Abstract
The hereditary spastic paraplegias (HSPs) are genetic conditions characterized by distal axonopathy of the longest corticospinal tract axons, and so their study provides an important opportunity to understand mechanisms involved in axonal maintenance and degeneration. A group of HSP genes encode proteins that localize to endosomes. One of these is NIPA1 (non-imprinted in Prader-Willi/Angelman syndrome 1) and we have shown recently that its Drosophila homologue spichthyin inhibits bone morphogenic protein (BMP) signalling, although the relevance of this finding to the mammalian protein was not known. We show here that mammalian NIPA1 is also an inhibitor of BMP signalling. NIPA1 physically interacts with the type II BMP receptor (BMPRII) and we demonstrate that this interaction does not require the cytoplasmic tail of BMPRII. We show that the mechanism by which NIPA1 inhibits BMP signalling involves downregulation of BMP receptors by promoting their endocytosis and lysosomal degradation. Disease-associated mutant versions of NIPA1 alter the trafficking of BMPRII and are less efficient at promoting BMPRII degradation than wild-type NIPA1. In addition, we demonstrate that two other members of the endosomal group of HSP proteins, spastin and spartin, are inhibitors of BMP signalling. Since BMP signalling is important for distal axonal function, we propose that dysregulation of BMP signalling could be a unifying pathological component in this endosomal group of HSPs, and perhaps of importance in other conditions in which distal axonal degeneration is found.
INTRODUCTION
The hereditary spastic paraplegias (HSPs) are genetic disorders characterized by distal axonopathy involving the longest axons of the motor neurons of the corticospinal tract (1–3). Their study provides an opportunity to understand molecular cellular mechanisms involved in axonal maintenance and in ‘dying-back’ axonopathy. Since a similar dying-back axonopathy is seen in some common neurological conditions (4,5), understanding its cause may have broad clinical relevance.
Numerous genes mutated in HSPs have been identified (2,3,6). An important subgroup of the proteins they encode localize to the endosomal membrane traffic compartment, suggesting that the long axons of the corticospinal tract may be especially vulnerable to endosomal dysfunction. This endosomal group includes spastin, spartin and NIPA1 (non-imprinted in Prader-Willi/Angelman syndrome 1), as well as others including maspardin and spastizin (2,3,7). Mutations in the spastin gene are the most frequent cause of HSP and in most cases are likely to act via a haploinsufficiency mechanism (8–11). Spastin is a microtubule-severing enzyme and we have recently shown that it can be recruited to a number of membrane sites, including endosomes, where it couples membrane traffic processes to microtubule remodelling (12). Mutation of the gene encoding spartin causes Troyer syndrome, an autosomal recessive HSP first identified in the Old Order Amish population, where the causative mutation is a frameshift likely to cause loss of the protein (13,14). Spartin can be recruited to endosomes, and endogenous spartin is required for efficient endosomal degradation of the EGF receptor (15,16).
Mutations in the gene that encodes NIPA1 cause an autosomal dominant HSP (17). All of the disease-causing mutations reported so far have been missense mutations (18), which affect the trafficking of the protein through the biosynthetic pathway by causing its trapping in the endoplasmic reticulum (19,20). It has been argued, based on data from overexpression systems in mammalian cells and Caenorhabditis elegans, that this could cause the disease via a gain-of-function induction of ER stress and the unfolded protein response (UPR) (19). An alternative hypothesis arising from our study of spichthyin, the Drosophila homologue of NIPA1, is that bone morphogenic protein (BMP) signalling could be involved, since spichthyin is an inhibitor of BMP signalling (21).
In Drosophila, enhanced BMP signalling causes an abnormal distal axonal overgrowth phenotype at the presynaptic neuromuscular junction (NMJ) (21–23), and indeed larvae lacking spichthyin have a distal axonal abnormality that includes synaptic overgrowth at the NMJ (21). The distal axonal phenotype seen in flies lacking spichthyin is rescued by genetic manipulations blocking BMP signalling, indicating that enhanced BMP signalling is critical to its development. The observations in the fly system therefore suggest that abnormal BMP signalling is a potential candidate mechanism by which abnormality of NIPA1 could cause axonopathy in humans.
In this study, we sought first to test whether NIPA1 regulates BMP signalling in mammalian cells. Using overexpression and depletion systems, we show that NIPA1 is an inhibitor of BMP signalling in neuronal and non-neuronal cells. We elucidate the mechanism of this effect by demonstrating that the type II BMP receptor (BMPRII) is degraded in lysosomes and that NIPA1 enhances this degradation by physically interacting with BMPRII and promoting its endocytosis and trafficking to lysosomes. We also show that mutant versions of NIPA1 are trapped in the ER, but in contrast to previous results, we find no increase in ER stress or induction of UPR on expression of NIPA1 mutants. However, we show that mutant NIPA1 does alter the trafficking of BMPRII, such that degradation of BMPRII is less efficient than when only wild-type NIPA1 is present. In addition, we show by depletion experiments that two other endosomal HSP proteins, spastin and spartin, are inhibitors of BMP signalling. As well as elucidating molecular mechanisms that regulate mammalian BMP receptor trafficking, our results suggest that dysregulation of BMP signalling could be an important unifying mechanism causing axonopathy in the endosomal group of HSPs.
RESULTS
NIPA1 is present on endosomes and at the plasma membrane, where it is subject to clathrin-dependent endocytosis
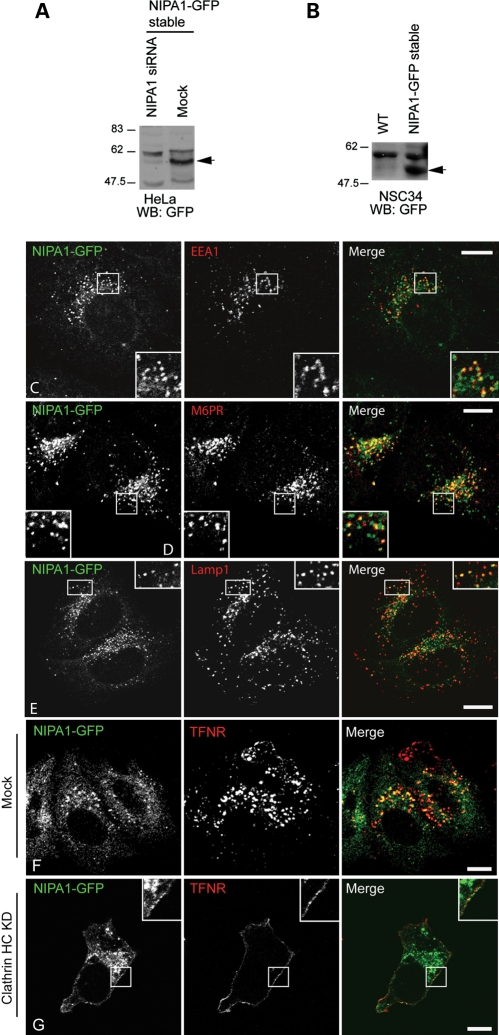
In HeLa and NSC34 (a murine hybrid neuroblastoma/motor neuronal cell line (24)) cell lines stably expressing NIPA1-GFP (Fig. 1A and B), we observed a predominantly punctate/vesicular cytoplasmic localization of NIPA1. These puncta showed partial co-localization with a number of endosomal pathway markers, including markers of early, late and recycling endosomes and of lysosomes, but did not appear to show preferential co-localization in any specific endosomal compartment that we examined (Fig. 1C–E; Supplementary Material, Fig. S1A–D). In NSC34 cells, the NIPA1 staining extended into neurites (Supplementary Material, Fig. S1C). We saw no co-localization with Golgi markers (data not shown). In some cells weak plasma membrane staining was seen. This plasma membrane staining was enhanced by clathrin heavy chain depletion (Fig. 1F and G), indicating that plasma membrane NIPA1-GFP is subject to clathrin-mediated endocytosis. Our data are therefore consistent with previous reports that endogenous NIPA1 localizes to endosomes and to the plasma membrane in cultured mammalian cells (19,20), but inconsistent with the additional localization of NIPA1 to the Golgi apparatus reported by Zhao et al. (20).
Figure 1.
NIPA1-GFP localizes to endosomes. (A) Immunoblot showing expression of a band (arrow) of approximately the size predicted for NIPA1-GFP (∼61 kDa) in the NIPA1-GFP HeLa stable cell line, but not in similar cells transfected with pooled siRNA against NIPA1. (B) Immunoblot showing expression of a band of approximately the size predicted for NIPA1-GFP (arrow) in the NIPA1-GFP NSC34 stable cell line, but not in wild-type (WT) NSC34 cells. (C–E) Stably expressed NIPA1-GFP co-localizes in HeLa cells with early endosome antigen (EEA1; early endosomes; C), mannose 6-phosphate receptor (M6PR; late endosomes; D) and Lamp1 (lysosomes; E). Although results are shown for a mixed stable cell line, similar findings were obtained with individual clonal cell lines (data not shown). (F and G) Clathrin heavy chain (HC) knock-down redistributed NIPA1-GFP to the plasma membrane. (F) Control cells stably expressing NIPA1-GFP in which mock siRNA transfections were carried out. Minimal NIPA1-GFP or transferrin receptor (TFNR) is present at the plasma membrane. However, when clathrin HC is depleted by siRNA knock-down (G), TFNR was redistributed, as expected, to the plasma membrane. NIPA1-GFP was also partially redistributed to the plasma membrane (see inset higher magnification box). In these and subsequent micrographs, the right-hand panels show the merged images; the colour of each marker in the merged image is shown by the colour of its lettering in the non-merged panels. Scale bars: 10 µm in these and subsequent micrographs.
