Abstract
The primary non-motile cilium, a membrane-ensheathed, microtubule-bundled organelle, extends from virtually all cells and is important for development. Normal functioning of the cilium requires proper axoneme assembly, membrane biogenesis and ciliary protein localization, in tight coordination with the intraflagellar transport system and vesicular trafficking. Disruptions at any level can induce severe alterations in cell function, giving rise to a myriad of human genetic diseases known as ciliopathies. Here we show that the Abelson helper integration site 1 (Ahi1) gene, whose human ortholog is mutated in Joubert syndrome, regulates cilium formation via its interaction with Rab8a, a small GTPase critical for polarized membrane trafficking. We find that the Ahi1 protein localizes to a single centriole, the mother centriole, which becomes the basal body of the primary cilium. In order to determine whether Ahi1 functions in ciliogenesis, loss of function analysis of Ahi1 was performed in cell culture models of ciliogenesis. Knockdown of Ahi1 expression by shRNAi in cells or targeted deletion of Ahi1 (Ahi1 knockout mouse) leads to impairments in ciliogenesis. In Ahi1-knockdown cells, Rab8a is destabilized and does not properly localize to the basal body. Since Rab8a is implicated in vesicular trafficking, we next examined this process in Ahi1-knockdown cells. Defects in the trafficking of endocytic vesicles from the plasma membrane to the Golgi and back to the plasma membrane were observed in Ahi1-knockdown cells. Overall, our data indicate that the distribution and functioning of Rab8a is regulated by Ahi1, not only affecting cilium formation, but also vesicle transport.
INTRODUCTION
The primary cilium is a conserved cell structure extending from the apical cell surface into the extracellular space (1). It extends from virtually all cells and is important for normal development (1–6). The cilium is thought to act as a sensory organelle for the cell (7). For instance, the formation and functioning of cilia have been implicated in the maintenance of the sonic hedgehog (Shh) and Wnt signalling pathways, leading to their critical roles in the patterning of the vertebrate body plan (2–5,8). An emerging body of evidence has implicated the cilium in a range of human developmental disorders. Such diseases are now referred to as ciliopathies (9), reflecting the fact that the proper assembly and maintenance of the cilium are critical for normal development and cell signalling (2–5,8).
Ciliogenesis and cilium function require axoneme assembly, intraflagellar transport (IFT), membrane biogenesis and proper compartmentation of signalling proteins (1). One important cellular and ciliary process is the coordination of polarized vesicle trafficking, a process modulated by members of the Rab and Arf small GTPase protein families (10,11). Rab proteins are small, monomeric GTPases that comprise the largest family of proteins within the Ras superfamily. Like other GTPases, Rab activity is regulated by guanine nucleotide exchange and modulation of GTP hydrolysis (12). Inactive GDP-bound Rab is activated by the exchange of GDP for GTP. Rabs are tightly controlled by a host of effector proteins including GDP/GTP exchange factors (GEFs), GTPase activating proteins (GAPs) and GDP dissociation inhibitors (GDIs) (12,13). Rabs are critical mediators of both exocytotic and endocytotic membrane trafficking, facilitating the formation of protein complexes that are involved in vesicle targeting, transport and fusion (12,14–16). Rab5, 8a and 11 are involved in vesicular trafficking between the Golgi network and the plasma membrane (17). Interestingly, Rab8a also has been found in the primary cilium, where it plays important roles in ciliogenesis and ciliary function (11). Alterations to the Rab8a pathway have a significant impact on ciliary membrane biogenesis (11,18–20); however, the mechanism by which Rab8a is recruited to the basal body, which is necessary for its effects on ciliogenesis, remains unclear.
