C., Blessing M., et al. A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J. Clin. Invest. 2004;113:1456–1464. doi: 10.1172/JCI20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seals D.F., Courtneidge S.A. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 28.Van Wart H.E., Birkedal-Hansen H. The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc. Natl Acad. Sci. USA. 1990;87:5578–5582. doi: 10.1073/pnas.87.14.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders A., Gilbert S., Garten W., Postina R., Fahrenholz F. Regulation of the alpha-secretase ADAM10 by its prodomain and proprotein convertases. FASEB J. 2001;15:1837–1839. doi: 10.1096/fj.01-0007fje. [DOI] [PubMed] [Google Scholar]
- 30.Bertram L., McQueen M.B., Mullin K., Blacker D., Tanzi R.E. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat. Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 31.Bertram L., Lange C., Mullin K., Parkinson M., Hsiao M., Hogan M.F., Schjeide B.M., Hooli B., Divito J., Ionita I., et al. Genome-wide association analysis reveals putative Alzheimer's disease susceptibility loci in addition to APOE. Am. J. Hum. Genet. 2008;83:623–632. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blacker D., Haines J.L., Rodes L., Terwedow H., Go R.C., Harrell L.E., Perry R.T., Bassett S.S., Chase G., Meyers D., et al. ApoE-4 and age at onset of Alzheimer's disease: the NIMH genetics initiative. Neurology. 1997;48:139–147. doi: 10.1212/wnl.48.1.139. [DOI] [PubMed] [Google Scholar]
- 33.Bertram L., Hiltunen M., Parkinson M., Ingelsson M., Lange C., Ramasamy K., Mullin K., Menon R., Sampson A.J., Hsiao M.Y., et al. Family-based association between Alzheimer's disease and variants in UBQLN1. N. Engl. J. Med. 2005;352:884–894. doi: 10.1056/NEJMoa042765. [DOI] [PubMed] [Google Scholar]
- 34.Levy-Lahad E., Wasco W., Poorkaj P., Romano D.M., Oshima J., Pettingell W.H., Yu C.E., Jondro P.D., Schmidt S.D., Wang K., et al. Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 35.Kauwe J.S., Jacquart S., Chakraverty S., Wang J., Mayo K., Fagan A.M., Holtzman D.M., Morris J.C., Goate A.M. Extreme cerebrospinal fluid amyloid beta levels identify family with late-onset Alzheimer's disease presenilin 1 mutation. Ann. Neurol. 2007;61:446–453. doi: 10.1002/ana.21099. [DOI] [PubMed] [Google Scholar]
- 36.Deuss M., Reiss K., Hartmann D. Part-time alpha-secretases: the functional biology of ADAM 9, 10 and 17. Curr. Alzheimer Res. 2008;5:187–201. doi: 10.2174/156720508783954686. [DOI] [PubMed] [Google Scholar]
- 37.Clement A.B., Hanstein R., Schroder A., Nagel H., Endres K., Fahrenholz F., Behl C. Effects of neuron-specific ADAM10 modulation in an in vivo model of acute excitotoxic stress. Neuroscience. 2008;152:459–468. doi: 10.1016/j.neuroscience.2007.10.060. [DOI] [PubMed] [Google Scholar]
- 38.Skovronsky D.M., Moore D.B., Milla M.E., Doms R.W., Lee V.M. Protein kinase C-dependent alpha-secretase competes with beta-secretase for cleavage of amyloid-beta precursor protein in the trans-golgi network. J. Biol. Chem. 2000;275:2568–2575. doi: 10.1074/jbc.275.4.2568. [DOI] [PubMed] [Google Scholar]
- 39.Loechel F., Overgaard M.T., Oxvig C., Albrechtsen R., Wewer U.M. Regulation of human ADAM 12 protease by the prodomain. Evidence for a functional cysteine switch. J. Biol. Chem. 1999;274:13427–13433. doi: 10.1074/jbc.274.19.13427. [DOI] [PubMed] [Google Scholar]
- 40.Milla M.E., Leesnitzer M.A., Moss M.L., Clay W.C., Carter H.L., Miller A.B., Su J.L., Lambert M.H., Willard D.H., Sheeley D.M., et al. Specific sequence elements are required for the expression of functional tumor necrosis factor-alpha-converting enzyme (TACE) J. Biol. Chem. 1999;274:30563–30570. doi: 10.1074/jbc.274.43.30563. [DOI] [PubMed] [Google Scholar]
- 41.Leonard J.D., Lin F., Milla M.E. Chaperone-like properties of the prodomain of TNFalpha-converting enzyme (TACE) and the functional role of its cysteine switch. Biochem. J. 2005;387:797–805. doi: 10.1042/BJ20041727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 43.Blacker D., Albert M.S., Bassett S.S., Go R.C., Harrell L.E., Folstein M.F. Reliability and validity of NINCDS-ADRDA criteria for Alzheimer's disease. The National Institute of Mental Health Genetics Initiative. Arch. Neurol. 1994;51:1198–1204. doi: 10.1001/archneur.1994.00540240042014. [DOI] [PubMed] [Google Scholar]
- 44.Blacker D., Bertram L., Saunders A.J., Moscarillo T.J., Albert M.S., Wiener H., Perry R.T., Collins J.S., Harrell L.E., Go R.C., et al. Results of a high-resolution genome screen of 437 Alzheimer's disease families. Hum. Mol. Genet. 2003;12:23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- 45.Schjeide B.M., McQueen M.B., Mullin K., Divito J., Hogan M.F., Parkinson M., Hooli B., Lange C., Blacker D., Tanzi R.E., et al. Assessment of Alzheimer's disease case-control associations using family-based methods. Neurogenetics. 2009;10:19–25. doi: 10.1007/s10048-008-0151-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barrett C.P., Noble M.E. Dynamite extended: two new services to simplify protein dynamic analysis. Bioinformatics. 2005;21:3174–3175. doi: 10.1093/bioinformatics/bti464. [DOI] [PubMed] [Google Scholar]
- 47.Fisher R.A. Statistical Methods for Research Workers. Edinburgh: Oliver and Boyd; 1925. [Google Scholar]
- 48.Lichtenthaler S.F., Dominguez D.I., Westmeyer G.G., Reiss K., Haass C., Saftig P., De Strooper B., Seed B. The cell adhesion protein P-selectin glycoprotein ligand-1 is a substrate for the aspartyl protease BACE1. J. Biol. Chem. 2003;278:48713–48719. doi: 10.1074/jbc.M303861200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.