NIPA1 is an inhibitor of BMP signalling in neuronal and non-neuronal mammalian cells
We next examined whether NIPA1 regulates BMP signalling. In the BMP signalling pathway, binding of BMP ligand to the serine/threonine kinase BMPRII activates type I BMP receptors, which in turn phosphorylate the intracellular signalling molecules Smad 1, 5 and 8. These phosphorylated Smads (pSmads) bind Smad 4 and then the Smad complex translocates to the nucleus to regulate target gene transcription, notably of the Id gene family (21).
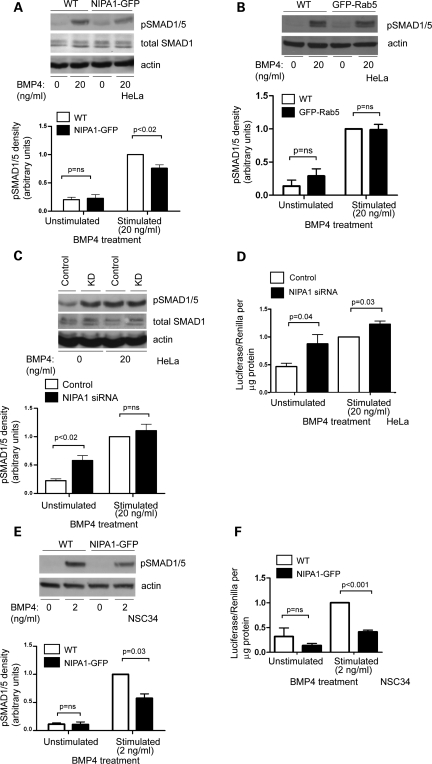
We first characterized the effect of NIPA1 overexpression or depletion on BMPR-mediated phosphorylation of Smads 1 and 5. In a mixed stable HeLa cell line, expression of NIPA1-GFP resulted in a diminished pSmad1/5 response to BMP4 stimulation, compared with untransfected cells and cells expressing another endosomal protein, GFP-Rab5 (Fig. 2A and B). Similar statistically significant effects were found with two clonal NIPA1-GFP HeLa cell lines (data not shown). The attenuation of the pSmad1/5 response by NIPA1 was slightly less than that resulting from BMPRII knock-down (Supplementary Material, Fig. S2A). Conversely, in HeLa cells depleted of NIPA1 using an siRNA pool of four oligonucleotides, the concentration of pSmad1/5 significantly increased in unstimulated cells and showed a slight but not significant increase in cells stimulated with BMP4 (Fig. 2C). A similar increase in pSmad1/5 concentration in unstimulated cells was seen when two siRNA oligonucleotides from the pool were used individually (Supplementary Material, Fig. S2B), strongly suggesting that this was not an off-target effect.
Figure 2.
NIPA1 regulates BMP signalling. (A) A representative immunoblot examining the effect of BMP4 stimulation on the amount of pSmad1/5, in wild-type (WT) HeLa cells or in HeLa cells stably expressing NIPA1-GFP. Immunoblotting for total Smad1 (which did not differ in concentration between WT and NIPA1-GFP-expressing cells) is also shown. The histogram shows quantitation of pSmad1/5 immunoblot band density in five such experiments. (B) A representative immunoblot of the effect of BMP4 stimulation on the amount of pSmad1/5, in WT HeLa cells or in HeLa cells stably expressing GFP-Rab5. The corresponding histogram shows quantitation of pSmad1/5 immunoblot band density in three such experiments. GFP-Rab5 expression in this line was similar to the expression level of NIPA1-GFP in the NIPA1-GFP stable line (data not shown). (C) Representative immunoblot examining the amount of pSmad1/5 and total Smad1 in HeLa cells depleted of NIPA1 by siRNA knock-down (KD) using an oligonucleotide pool, and in control cells. Results are shown for unstimulated cells or cells stimulated with BMP4. The pSmad1/5 immunoblotting results of four such experiments are quantified in the corresponding histogram. (D) The effect of NIPA1 depletion by siRNA KD using an siRNA pool on Id luciferase reporter gene activation, normalized to Renilla luciferase activity and protein mass (n = 6). (E) A representative immunoblot examining the effect of BMP4 stimulation on the amount of pSmad1/5, in WT NSC34 cells or in NSC34 cells stably expressing NIPA1-GFP. The histogram shows quantitation of pSmad1/5 immunoblot band density in three such experiments. (F) Id luciferase reporter gene activity, normalized to Renilla luciferase activity and protein mass, in WT NSC34 cells or in NSC34 cells stably expressing NIPA1-GFP, either unstimulated or stimulated with BMP4 (n = 4). (A–C and E) Actin immunoblotting verified equal loading. In all histograms, values were normalized by dividing by the value for stimulated WT or control. Knock-down control samples were either mock-transfected or transfected with a non-targeting siRNA. Control experiments (data not shown) demonstrated that the effect of mock-transfection or transfection with control siRNA on pSmad1/5 and id luciferase assays was identical, so these controls were used interchangeably. Error bars: SEM. P-values were calculated using two-tailed paired t-tests.
We assessed the effects of NIPA1 depletion on the downstream BMP pathway transcriptional response by using a reporter gene assay with a construct containing a BMP-responsive element derived from an Id gene promoter, fused to the firefly luciferase gene (26). Results were normalized against Renilla luciferase activity to control for transfection efficiency and cell viability. Depletion of NIPA1 using an siRNA pool resulted in increased reporter activity in unstimulated HeLa cells and cells stimulated with BMP4, compared with control cells (Fig. 2D). Similar effects were found when two oligonucleotides from the pool were used individually (Supplementary Material, Fig. S2C). This assay performed as expected following depletion of BMPRII, which significantly attenuated the response to BMP stimulation (Supplementary Material, Fig. S2D).
NSC34 cells are a motor neuron-derived line relevant to the HSP phenotype and so we also used them to examine the role of NIPA1 in BMP signalling. Since we were unable to obtain efficient siRNA-mediated knock-down of NIPA1 in this line, we examined the effects of stable expression of NIPA1-GFP. This lowered both the pSmad1/5 response to BMP stimulation (Fig. 2E), and Id promoter reporter transcription (Fig. 2F) in cells stimulated with BMP4, compared to wild-type NSC34 cells or NSC34 cells stably transfected with GFP-Rab5 (Supplementary Material, Fig. S2E and F).
We concluded that NIPA1 is an inhibitor of BMP signalling in mammalian neuronal and non-neuronal cells.
NIPA1 redistributes BMPRII from the plasma membrane to endosomes and lysosomes
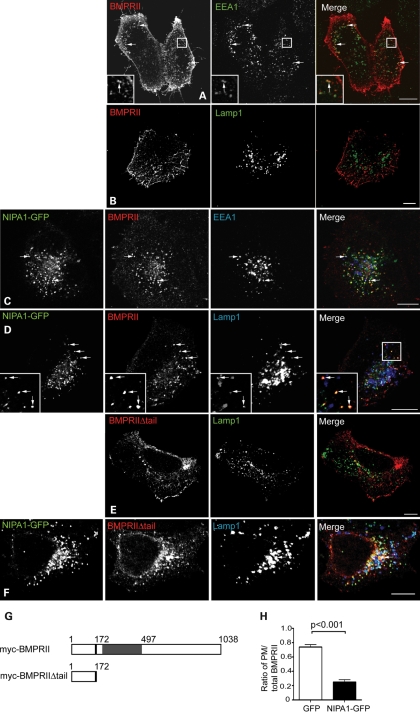
We next examined whether the inhibition of BMP signalling caused by NIPA1 was associated with altered BMP receptor trafficking. In the Drosophila system, expression of spichthyin results in internalization of the BMPRII receptor to an early endosomal compartment (21). For the mammalian cell experiments, we concentrated on HeLa cells to facilitate morphological examination. In transfected wild-type HeLa cells, myc-tagged BMPRII is mainly expressed on the cell surface, with a small amount of staining in sparse cytoplasmic vesicles. These co-label with endogenous markers of recycling and early endosomes, but not with late endosomes and lysosomes (Fig. 3A and B; Supplementary Material, Fig. S3). However, in HeLa cells stably expressing NIPA1-GFP, myc-BMPRII was strikingly internalized from the plasma membrane and co-localized with NIPA1-GFP not only in early and recycling endosomes, but also in late endosomes and lysosomes (Fig. 3C and D; Supplementary Material, Fig. S3). A similar NIPA1-induced redistribution from the plasma membrane to internal vesicles, including lysosomes, was observed with a form of myc-BMPRII (myc-BMPRIIΔtail) deleted for the intracellular tail (Fig. 3E–G). Quantitation of the effect of NIPA1-GFP indicated that 75% of total full-length myc-BMPRII fluorescence was at the plasma membrane in cells expressing GFP alone, whereas only 25% of BMPRII fluorescence was at the plasma membrane in cells expressing NIPA1-GFP (Fig. 3H). There was no effect on the distribution of the epidermal growth factor receptor (EGFR) in the NIPA1-GFP cells (Supplementary Material, Fig. S4), nor did expression of GFP alone affect the distribution of BMPRII compared with untransfected cells (data not shown).
Figure 3.