Joubert syndrome (JBTS) is an autosomal recessive neurodevelopmental disorder in humans characterized by retinal dystrophy and by defects in the development of the hindbrain (21). The Abelson helper integration site 1 (AHI1) gene is one of seven identified genes [NPHP1 (22), NPHP6/CEP290 (23,24), MKS3 (25), RPGRIP1L/NPHP8 (26,27), ARL13B (28) and CC2D2A (29)] implicated in JBTS, with 10–15% of all individuals with JBTS having mutations in AHI1 (30–32). AHI1 is a cytoplasmic multidomain protein composed of an N-terminal coiled-coil domain, WD40 repeats and a C-terminal SH3 domain (31,33). Given these domains, it is presumed that AHI1 acts as a scaffolding protein (31). AHI1 is expressed in the neurons of the hindbrain, midbrain and ventral forebrain; but it is also present in the pituitary, testis and kidney (34). Little is known about the function of AHI1/Ahi1 or how mutations in AHI1 lead to JBTS, but JBTS is considered to be a disease of the primary cilium, based on clinical-symptom similarities with other ciliopathies (9,35). Many of the proteins implicated in JBTS appear to be cytoplasmic proteins that are not membrane-associated, suggesting that these proteins may be involved in the signal transduction events that occur following receptor activation. Whether AHI1 shares similar properties and can be grouped with these proteins, as a cause of disorders now being termed ciliopathies, and/or whether AHI1 defines a separate pathway involved in manifesting this phenotype is currently unknown. Elucidation of the role of AHI1 in ciliary function is critical not only for a better understanding of JBTS, but also for identifying the role of the cilium in development and cell function.
Given the overlap between JBTS and other ciliopathies, we postulated that Ahi1 may be necessary for proper primary cilium formation or function. We tested our hypothesis by examining whether inner medullary collecting duct cells (IMCD3) that had shRNAi knockdown of Ahi1 had normal primary cilium formation, and whether any defects in ciliogenesis were caused by altered Rab8a function or vesicular trafficking defects. Here we show that Ahi1 is found at both the mother centriole and the basal body of the primary cilium. Knockdown of Ahi1 expression by shRNAi leads to (i) loss of primary non-motile cilia, (ii) defects in vesicular trafficking and (iii) disruption of Rab8a function. In Ahi1-knockdown cells, endocytic vesicles are formed, but do not properly traffic. Vesicles, transporting the transferrin receptor, form and traffic centripetally, but the transferrin receptor does not recycle to the plasma membrane. In cells with knockdown of Ahi1, Rab8a is destabilized leading to decreases in its expression levels, and cannot properly localize to the basal body. Lastly, in further support for a role of Ahi1 in ciliogenesis, we found that mouse embryonic fibroblasts obtained from mice with a targeted deletion of Ahi1 have significantly decreased numbers of cells with primary non-motile cilia. Together, these data indicate a role for Ahi1 in the process of ciliogenesis as well as in the coordination of polarized vesicle trafficking. Our results begin to clarify the role of Ahi1 in JBTS in addition to further implicating the primary cilium in the pathogenesis observed in JBTS.
RESULTS
Ahi1 localizes to the mother centriole
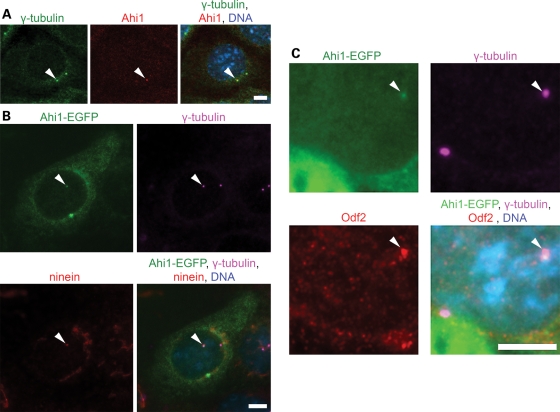
We find that Ahi1, the mouse ortholog of AHI1, is found at the centrosome as demonstrated by co-localization with the centrosomal marker, γ-tubulin, in mouse renal inner medullary collecting duct (IMCD3) cells (Fig. 1A), NIH/3T3 cells (mouse fibroblast cell line; Supplementary Material, Fig. S1A) and spermatogonia cells (Supplementary Material, Fig. S1B); all cell lines containing a primary non-motile cilium. Analysis of the distribution of Ahi1 at each phase of the cell cycle demonstrated that Ahi1 is present in the centrioles at all stages (Supplementary Material, Fig. S2). We further noticed that only one of the centrioles shows Ahi1 immunostaining (Fig. 1A). To determine whether Ahi1 is present in the mother or the daughter centriole (36), we performed co-staining of γ-tubulin with two markers of the mother centriole, ninein and Odf2 (37,38), in IMCD3 cells transfected with an enhanced green fluorescent protein tagged mouse Ahi1 plasmid (Ahi1-EGFP). In agreement with the results of the Ahi1 immunostaining experiments, Ahi1-EGFP co-localizes with γ-tubulin (Fig. 1B). Ninein has been found to label both the mother and the daughter centriole, with ninein having greater localization at the mother centriole than the daughter centriole (38). We find that Ahi1-EGFP is found in association with the centriole having higher localization of ninein, the mother centriole (Fig. 1B). In further support, Ahi1-EGFP is also co-localized only at the Odf2-positive centriole (Fig. 1C). These data indicate that Ahi1 associates with a single centriole, the Odf2- and ninein-positive mother centriole, in cycling cells.