NIPA1 promotes traffic of BMPRII to a lysosomal compartment in HeLa cells. Wild-type HeLa cells (A, B and E) or cells stably expressing NIPA1-GFP (C, D and F) were transiently transfected with myc-BMPRII (A–D) or myc-BMPRIIΔtail (E and F) and labelled with the early endosomal marker EEA1 (A and C) or the lysosomal marker Lamp1 (B and D–F). In the absence of NIPA1, there is minimal co-localization between BMPRII and EEA1 (A) and almost no co-localization between BMPRII and Lamp1 (B and E). In the cells expressing NIPA1-GFP (C, D and F), note the striking internalization of both full-length and truncated BMPRII from the plasma membrane, the appearance of co-localization between NIPA1-GFP and BMPRII (which appears yellow in the merged images) and of three-way co-localization between NIPA1, BMPRII and EEA1/Lamp1. In (A), (C), (D) and (F), arrows indicate discrete puncta showing co-localization. (G) Schematic diagram illustrating the two N-terminal myc-tagged constructs used in experiments. The constructs encoded either full-length BMPRII (myc-BMPRII) or a truncated form of BMPRII lacking the intracellular tail (myc-BMPRIIΔtail). The transmembrane region is shown in black, with the serine/threonine kinase domain in grey. (H) Representative experiment showing quantitation of plasma membrane anti-myc-BMPRII fluorescence versus total BMPRII fluorescence, in HeLa cells transiently expressing GFP alone (32 cells), or stably expressing NIPA1-GFP (35 cells). Error bars: SEM. P-value was calculated using unpaired t-test.
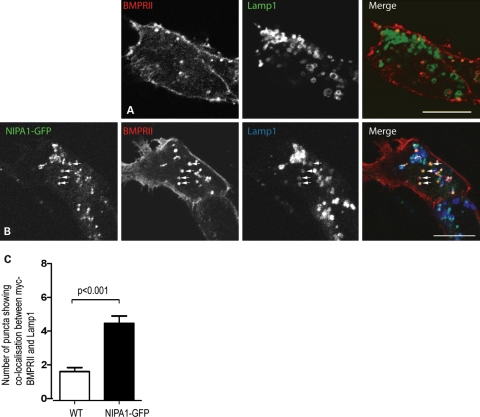
In HeLa cells depleted of NIPA1, BMPRII remained predominantly at the plasma membrane (data not shown). In NSC34 cells stably expressing NIPA1-GFP, the reduction of myc-BMPRII at the plasma membrane was not as dramatic as seen in HeLa NIPA1-GFP stable cells. Nevertheless, expression of NIPA1 resulted in the appearance of much stronger co-localization between BMPRII and lysosomes than was seen in wild-type NSC34 cells (Fig. 4A–C). Thus, expression of NIPA1 caused redistribution of BMPRII from the plasma membrane to endosomal and lysosomal compartments, in neuronal and non-neuronal cells, and this redistribution did not depend on BMPRII tail sequences.
Figure 4.
NIPA1 promotes traffic of BMPRII to a lysosomal compartment in neuronal cells. (A and B) Wild-type NSC34 cells (A) or NSC34 cells stably expressing NIPA1-GFP (B) were transiently transfected with myc-BMPRII and labelled with the lysosomal marker Lamp1. Note the appearance of strong co-localization between BMPRII and Lamp1 in the NIPA1-GFP-expressing cell. Arrows indicate selected individual vesicles that show co-localization. (C) Count of vesicles doubly positive for myc-BMPRII and Lamp1 in wild-type NSC34 cells (32 cells) or stably expressing NIPA1-GFP (35 cells). Note the increase in the number of puncta showing co-localization between Lamp1 and myc-BMPRII in the NIPA1-stable cells. Error bars: SEM. P-value was calculated using unpaired t-test.
NIPA1 enhances lysosomal degradation of endogenous BMPRII
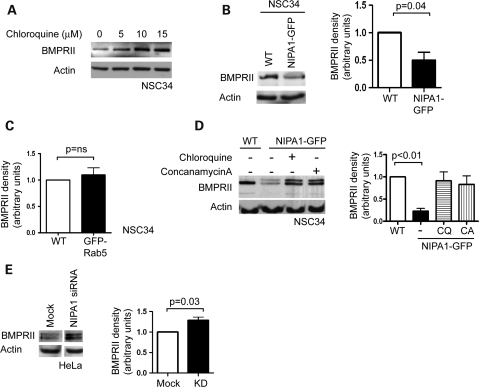
We next assessed whether the NIPA1-mediated redistribution of BMPRII to lysosomes results in increased degradation of BMPRII. We primarily examined NSC34 cells, as a cell line relevant to HSP. We first showed that the inhibition of lysosomal function by treatment with chloroquine (a weak base which accumulates in endosomes and lysosomes, increasing their pH and thus inactivating lysosomal proteases) increased the amount of endogenous BMPRII, suggesting that BMPRII normally undergoes significant lysosomal degradation (Fig. 5A). We then examined the effect of NIPA1-GFP on endogenous BMPRII and found significantly less in cells stably expressing NIPA1-GFP versus wild-type cells or cells expressing GFP-Rab5 (Fig. 5B and C). Similar effects were seen in HeLa cells (data not shown). This effect could be rescued by treatment with the lysosomal inhibitor chloroquine or concanomycin A (an inhibitor of the vacuolar ATPase (27)) (Fig. 5D). Interestingly, on immunoblotting to detect endogenous BMPRII, we typically saw the appearance of a higher molecular weight band in NSC34 cells stably expressing NIPA1-GFP (Fig. 5D), suggesting that NIPA1 promotes or stabilizes a post-translational modification of BMPRII. In HeLa cells this higher molecular weight band was constitutively present (Supplementary Material, Fig. S2A).
Figure 5.
NIPA1 enhances lysosomal degradation of BMPRII. (A) Representative experiment showing endogenous BMPRII immunoblotting in NSC34 cells treated with the indicated amount of chloroquine. (B) Representative experiment in NSC34 cells showing the effects of stable NIPA1-GFP expression on endogenous BMPRII. Quantitation of immunoblot band density in four such experiments is shown in the histogram. Stable expression of GFP-Rab5 did not affect endogenous BMPRII levels, as shown in (C). (D) Representative experiment showing rescue of endogenous BMPRII levels by the lysosomal inhibitors chloroquine (CQ) or concanamycin A (CA) in NSC34 cells stably expressing NIPA1-GFP. Note that an additional higher molecular weight band is detected by BMPRII immunoblotting in the stable cell line. Quantitation of immunoblot band density in three such experiments is shown in the histogram. (E) Representative immunoblot showing the effect on endogenous BMPRII of NIPA1 depletion by siRNA. The images were taken from the same immunoblot, and spurious lanes have been spliced out. The histogram shows quantitation of BMPRII immunoblot density in three such experiments. (A, B, D and E) Actin immunoblotting verified equal loading. In the histograms, values were normalized to the relevant wild-type or mock value. Error bars: SEM. P-values were calculated using two-tailed paired t-tests.
We then went on to examine the effects of NIPA1 depletion on the amount of endogenous BMPRII, using HeLa cells, where we could efficiently knock-down NIPA1. We found more BMPRII in cells depleted of NIPA1 after transfection with pooled NIPA1 oligonucleotides (Fig. 5E). Similar effects were seen on depletion using two individual oligonucleotides from the pool, strongly suggesting that this was not an off-target effect (Supplementary Material, Fig. S5). Thus, overexpression and depletion of NIPA1 had converse effects. This effect showed some specificity, since the amount of EGFR was not altered by NIPA1-GFP expression (Supplementary Material, Fig. S4). Together with the trafficking data, our results suggest that NIPA1 promotes internalization of BMPRII and enhances its transport to lysosomes, where it is degraded. This is likely to be the mechanism underlying the inhibitory effect of NIPA1 on BMP signalling.
NIPA1 physically interacts with endogenous BMPRII
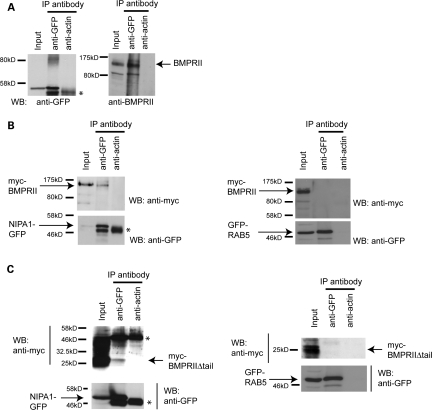
We next examined whether there is an interaction between NIPA1 and BMPRII. We first immunoprecipitated NIPA1-GFP from NSC34 cells stably expressing the protein, using anti-GFP, or anti-actin as a spurious control antibody, and immunoblotted with anti-BMPRII. In samples immunoprecipitated with anti-GFP, but not anti-actin, we observed faint immunoblot bands corresponding to the size of BMPRII (data not shown). In order to more clearly demonstrate this co-immunoprecipitation, we treated the NIPA1-GFP-expressing cells with chloroquine, to increase BMPRII concentration. In this situation, we found clear co-immunoprecipitation of endogenous BMPRII in cells immunoprecipitated with anti-GFP, but not with the anti-actin control antibody (Fig. 6A).
Figure 6.
NIPA1 co-immunoprecipitates with BMPRII. (A) NSC34 cells stably expressing NIPA1-GFP were first treated with chloroquine to increase the concentration of endogenous BMPRII and used in immunoprecipitation experiments with anti-GFP, or anti-actin as a control antibody. Total cell lysates (input lane) and the immunoprecipitated samples were then immunoblotted with anti-GFP or anti-BMPRII antibodies. The expected size of full-length endogenous BMPRII is indicated. (B and C) HeLa cells stably expressing NIPA1-GFP and transiently transfected with either myc-BMPRII (B) or myc-BMPRIIΔtail (C) were used in immunoprecipitation experiments with anti-GFP, or anti-actin as a control antibody. Total cell lysates (input lanes) and immunoprecipitated samples were then immunoblotted with anti-GFP or anti-myc. Immunoglobulin bands are marked ‘*’. Control experiments in which the immunoprecipitation was carried out in cells stably expressing GFP-Rab5 are shown on the right-hand side of (B) and (C).