Figure 1.
Ahi1 localizes to the mother centriole. (A) IMCD3 cells were immunostained for the centrosomal marker γ-tubulin (green), and for endogenous Ahi1 (red). Arrowheads indicate the co-localization of γ-tubulin and Ahi1 to only one of the paired centrioles. (B) Ahi1-EGFP (green) transfected IMCD3 cells were stained with γ-tubulin (violet) and with ninein (red), a marker for the mother centriole. Arrowheads indicate the co-localization of Ahi1, γ-tubulin and ninein in the mother centriole. (C) Ahi1-EGFP (green) transfected IMCD3 cells were stained with γ-tubulin (violet) and with Odf2 (red), a mother centriole marker. Arrowheads indicate the co-localization of Ahi1, γ-tubulin, and Odf2 only in the mother centriole. DNA is visualized with Hoechst 33258 (blue). Scale bars = 5 µm.
Ahi1 localizes to the basal body of the primary cilium
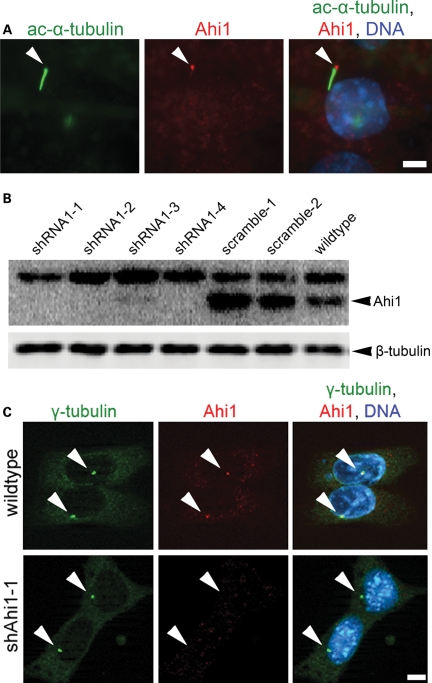
While Ahi1 was found throughout the cytoplasm, very little expression was observed in either the juxtamembrane domain or in the nucleus of confluent cells. Since the mother centriole is the origin of the basal body of the primary cilium (1,39), we sought to determine whether Ahi1 is directly associated with the basal body. We performed Ahi1 immunostaining on IMCD3 cells after serum deprivation, a treatment that induces the formation of primary non-motile cilia (40,41). As expected, we observe a single elongated primary non-motile cilium, as determined by acetylated α-tubulin immunostaining, in the vast majority of the serum-starved cells (Fig. 2A). Ahi1 immunostaining mapped to the base of the primary non-motile cilium, specifically the basal body (Fig. 2A) (42). The mapping of Ahi1 to the basal body of the cilium was also observed in other mouse cell lines derived from brain, skin and gonadal tissue (Supplementary Material, Fig. S3). In some cilia, we observed staining for Ahi1, not only in the basal body, but also faintly in the cilium axoneme (Fig. 2A); nevertheless, Ahi1 staining was always most pronounced in the basal body. These results indicate that Ahi1 associates with the mother centriole-derived organelle, the basal body, in cells that have Ahi1 expression and a primary non-motile cilium (42).
Figure 2.
Ahi1 localizes to the basal body of the primary non-motile cilium and its expression is lowered in Ahi1-knockdown IMCD3 cells. (A) Serum-deprived IMCD3 cells have robust cilium formation (one/cell). Ciliated IMCD3 cells were stained with acetylated α-tubulin (green), a ciliary axoneme marker, and for Ahi1 (red). Arrowheads indicate the localization of Ahi1 to the base of a primary cilium. DNA is visualized with Hoechst 33258 (blue). For evaluation of knockdown efficiency of Ahi1, IMCD3 cells stably expressing shRNAi against Ahi1 were established and analysed for Ahi1 protein expression by (B) western blotting and (C) immunostaining. (B) Protein levels of Ahi1 in four independent Ahi1-knockdown IMCD3 stable clones, two control shRNAi scramble stable clones and wild-type cells were analysed by western blotting. Ahi1 (130 kDa, lower band) can be detected in scramble control and wild-type cells, but not in cells expressing a shRNAi against Ahi1. The top band is considered to be a non-specific band (34). (C) Immunostaining for γ-tubulin (green), for Ahi1 (red) and for DNA (Hoechst 33258 (blue)) was performed in wild-type IMCD3 cells and in Ahi1-knockdown cell line 1 (shAhi1-1). Scale bars = 5 µm.