We examined whether the interaction between NIPA1 and BMPRII is mediated by sequences on the cytoplasmic tail of BMPRII by transfecting full-length myc-BMPRII or myc-BMPRIIΔtail (Fig. 3G) into HeLa cells stably expressing NIPA1-GFP, and then immunoprecipitating as described earlier. We did not employ chloroquine treatment in these experiments. We found co-immunoprecipitation of both myc-BMPRII constructs with NIPA1-GFP, but not with a GFP-Rab5 expressing control cell line (Fig. 6B–E). Thus, mammalian NIPA1 interacts with BMPRII, and this interaction is not dependent on intracellular tail sequences of BMPRII. These results are consistent with our finding that internalization of BMPRII from the plasma membrane to endosomes is independent of the BMPRII cytoplasmic tail (Fig. 3E and F).
Stable expression of mutant NIPA1 results in ER trapping but does not provoke increased ER stress compared with wild-type NIPA1
Reports of transient transfection experiments in mammalian cells have indicated that mutant NIPA1 is subject to increased ER trapping compared with wild-type NIPA1, although in at least some published experiments this was in the context of cells cultures showing extensive apoptotic cell death caused by expression of the mutant protein (19,20). We generated mixed stable HeLa cell lines expressing T45R or G106R NIPA1-GFP. These cell lines maintained expression of the mutant NIPA1-GFP well, and we did not see any evidence of an increase in cells with apoptotic nuclei. We therefore used these cellular models to examine whether, even in the absence of increased cell death, mutant NIPA1 is subject to increased ER trapping.
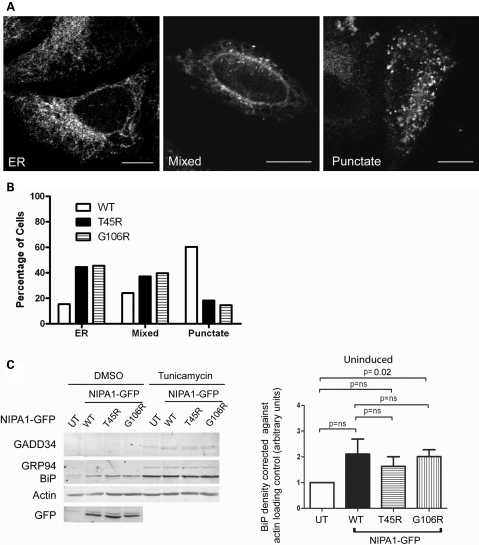
In both mutant lines, a proportion of cells expressed NIPA1 in a reticular pattern that co-localized with ER markers, whereas in other cells, NIPA1 was punctate and co-localized with endosomal markers, including EEA1, M6PR and Lamp 1. Other cells showed an overlapping pattern between these two extremes (Fig. 7A; Supplementary Material, Fig. S6; data not shown). The ER/reticular pattern was much more frequent in the mutant cell lines (>40% of cells) than in the wild-type NIPA1 line (<20% of cells). Conversely, the punctate expression pattern was much less frequent in the mutant cell lines (<20% of cells compared with >60% of cells in the wild-type line). Our results are therefore consistent with previous studies which showed that mutant NIPA1 is subject to increased trapping in the ER compared with wild-type NIPA1.
Figure 7.
Mutant NIPA1 is prone to ER trapping but does not induce ER stress or UPR more strongly than wild-type NIPA1. (A) When stably expressed in HeLa cells, the T45R-NIPA1 showed an ER-like reticular distribution in some cells (left panel), a punctate distribution in others (right panel) and a mixed distribution in a third group (middle panel). (B) Quantitation of these three distribution patterns. n = 298, 312 and 310 cells of the WT NIPA1 line, the T45R mutant line and the G106R mutant line, respectively. (C) Representative immunoblots show GADD34, BiP and GRP94 concentration in untransfected HeLa cells (UT) and HeLa cells stably expressing wild-type (WT) or mutant forms of NIPA1-GFP, in cells treated with vehicle (DMSO) or tunicamycin. Actin immunoblotting verifies equal loading, and GFP immunoblotting verifies equal expression levels of WT and mutant NIPA1-GFP in respective cell lines. (D) Histogram shows quantitation of BiP expression in uninduced (i.e. no tunicamycin) samples from five such experiments. Graphed values were corrected for actin loading and then normalized against the value for the untransfected (UT) samples. Error bars: SEM. P-values were calculated using two-tailed paired t-tests.
Zhao et al. (20) suggested that transient expression in mammalian cells of mutant NIPA1, but not wild-type NIPA1, led to ER stress and UPR, although this was in the context of cultures showing extensive cell death. We therefore examined whether we could detect an increased ER stress response in our stable cell lines. We assessed expression levels of GADD34, BiP and GRP94. GADD34 is strongly induced during ER stress, and induction of the ER chaperone BiP has been used extensively as an indicator of the onset of UPR (28). GRP94 is an ER chaperone which is in complex with BiP (29). We saw no evidence of any induction of GADD34 or GRP94 in either the wild-type or mutant cell lines. Compared with untransfected cells, there was a trend for BiP concentration to increase in the stable cell lines, but this was no greater in the mutant lines than in the wild-type NIPA1 line (Fig. 7C). As a positive control, we examined the effect of tunicamycin on the cell lines. Correct protein folding of many ER client proteins relies on N-linked glycosylation and exposure to the glycosylation inhibitor tunicamycin leads to increased protein misfolding and ER stress (28). In response to tunicamycin GADD34, GRP94 and BiP were all induced to a similar extent in untransfected cells and in the wild-type and mutant stable lines, confirming that these lines are capable of showing ER stress and UPR (Fig. 7C). In conclusion, we found no evidence of ER stress or UPR in NIPA1 mutant cell lines, above that seen in cells expressing wild-type NIPA1.
Mutant NIPA1 alters the trafficking of BMPRII and downregulates BMPRII less efficiently than wild-type NIPA1
Since our results did not provide support for a model in which mutant NIPA1-induced ER stress is a mechanism for the disease, we asked whether mutant NIPA1 might differ from wild-type NIPA1 in its role in the regulation of BMP signalling or BMPRII traffic.
First, we examined whether there was any abnormality of BMP signalling in cell lines stably expressing mutant NIPA1. We carried out pSMAD1/5 and id luciferase assays in untransfected cells and in NSC34 cell lines stably expressing wild-type or mutant NIPA1. We used motor neuron-derived NSC34 cells to more closely reflect the pathological target cells of HSP. We found that both of the mutant proteins and the wild-type protein inhibited the response to BMP4 stimulation to a similar degree (Supplementary Material, Fig. S7).
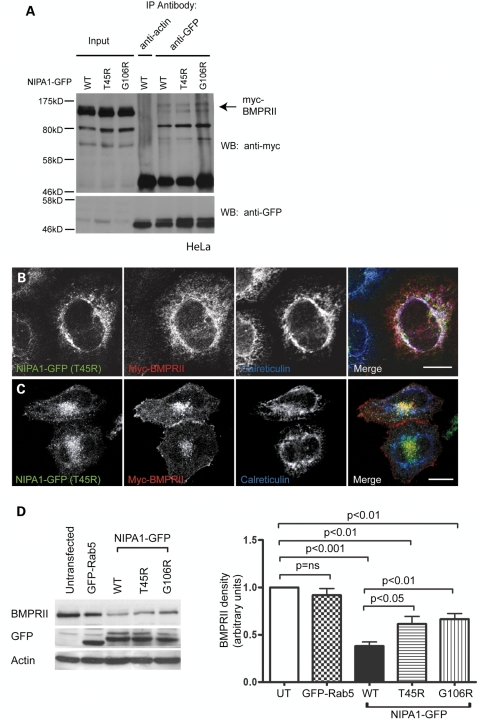
We next examined whether the NIPA1 mutants influenced BMPRII traffic. We began by showing that both the T45R and G106R mutants are, like wild-type NIPA1, capable of interacting with BMPRII in co-immunoprecipitation experiments (Fig. 8A). We then determined whether the steady-state distribution of myc-BMPRII was altered in cells expressing mutant NIPA1. We used HeLa cells stably transfected with NIPA1 for these assays, to facilitate examination of subcellular morphology. In cells showing an ER-like pattern of NIPA1-mutant distribution, we found that myc-BMPRII remained in the ER and co-localized with the mutant NIPA1 (Fig. 8B). However, in cells in which mutant NIPA1 was present in a punctate endosomal pattern, BMPRII was also present on the puncta (Fig. 8C). Similar results were found for both mutants (G106R data not shown). We concluded that if it leaves the ER, mutant NIPA1 is trafficked in a similar fashion to wild-type NIPA1 and is capable of redistributing BMPRII from the cell surface to endosomes. In contrast, if mutant NIPA1 is trapped in the ER, it causes retention of BMPRII there also.
Figure 8.