Ahi1 is required for primary cilium formation
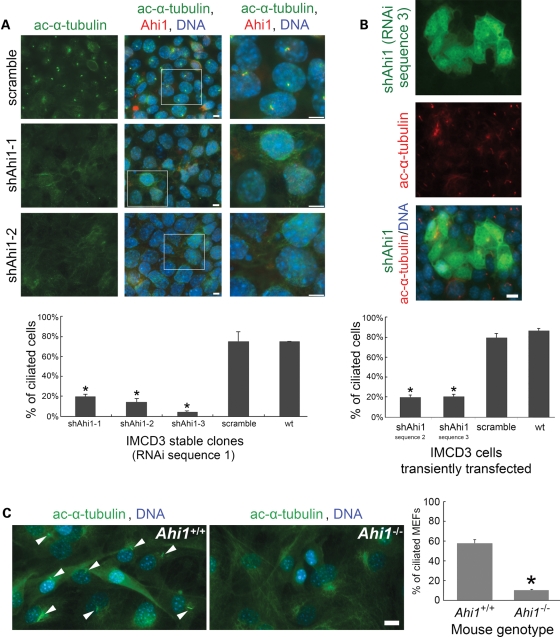
Given the mapping of Ahi1 to the ciliary basal body, we examined whether Ahi1 is necessary for cilium formation. Knockdown of Ahi1 in IMCD3 cells (shAhi1), by either stable integration or transient transfection of three independent shRNAi constructs targeted for Ahi1, produced a significant decrease in Ahi1 expression when compared with wild-type or scramble shRNAi-transfected cells [Fig. 2B (RNAi sequence 1: >90% decrease in Ahi1 expression); data not shown (RNAi sequences 2 and 3)]. No Ahi1 expression to the centriole or basal body was seen in shAhi1 cells [Fig. 2C (RNAi sequence 1); data not shown (RNAi sequences 2 and 3)]. Following serum deprivation, both scramble shRNAi cells and wild-type cells have robust cilium outgrowth in nearly every cell, as assessed by acetylated α-tubulin (Fig. 3A) and Arl13b [an additional marker for the primary cilium (3); data not shown] immunostaining. However, in Ahi1-knockdown cells, we observed a dramatic reduction in cilium formation. Greater than 80% of Ahi1-knockdown cells lacked a primary cilium (Fig. 3A and B). These results suggest that Ahi1 is necessary for the formation of the primary non-motile cilium.
Figure 3.
Ahi1 is required for the normal formation of primary non-motile cilium. (A) Control cells (scramble shRNAi), Ahi1-knockdown cell line 1 (shAhi1-1) and Ahi1-knockdown cell line 2 (shAhi1-2) (both shAhi1-1 and shAhi1-2 were RNAi sequence 1) were cultured under serum-deprived conditions, for initiation of robust cilium formation. Cells were immunostained for acetylated α-tubulin (green; marker of the primary cilium) and for Ahi1 (red). Note that the acetylated α-tubulin staining was actually acquired on a Cy5 channel and has been pseudocoloured green (the true FITC channel visualizes the shAhi1 construct that is GFP tagged and is not shown in this image). Higher magnification images of the boxed regions are shown in the right column. The histogram shows the percentage of cells having a primary cilium based on counts of cells with acetylated α-tubulin-positive cilia, for Ahi1-knockdown cells (shAhi1-1, -2 and -3), control-scramble cells and wild-type cells. Ten randomly acquired fields (a minimum of 500 cells were counted for each clone) were analysed. The error bars represent the standard error of the mean. Asterisks indicate significance from scramble and wild-type cells (P < 0.0001) using Chi-square analyses. (B) Ahi1-knockdown cells that had been transiently transfected with RNAi sequence 3 were cultured under serum-deprived conditions, for initiation of robust cilium formation. Cells were immunostained for acetylated α-tubulin (red) with the green channel highlighting cells that had been transfected with the shRNAi construct. The histogram shows the percentage of cells having a primary cilium based on counts of cells with acetylated α-tubulin-positive cilia, for Ahi1-knockdown cells that were transfected with shRNAi constructs against Ahi1 (RNAi sequences 2 and 3), a scramble shRNAi construct or no construct (wild-type cells). Three randomly acquired fields (a minimum of 200 cells were counted for each clone) were analysed. The error bars represent the standard error of the mean. Asterisks indicate significance from scramble and wild-type cells (P < 0.0001) using Chi-square analyses. (C) Mouse embryonic fibroblasts (MEFs) cultured from wild-type (Ahi1+/+) and Ahi1 knockout (Ahi1−/−) mouse embryos. MEFs were immunostained for acetylated α-tubulin (green; marker of the primary cilium). The histogram shows the percentage of Ahi1+/+ and Ahi1−/− MEFs having a primary cilium based on counts of cells with an acetylated α-tubulin-positive cilium. Three randomly acquired fields (a minimum of 100 cells were counted for each field) were analysed for each mouse. The error bars represent the standard error of the mean. Asterisk indicates significance from wild-type (Ahi1+/+) MEFs (P < 0.0001) using Chi-square analysis. Scale bars = 5 µm.