Mutant NIPA1 interacts with BMPRII and alters its trafficking. (A) HeLa cells stably expressing wild-type or mutant forms of NIPA1-GFP and transiently transfected with myc-BMPRII were used in immunoprecipitation experiments with anti-GFP. Anti-actin was used as a control antibody in immunoprecipitation of the WT NIPA1-GFP sample. Total cell lysates (input lanes) and immunoprecipitated samples were then immunoblotted with anti-GFP or anti-myc. (B and C) HeLa cells stably expressing T45R NIPA1-GFP were transiently transfected with myc-BMPRII and labelled with the ER marker calreticulin. (B) A cell in which the mutant NIPA1, BMPRII and calreticulin show three-way co-localization. (C) A cell in which the BMPRII co-localizes with mutant NIPA1 in perinuclear puncta. (D) Representative experiment showing the effects of stable expression of WT or mutant forms of NIPA1-GFP on endogenous BMPRII concentration. A HeLa cell line stably expressing GFP-Rab5 was used as a control. GFP immunoblotting verified approximately equal expression levels of GFP-tagged proteins in respective cell lines, and actin immunoblotting verified equal loading. Quantitation of immunoblot band density in four such experiments is shown in the histogram. In the histograms, values were normalized to the value for the untransfected sample. Error bars: SEM. P-values were calculated using two-tailed paired t-tests.
We then asked whether mutant NIPA1 could downregulate BMPRII expression level in the same way as wild-type NIPA1 did. We examined the concentration of endogenous BMPRII in the HeLa stable cell lines expressing wild-type NIPA1, versus the cell lines expressing the T45R or G106R mutant protein. Although downregulation of BMPRII did occur in the mutant cell lines, it was significantly less than that seen in cells expressing the wild-type protein (Fig. 8D). This may be because BMPRII reaches lysosomes less efficiently in the mutant lines, either because it is sequestered in the ER by mutant NIPA1, or because endosomal NIPA1 levels were insufficient to promote this.
We concluded that mutant NIPA1 is prone to ER trapping, and that this can influence the trafficking and degradation of BMPRII.
Spastin and spartin are inhibitors of BMP signalling
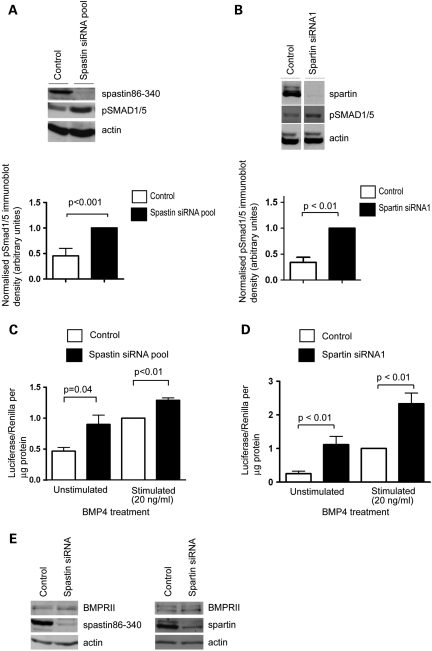
Finally, we examined whether regulation of BMP signalling might be a function common to other endosomal HSP proteins, focusing on spastin and spartin. The main pathological mechanism associated with these proteins is loss of, or reduced, normal function, for spastin by haploinsufficiency (8–11) and for spartin by absence of the protein (13,14), so cells depleted of the proteins are therefore appropriate disease models. Remarkably, depletion of spastin or spartin in HeLa cells was associated with increased pSmad1/5 concentration in non-BMP4-stimulated cells (Fig. 9A and B) and increased Id luciferase reporter activity in unstimulated and BMP4-stimulated cells (Fig. 9C and D). For each protein, these results were verified with two individual siRNA oligonucleotides, strongly suggesting that they were not caused by an off-target effect (Supplementary Material, Fig. S8). In contrast, the use of a control non-targeting siRNA did not affect basal pSmad1/5 levels in unstimulated cells or have any significant effect on the Id luciferase reporter assay, compared with mock-transfected cells (data not shown). The effect of spastin and spartin depletion on the pSmad and Id luciferase assays is therefore strikingly similar to that seen with NIPA1 depletion. These results indicate that all the three of the endosomal HSP proteins that we have tested are inhibitors of BMP signalling. However, in the case of spastin and spartin, the mechanism of this effect does not appear to be via regulation of BMPRII degradation, since depletion of spastin or spartin had no effect on BMPRII concentration (Fig. 9E).
Figure 9.
Spastin and spartin are inhibitors of BMP signalling. (A) Representative experiment examining the effect of spastin siRNA knock-down (KD) using a pool of four oligonucleotides, on the amount of pSmad1/5 in HeLa cells, in the absence or presence of BMP4 stimulation. The histogram shows quantitation of the pSmad1/5 immunoblot band density in seven such experiments, normalized against the spastin KD value. (B) Equivalent experiments to (A), but examining the effects of spartin depletion using spartin siRNA1 (n = 4). (C) The effect of spastin KD on Id luciferase reporter gene assays, in unstimulated HeLa cells or in cells stimulated with BMP4 (n = 3). (D) An equivalent experiment to (C), but examining the effects of spartin depletion (n = 5 unstimulated, n = 7 stimulated). Histogram results were normalized against the control-stimulated value. (E) Immunoblotting of BMPRII in control cells or cells depleted, by siRNA, of spastin or spartin. Control samples were either mock-transfected or transfected with a non-targeting siRNA. Error bars: SEM. P-values were calculated using two-tailed paired t-tests.
DISCUSSION
We have found that depletion of endogenous spastin, spartin or NIPA1 enhanced pre- and post-transcriptional BMP signalling, since it resulted in increased concentrations of pSmad1/5 and increased transcriptional activity of a reporter gene construct containing a BMP-responsive promotor. In each case these results were verified with at least two individual siRNA oligonucleotides, indicating that they are highly unlikely to be due to off-target effects. Remarkably, our study therefore indicates that all three of these endosomal HSP proteins are inhibitors of BMP signalling.
How could endosomal proteins influence BMP signalling? In signalling pathways, there is often a close relationship between signal transduction and endocytic membrane traffic (30–32). For many cell surface receptors, e.g. EGFR, plasma membrane receptor levels and the consequent signalling response can be regulated by sorting decisions made at the early endosome, for example between recycling of the receptor back to the plasma membrane, or degradation in the lysosome (33). In addition, as well as modulating receptor levels, endocytic processes can be important in maintaining or attenuating the signalling response (30–32). Such a relationship between signalling and endocytosis is known to be important in the BMP pathway in flies, where there is an emerging recognition that endocytic processes provide a mechanism to fine-tune BMP signalling. Drosophila lacking a number of endosomal proteins, such as spichthyin, nervous wreck, Dap160 and spinster, show upregulated BMP signalling (22,23,34). At least some of these endosomal proteins, including spichthyin, have been implicated in regulating endosomal BMPR traffic (21,23). Thus, spichthyin binds the Drosophila BMPRII, and in Drosophila S2 cells, expression of spichthyin promotes endocytosis of BMPRII from the plasma membrane (21), although the mechanism by which this, in turn, downregulates BMP signalling in flies is not yet clear.
In mammals, although it is known that endocytosis is important for BMP signalling (35,36), neither the endocytic trafficking itinerary of BMPRs nor the molecular mechanisms regulating endocytic BMPR trafficking have been fully characterized. We therefore analysed whether mammalian NIPA1 plays a similar role to Drosophila spichthyin. We found that NIPA1 resides on the plasma membrane and endosomes, is an inhibitor of BMP signalling and promotes internalization of BMPRII from the plasma membrane to an endocytic compartment. In these respects, it behaves strikingly like spichthyin. In addition, our results have given novel insights into the mechanism by which NIPA1 regulates BMP signalling. We have shown that the lysosome makes a significant contribution to BMPRII degradation and that NIPA1 enhances this by binding to BMPRII and promoting its traffic from the plasma membrane towards the lysosomal compartment. Outside of the serine/threonine kinase domain, there is little sequence homology between the tail of mammalian and Drosophila BMPRII (37,38), and neither binding of NIPA1 to BMPRII nor the effect of NIPA1 in promoting endocytosis of BMPRII from the plasma membrane required sequences in the BMPRII tail. Since NIPA1 is strongly hydrophobic and has at least seven transmembrane regions, we speculate that the binding between BMPRII and NIPA1 may be mediated via a transmembrane interaction. Interestingly, there are several transmembrane sequences that show strong conservation between NIPA1 and spichthyin, which could be candidate binding domains.
BMP signalling is important for axonal function. In Drosophila, BMP signalling is a main determinant of synaptic growth at the NMJ, and enhanced BMP signalling, for example as found in flies lacking spichthyin or the other endosomal proteins mentioned earlier, causes an abnormal distal axonal overgrowth phenotype (21–23). During mammalian development, BMP signalling regulates axonal growth, guidance and differentiation (39–41); for example Id2, a member of the Id family of BMP-responsive transcription factors, controls axonal differentiation by regulating a number of axonal inhibitory pathways. These include the Nogo pathway, which is involved in the inhibition of synaptic plasticity and the prevention of CNS axonal regeneration after injury (41). Moreover, expression of BMPs is upregulated after corticospinal tract injury, and the inhibition of this upregulated BMP signalling promotes corticospinal axon regeneration (42).
Against this background, we have found that three endosomal HSP proteins are involved in the regulation of BMP signalling. Are our findings relevant to the molecular pathology of the disease? For spastin and spartin, where haploinsufficiency or complete loss of the protein are pathological mechanisms (8–11,14), the cellular depletion assays that we examined are pathologically relevant. Results from these depletion experiments therefore suggest that upregulation of BMP signalling could have a role in the development of the phenotype. The situation is more complex with NIPA1. We modelled the disease here by generating stable cell lines expressing disease-associated mutants. In keeping with previous studies, we found that the mutant protein is more prone to ER retention than the wild-type protein, and that this retention affected BMPRII trafficking. Thus, when mutant NIPA1 was retained in the ER, BMPRII was retained with it, whereas when mutant NIPA1 exited the ER and was endosomal/lysosomal, so too was BMRPII. Mutant NIPA1 was less efficient at promoting BMPRII degradation than wild-type NIPA1, perhaps because BMPRII sequestered in the ER is protected from lysosomal degradation, or because insufficient NIPA1 was available at endosomes to promote efficient degradation of BMPRII. However, despite the obvious effect of mutant NIPA1 on BMPRII traffic, we did not find any effect of expression of the mutant protein on BMP signalling assays. The end result of expression of mutant NIPA1 on the BMP signalling pathway is likely to be determined by an interplay between how much NIPA1 is retained in the ER, how much BMPRII is retained there with it and how much NIPA1 escapes the ER to be active in promoting BMPRII degradation at lysosomes. An overexpression system such as the one we used is unlikely to be able to model all of these factors correctly, and this may explain why we did not see any effects on BMP signalling. The development of models expressing physiological levels of the mutant protein (e.g. knock-in mice) will be required to fully address this issue.
Recently, Zhao et al. (20) have proposed that the molecular pathological mechanism of NIPA1 HSP is a toxic gain-of-function. In mammalian cells, they found that transient transfection of mutant NIPA1 triggered ER stress and UPR, caused by accumulation of the mutant protein in the ER. However, their transient transfection experiments were associated with massive cell death (>90% cell death 48 h after transfection), so it would appear that this system was highly non-physiological. Zhao et al. also presented data to suggest that UPR played a significant role in a motor phenotype seen in C. elegans overexpressing disease-associated mutant versions of NIPA1. In contrast, we saw no evidence of increased cell death in our stable cell lines expressing mutant NIPA1, nor did we see any evidence of ER stress or UPR above that seen in cell lines expressing equivalent amounts of the wild-type protein. Thus, while not excluding the possibility that ER stress may make a contribution to the NIPA1-HSP phenotype, our results do not support it.
In summary, we show that three endosomal HSP proteins are inhibitors of BMP signalling. While not excluding involvement of other signalling pathways and ER stress, we propose that upregulation of BMP signalling, caused in at least some cases by altered endocytic BMPR traffic, is an important candidate pathological component of the axonal degeneration seen in the endosomal group of HSPs, and may be relevant in other conditions showing distal axonopathy.
MATERIALS AND METHODS
Antibodies
Mouse monoclonal anti-EEA1 and anti-BMPRII were from BD Transduction Laboratories. Mouse monoclonal anti-Lamp1 (clone H4A3), rabbit polyclonal anti-GADD34 (C-19) and rabbit polyclonal anti-EGFR (1005; used in immunoblotting) were from Santa Cruz Biotechnology, Inc. Mouse monoclonal anti-transferrin receptor was from LGC Promochem. Mouse monoclonal anti-EGFR (Ab-5; used for immunofluorescence) was from Calbiochem. Rabbit polyclonal anti-pSmad1/5 and anti-total Smad1 were from Cell Signaling. Mouse monoclonal anti-myc (4A6) was from Upstate Biotechnology, Inc. Rabbit polyclonal anti-GFP (66,57–26) was from AbCam. Mouse monoclonal anti-KDEL (used to detect BiP and Grp94 in immunoblotting) was from Stressgen. Rabbit polyclonal anti-actin was from Sigma. Rabbit polyclonal anti-M6PR and anti-spastin86–340 were as described previously (12,43). Rabbit polyclonal anti-spartin was raised (Harlan SeraLabs) against a GST full-length spartin-fusion protein. Rabbit polyclonal anti-VPS26 was a kind gift of Matthew Seaman (Cambridge), and the anti-clathrin heavy chain was a kind gift of Margaret S. Robinson (Cambridge). Peroxidase-conjugated secondary antibodies for western blotting were obtained from Sigma. Alexafluor-labelled secondary antibodies for immunofluorescence were obtained from Invitrogen Molecular Probes.
Constructs
The full-length cDNA for NIPA1 was amplified by PCR from IMAGE clone 7967979 and cloned into the pEGFP-N3 vector. The sequence of the inserts and insertion sites was verified by sequencing (Geneservice). For production of stable cell lines, the NIPA1-GFP coding cassette was excised from the pEGFP vector using NheI and NotI, and then ligated into compatible sites in the pIRESneo2 vector. Site-directed mutagenesis to generate disease-associated mutant versions of NIPA1 was carried out using the Phusion Site-Directed Mutagenesis Kit (Finnzymes). The Id Firefly Luciferase construct (containing a BMP-responsive element from mouse Id1, coupled to firefly luciferase) was a kind gift of P. ten Dijke (Amsterdam) (26). Renilla luciferase vector phRL-TK(Int-) was purchased from Promega. The Myc-BMPRII construct was based on a pcDNA3 vector, into which a c-Myc sequence 5′ to and in frame with the coding sequence of full-length human BMPRII was cloned.
Cell lines
The HeLa cell line used in this study was HeLaM (44). HeLa and NSC34 (24) cells were maintained in culture as described previously (12). Medium was supplemented with 500 µg/ml G418 (Sigma) as a selective agent for stable cell lines.
NIPA1-GFP wild-type and mutant stable HeLa and NSC34 cell lines were made by transfecting the relevant cell type with pIRESneo2 NIPA1-GFP constructs. Two days after transfection, transfected cells were placed under selection, by supplementing normal medium with G418, as described earlier. After 2 weeks, surviving clones were trypsinized and pooled, to constitute a stable cell line of mixed clonality, referred to subsequently as a mixed stable cell line. The GFP-Rab5-expressing stable cell line was a kind gift of Matthew Seaman (Cambridge, UK). The NSC34 cell line was a kind gift of Dr Neil Cashman (Toronto, Canada).
Transient transfection of plasmid DNA and immunofluorescence microscopy
Transient transfections of most plasmid DNA constructs were carried out using the Effectene® transfection reagent (Qiagen), according to the manufacturer's instructions and as described previously (12,45). Following transfection, cells were incubated for 24 or 48 h. Transient transfections of Id luciferase and Renilla luciferase constructs were carried out using HeLa Monster (Mirus) transfection reagent, according to the manufacturer's instructions.
Coverslips were processed for immunofluorescence microscopy as described previously (12,45). For anti-M6PR labelling, cells were fixed and permeabilized in methanol; with other antibodies, 3.8% formaldehyde in PBS was used. Coverslips were mounted in anti-Fade Gold medium (Invitrogen Molecular Probes) on a glass slide. Stained samples were analysed on a Zeiss 510 Meta confocal microscope. Images were recorded using LSM Image Analyzer software, and data were subsequently processed using Adobe Photoshop and Illustrator programs, or Volocity software (Improvision) for quantitation of plasma membrane fluorescence.
Protein depletion by siRNA knock-down
ON-TARGET or ON-TARGETplus siRNA oligonucleotides targeting NIPA1 (catalogue numbers J-106270-05 to 08, directed against NM_144599, termed NIPA1 siRNA1-4) were obtained from Dharmacon and had the following sequences: siRNA1, GCGAGGUACUUCCUAUUUAUU; siRNA2, CCGAUGAUCUUCUCUAUAAUU; siRNA3, GUUAGAAUCUUUCAGGUUUUU; siRNA4, GAAUACAUGUGGCUAACAAUU. An siRNA oligonucleotide to BMPRII (005309-02, directed against NM_001204) transcripts was also obtained from Dharmacon, Inc. Spartin siRNA oligonucleotides were manufactured by Invitrogen and had the following sequences obtained from (15): siRNA1, 5′-GGCAAGGAUUGGAAUGUGCAGCUAA; siRNA2, 5′-CAAAUACGGAUAUAAUGCAGGAGAAUU. Human spastin (SPG4; catalogue numbers D-014070-01–04) individual siGENOME duplexes were obtained from Dharmacon, Inc. and had the following sequences: siRNA1, 5′-UUAUAGAAGGUUGAAGUUCUU; siRNA2, 5′-UCAUUAUAGACGUCCGUUUUU; siRNA3, 5′-UAAACUUGCAGCACUUAUAUU; siRNA4, 5′-UUAGCCAGCAUUGUCUUCCUU. Non-targeting control siRNA (1, catalogue number D-001210-01) was obtained from Dharmacon, Inc. SiRNA directed against clathrin heavy chain had the sequence UAAUCCAAUUCGAAGACCAAU, obtained from Motley et al. (46).
A ‘double hit’ oligonucleotide transfection protocol using oligofectamine transfection reagent (Invitrogen) was employed for siRNA transfection, as described previously (12), except prior to luciferase assays, where a ‘single hit’ protocol was used. NIPA1 oligonucleotides 1–4 were used in a pool at 50 nm total concentration, or NIPA1 siRNA 1 and 2 were used individually at 50 nm. Spastin oligonucleotides 1–4 were used either in a pool or singly, both at 5 nm total concentration. The BMPRII and spartin oligonucleotides were used at 10 nm and the clathrin heavy chain oligonucleotide at 5 nm. In the case of BMPRII, clathrin heavy chain, spastin and spartin, protein depletion was confirmed in each experiment by immunoblotting using specific antibodies. Since we did not have an NIPA1 antibody that worked in immunoblotting, depletion of this protein was monitored in one of two ways. First, we carried out siRNA transfections on HeLa cells stably expressing NIPA1-GFP in parallel with, and using the same pool of reagents as, the experiments on wild-type HeLa cells. Loss of NIPA1-GFP was subsequently confirmed by immunofluorescence or immunoblotting (Fig. 1A; Supplementary Material, Fig. S9) and used as a reporter for NIPA1 depletion in the wild-type cells transfected with the same reagents. Alternatively or in addition, we used RT–PCR to demonstrate depletion of the NIPA1 mRNA in wild-type HeLa cells transfected with NIPA1 siRNA. RNA was extracted from cells using the RNeasy Kit (Qiagen), DNAse-treated (Promega) and reverse-transcribed using Superscript II (Invitrogen), according to the manufacturer's instructions. TaqMan real-time PCR for NIPA and an endogenous control protein (hypoxanthine phosphoribosyltransferase 1) was then carried out in triplicate, using TaqMan®Gene Expression Assays (Applied Biosystems, NIPA1: Assay ID Hs00895258_m1; hypoxanthine phosphoribosyltransferase 1, Assay ID Hs99999909_m1) on a MicroAmp™ Optical 96-Well Reaction Plate with Barcode (Applied Biosystems). Relative RNA concentrations in the mock- and siRNA-transfected samples were calculated using the comparative CT method, according to the manufacturer's instructions (Applied Biosystems, real-time PCR system) (Supplementary Material, Fig. S9).
pSMAD1/5 assays
Cells cultured in a six-well plate were washed with PBS and then placed in serum-free medium overnight. In experiments on knocked-down cells, this step happened on the day following the second siRNA transfection. The next day, cells were washed in PBS and placed in serum-free medium for 1 h. In experiments involving BMP4 stimulation, this was supplemented with 2 ng/ml (for NSC34 cells) or 20 ng/ml (for HeLa cells) of BMP4 (R&D systems). Cells were washed in ice-cold PBS, and harvested on ice in lysis buffer containing 1% Triton X-100, in the presence of a complete protease inhibitor cocktail (Roche). Lysates were then centrifuged at 10 000g for 5 min at 4°C and the supernatant removed. An aliquot of the supernatant was used to assay protein concentration, determined by the Bio-Rad Protein Assay (Bio-Rad) based on the Bradford method. The remaining supernatant was added to 3× sodium dodecyl sulphate (SDS) sample buffer (188 mm TRIS, 6% SDS, 30% glycerol, 0.03% bromophenol blue, 10% beta-mercaptoethanol, pH 6.8). Samples containing equivalent protein masses were heated to 98°C for 5 min before running on SDS–PAGE and subsequent immunoblotting. For pSmad1/5 immunoblotting, membranes were blocked in PBS with 5% powdered milk and 5% bovine serum albumin for 3 h at room temperature, followed by incubation overnight at 4°C with the primary antibody in 5% BSA in Tris-buffered saline with 0.1% Tween (TBS-T), then by incubation with an appropriate secondary antibody in TBS-T containing 5% powdered milk and 5% BSA. Labelled bands were identified using enhanced chemo-luminescence (Amersham).
Immunoblot quantitation and statistical software
Immunoblot band density was typically quantified using a Geldoc GS-710 densitometer and Quantity One software (Bio-Rad). Statistical analyses and histogram construction were carried out in GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA, USA). Quantitation of BiP immunoblot density was carried out using fluorescent secondary antibodies and a Typhoon™ 9410 Variable Mode Imager (GE Healthcare, Amersham, UK), according to the manufacturer's instructions.
Inhibition of lysosomal function
Cells were first grown to the desired confluency in six-well plates. They were then washed in PBS, and fresh medium was added, supplemented with chloroquine (10 µm) or concanamycin A (5 nm) for 8 h, before being harvested into chilled lysis buffer containing 1% Triton X-100, as described earlier.
Luciferase assays
Id Firefly luciferase and Renilla luciferase constructs (3 and 0.3 µg DNA, respectively) were transfected using HeLa Monster transfection reagent (Mirus). In experiments on knocked-down cells, this was done on the day following transfection with the relevant siRNA. Cells were placed in a serum-free medium for the final 24 h of the DNA transfection, and in experiments involving BMP4 stimulation, this was supplemented with 2 (for NSC34 cells) or 20 ng/ml (for HeLa cells) of BMP4 (R&D systems). Firefly and Renilla luciferase activities were then assayed in duplicate using the Promega Dual Luciferase Assay Kit, with a Promega Glomax luminometer, according to the manufacturer's instructions. Firefly luciferase activity was corrected for transfection efficiency and cell viability by dividing by Renilla luciferase activity, and then the results were expressed per microgram of lysate protein.
Co-immunoprecipitation
In experiments involving co-immunoprecipitation of exogenous myc-BMPRII and NIPA1-GFP, HeLa cells stably expressing NIPA1-GFP were plated out into two 10 cm dishes at 50% confluence and then transfected with the relevant myc-BMPRII construct, with Effectene® transfection reagent, using 3 µg of vector DNA per dish. At 48 h post-transfection, cells were harvested with ice-cold digitonin lysis buffer (0.1 m MES NaOH pH6.5, 1 mm Mg acetate, 0.5 mm EGTA, 200 µm Na orthovanadate, 1% digitonin) containing complete protease inhibitors (Roche). A soluble fraction was then obtained by retaining the supernatant after centrifugation at 10 000g for 5 min at 4°C. Subsequent processing was carried out as described previously (45,47). Co-immunoprecipitates were suspended in 3× SDS sample buffer and then run on SDS–PAGE and immunoblotted with appropriate antibodies.
For co-immunoprecipitation of endogenous BMPRII with NIPA1-GFP, NSC34 cells stably expressing NIPA-GFP were plated out, then lysed and subsequently processed in the same way as described earlier. In some cases, the cells were treated with concanamycin A before lysis, as described earlier.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by the Wellcome Trust (a Senior Research Fellow in Clinical Science 082381 to E.R., a Strategic Award 079895 to Cambridge Institute for Medical Research); the Tom Wahlig Stiftung (to E.R.); and the Medical Research Council (G9310915 to J.P.L.). HTHT was supported by a Croucher Foundation Scholarship. HTHT and TLE were supported by UK government ORS awards and by the Cambridge Commonwealth Trust. TLE was supported by a Sackler Fellowship. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
ACKNOWLEDGEMENTS
We thank Nicholas Morrell and Anastasia Sobolewski for help and advice. We thank Stefan Marciniak for helpful discussions on ER stress and the UPR.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Harding A.E. Hereditary spastic paraplegias. Semin. Neurol. 1993;13:333–336. doi: 10.1055/s-2008-1041143. [DOI] [PubMed] [Google Scholar]
- 2.Reid E. Science in motion: common molecular pathological themes emerge in the hereditary spastic paraplegias. J. Med. Genet. 2003;40:81–86. doi: 10.1136/jmg.40.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soderblom C., Blackstone C. Traffic accidents: molecular genetic insights into the pathogenesis of the hereditary spastic paraplegias. Pharmacol. Ther. 2006;109:42–56. doi: 10.1016/j.pharmthera.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 4.DeLuca G.C., Ebers G.C., Esiri M.M. Axonal loss in multiple sclerosis: a pathological survey of the corticospinal and sensory tracts. Brain. 2004;127:1009–1018. doi: 10.1093/brain/awh118. [DOI] [PubMed] [Google Scholar]
- 5.Fischer L.R., Culver D.G., Tennant P., Davis A.A., Wang M., Castellano-Sanchez A., Khan J., Polak M.A., Glass J.D. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp. Neurol. 2004;185:232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Fink J.K. Hereditary spastic paraplegia. Curr. Neurol. Neurosci. Rep. 2006;6:65–76. doi: 10.1007/s11910-996-0011-1. [DOI] [PubMed] [Google Scholar]
- 7.Hanein S., Martin E., Boukhris A., Byrne P., Goizet C., Hamri A., Benomar A., Lossos A., Denora P., Fernandez J., et al. Identification of the SPG15 gene, encoding spastizin, as a frequent cause of complicated autosomal-recessive spastic paraplegia, including Kjellin syndrome. Am. J. Hum. Genet. 2008;82:992–1002. doi: 10.1016/j.ajhg.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hazan J., Fonknechten N., Mavel D., Paternotte C., Samson D., Artiguenave F., Davoine C.S., Cruaud C., Durr A., Wincker P., et al. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat. Genet. 1999;23:296–303. doi: 10.1038/15472. [DOI] [PubMed] [Google Scholar]
- 9.Fonknechten N., Mavel D., Byrne P., Davoine C.S., Cruaud C., Bonsch D., Samson D., Coutinho P., Hutchinson M., McMonagle P., et al. Spectrum of SPG4 mutations in autosomal dominant spastic paraplegia. Hum. Mol. Genet. 2000;9:637–644. doi: 10.1093/hmg/9.4.637. [DOI] [PubMed] [Google Scholar]
- 10.Depienne C., Fedirko E., Forlani S., Cazeneuve C., Ribai P., Feki I., Tallaksen C., Nguyen K., Stankoff B., Ruberg M., et al. Exon deletions of SPG4 are a frequent cause of hereditary spastic paraplegia. J. Med. Genet. 2007;44:281–284. doi: 10.1136/jmg.2006.046425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beetz C., Nygren A.O., Schickel J., Auer-Grumbach M., Burk K., Heide G., Kassubek J., Klimpe S., Klopstock T., Kreuz F., et al. High frequency of partial SPAST deletions in autosomal dominant hereditary spastic paraplegia. Neurology. 2006;67:1926–1930. doi: 10.1212/01.wnl.0000244413.49258.f5. [DOI] [PubMed] [Google Scholar]
- 12.Connell J., Lindon C., Luzio J., Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 2009;10:42–56. doi: 10.1111/j.1600-0854.2008.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel H., Cross H., Proukakis C., Hershberger R., Bork P., Ciccarelli F.D., Patton M.A., McKusick V.A., Crosby A.H. SPG20 is mutated in Troyer syndrome, an hereditary spastic paraplegia. Nat. Genet. 2002;31:347–348. doi: 10.1038/ng937. [DOI] [PubMed] [Google Scholar]
- 14.Bakowska J.C., Wang H., Xin B., Sumner C.J., Blackstone C. Lack of spartin protein in Troyer syndrome: a loss-of-function disease mechanism? Arch. Neurol. 2008;65:520–524. doi: 10.1001/archneur.65.4.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bakowska J.C., Jupille H., Fatheddin P., Puertollano R., Blackstone C. Troyer syndrome protein spartin is mono-ubiquitinated and functions in EGF receptor trafficking. Mol. Biol. Cell. 2007;18:1683–1692. doi: 10.1091/mbc.E06-09-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards T.L., Clowes V.E., Tsang H.T.G., Connell J.W., Sanderson C.S., Luzio J.P., Reid E. Endogenous spartin (SPG20) is recruited to endosomes and lipid droplets and interacts with the ubiquitin E3 ligases API4 and AIP5. Biochem. J. 2009 doi: 10.1042/BJ20082398. in press. doi:10.1042/BJ20082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rainier S., Chai J.H., Tokarz D., Nicholls R.D., Fink J.K. NIPA1 gene mutations cause autosomal dominant hereditary spastic paraplegia (SPG6) Am. J. Hum. Genet. 2003;73:967–971. doi: 10.1086/378817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beetz C., Schule R., Klebe S., Klimpe S., Klopstock T., Lacour A., Otto S., Sperfeld A.D., van de Warrenburg B., Schols L., et al. Screening of hereditary spastic paraplegia patients for alterations at NIPA1 mutational hotspots. J. Neurol. Sci. 2008;268:131–135. doi: 10.1016/j.jns.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 19.Goytain A., Hines R.M., El-Husseini A., Quamme G.A. NIPA1(SPG6), the basis for autosomal dominant form of hereditary spastic paraplegia, encodes a functional Mg2+ transporter. J. Biol. Chem. 2007;282:8060–8068. doi: 10.1074/jbc.M610314200. [DOI] [PubMed] [Google Scholar]
- 20.Zhao J., Matthies D.S., Botzolakis E.J., Macdonald R.L., Blakely R.D., Hedera P. Hereditary spastic paraplegia-associated mutations in the NIPA1 gene and its Caenorhabditis elegans homolog trigger neural degeneration in vitro and in vivo through a gain-of-function mechanism. J. Neurosci. 2008;28:13938–13951. doi: 10.1523/JNEUROSCI.4668-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X., Shaw W.R., Tsang H.T., Reid E., O'Kane C.J. Drosophila spichthyin inhibits BMP signaling and regulates synaptic growth and axonal microtubules. Nat. Neurosci. 2007;10:177–185. doi: 10.1038/nn1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keshishian H., Kim Y.S. Orchestrating development and function: retrograde BMP signaling in the Drosophila nervous system. Trends Neurosci. 2004;27:143–147. doi: 10.1016/j.tins.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 23.O'Connor-Giles K.M., Ho L.L., Ganetzky B. Nervous wreck interacts with thickveins and the endocytic machinery to attenuate retrograde BMP signaling during synaptic growth. Neuron. 2008;58:507–518. doi: 10.1016/j.neuron.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cashman N.R., Durham H.D., Blusztajn J.K., Oda K., Tabira T., Shaw I.T., Dahrouge S., Antel J.P. Neuroblastoma x spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev. Dyn. 1992;194:209–221. doi: 10.1002/aja.1001940306. [DOI] [PubMed] [Google Scholar]
- 25.Miyazono K., Maeda S., Imamura T. BMP receptor signaling: transcriptional targets, regulation of signals, and signaling cross-talk. Cytokine Growth Factor Rev. 2005;16:251–263. doi: 10.1016/j.cytogfr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 26.Korchynskyi O., ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 27.Drose S., Bindseil K.U., Bowman E.J., Siebers A., Zeeck A., Altendorf K. Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry. 1993;32:3902–3906. doi: 10.1021/bi00066a008. [DOI] [PubMed] [Google Scholar]
- 28.Marciniak S.J., Ron D. Endoplasmic reticulum stress signaling in disease. Physiol. Rev. 2006;86:1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- 29.Melnick J., Aviel S., Argon Y. The endoplasmic reticulum stress protein GRP94, in addition to BiP, associates with unassembled immunoglobulin chains. J. Biol. Chem. 1992;267:21303–21306. [PubMed] [Google Scholar]
- 30.Di Fiore P.P., De Camilli P. Endocytosis and signaling. An inseparable partnership. Cell. 2001;106:1–4. doi: 10.1016/s0092-8674(01)00428-7. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez-Gaitan M., Stenmark H. Endocytosis and signaling: a relationship under development. Cell. 2003;115:513–521. doi: 10.1016/s0092-8674(03)00932-2. [DOI] [PubMed] [Google Scholar]
- 32.Polo S., Di Fiore P.P. Endocytosis conducts the cell signaling orchestra. Cell. 2006;124:897–900. doi: 10.1016/j.cell.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 33.Le Roy C., Wrana J.L. Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat. Rev. Mol. Cell Biol. 2005;6:112–126. doi: 10.1038/nrm1571. [DOI] [PubMed] [Google Scholar]
- 34.Sweeney S.T., Davis G.W. Unrestricted synaptic growth in spinster—a late endosomal protein implicated in TGF-beta-mediated synaptic growth regulation. Neuron. 2002;36:403–416. doi: 10.1016/s0896-6273(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 35.Hartung A., Bitton-Worms K., Rechtman M.M., Wenzel V., Boergermann J.H., Hassel S., Henis Y.I., Knaus P. Different routes of bone morphogenic protein (BMP) receptor endocytosis influence BMP signaling. Mol. Cell. Biol. 2006;26:7791–7805. doi: 10.1128/MCB.00022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi W., Chang C., Nie S., Xie S., Wan M., Cao X. Endofin acts as a Smad anchor for receptor activation in BMP signaling. J. Cell Sci. 2007;120:1216–1224. doi: 10.1242/jcs.03400. [DOI] [PubMed] [Google Scholar]
- 37.Foletta V.C., Lim M.A., Soosairajah J., Kelly A.P., Stanley E.G., Shannon M., He W., Das S., Massague J., Bernard O. Direct signaling by the BMP type II receptor via the cytoskeletal regulator LIMK1. J. Cell Biol. 2003;162:1089–1098. doi: 10.1083/jcb.200212060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassel S., Eichner A., Yakymovych M., Hellman U., Knaus P., Souchelnytskyi S. Proteins associated with type II bone morphogenetic protein receptor (BMPR-II) and identified by two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2004;4:1346–1358. doi: 10.1002/pmic.200300770. [DOI] [PubMed] [Google Scholar]
- 39.Charron F., Tessier-Lavigne M. The Hedgehog, TGF-beta/BMP and Wnt families of morphogens in axon guidance. Adv. Exp. Med. Biol. 2007;621:116–133. doi: 10.1007/978-0-387-76715-4_9. [DOI] [PubMed] [Google Scholar]
- 40.Wen Z., Han L., Bamburg J.R., Shim S., Ming G.L., Zheng J.Q. BMP gradients steer nerve growth cones by a balancing act of LIM kinase and slingshot phosphatase on ADF/cofilin. J. Cell Biol. 2007;178:107–119. doi: 10.1083/jcb.200703055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lasorella A., Stegmuller J., Guardavaccaro D., Liu G., Carro M.S., Rothschild G., de la Torre-Ubieta L., Pagano M., Bonni A., Iavarone A. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature. 2006;442:471–474. doi: 10.1038/nature04895. [DOI] [PubMed] [Google Scholar]
- 42.Matsuura I., Taniguchi J., Hata K., Saeki N., Yamashita T. BMP inhibition enhances axonal growth and functional recovery after spinal cord injury. J. Neurochem. 2008;105:1471–1479. doi: 10.1111/j.1471-4159.2008.05251.x. [DOI] [PubMed] [Google Scholar]
- 43.Reaves B.J., Bright N.A., Mullock B.M., Luzio J.P. The effect of wortmannin on the localisation of lysosomal type I integral membrane glycoproteins suggests a role for phosphoinositide 3-kinase activity in regulating membrane traffic late in the endocytic pathway. J. Cell Sci. 1996;109:749–762. doi: 10.1242/jcs.109.4.749. [DOI] [PubMed] [Google Scholar]
- 44.Tiwari R.K., Kusari J., Sen G.C. Functional equivalents of interferon-mediated signals needed for induction of an mRNA can be generated by double-stranded RNA and growth factors. EMBO J. 1987;6:3373–3378. doi: 10.1002/j.1460-2075.1987.tb02659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanderson C.M., Connell J.W., Edwards T.L., Bright N.A., Duley S., Thompson A., Luzio J.P., Reid E. Spastin and atlastin, two proteins mutated in autosomal-dominant hereditary spastic paraplegia, are binding partners. Hum. Mol. Genet. 2006;15:307–318. doi: 10.1093/hmg/ddi447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Motley A., Bright N.A., Seaman M.N., Robinson M.S. Clathrin-mediated endocytosis in AP-2-depleted cells. J. Cell Biol. 2003;162:909–918. doi: 10.1083/jcb.200305145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reid E., Connell J., Edwards T.L., Duley S., Brown S.E., Sanderson C.M. The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum. Mol. Genet. 2005;14:19–38. doi: 10.1093/hmg/ddi003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.