In addition, we isolated mouse embryonic fibroblasts (MEFs) from mice with a targeted deletion of Ahi1 to determine whether cells from an Ahi1 knockout mouse lacked cilia (Supplementary Material, Fig. S4). Primary cilia were observed on 57% of all MEFs cultured from Ahi1+/+ embryos (Fig. 3C). However, there was a significant reduction in ciliated MEFs cultured from Ahi1−/− embryos with only 10% of the cells having a cilium (Fig. 3C). Overall, the lack of primary non-motile cilia in cells with either knockdown of Ahi1 or deletion of Ahi1 supports the involvement of Ahi1 in the process of ciliogenesis.
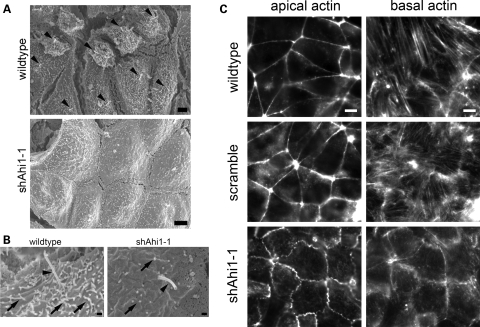
Scanning electron microscopy (SEM) was used to visualize the cilium in Ahi1-knockdown and control cell lines. A primary cilium was easily observable on the surface of the majority of wild-type and scramble shRNAi cells, but that was not the case for the Ahi1-knockdown cell lines (Fig. 4A). The few putative primary cilia that we observed in the Ahi1-knockdown cells were shorter than normal (Fig. 4B). We also noticed that the acetylated α-tubulin immunostaining in Ahi1-knockdown cells was diffusely distributed throughout the cytoplasm, unlike the pattern in control cells where α-tubulin was restricted mostly to the primary cilium (Fig. 3A and B; data not shown). Given the observed defects in tubulin, we also explored whether there were abnormalities in the actin cytoskeleton. We examined actin organization in control, scramble and Ahi1-knockdown cells using phalloidin, a marker for actin filaments (43). Unlike control cells, which had an organized actin filament pattern, the actin filaments in Ahi1-knockdown cells had significant disorganization of apical actin and notably decreased actin stress fibres (basal) (Fig. 4C), with the shAhi1-1 line being more severely affected than the shAhi1-2 line. Overall, these results suggest that Ahi1 is a protein that is critical for ciliogenesis, and may also affect the organization and formation of cytoskeletal networks (microtubules and actin) necessary for cilia formation.
Figure 4.
Ahi1 is required for the normal formation of primary cilia. (A) Low magnification scanning electron micrographic images of wild-type and Ahi1-knockdown (RNAi sequence 1) IMCD3 cells under conditions optimized for ciliogenesis. Wild-type cells (top) each clearly have an elongated and pronounced primary cilium (black arrowheads) on the cell surface. However, Ahi1-knockdown cells (bottom) have a striking lack of primary cilia. (B) Higher magnification scanning electron micrographs of the primary cilium (black arrowheads) and microvilli (black arrows) in wild-type (left) and Ahi1-knockdown IMCD3 (right) cells. Primary cilia were very difficult to find in Ahi1-knockdown IMCD3 cells; when putatively identified, they tended to be short and stumpy (right). (C) Apical and basal actin filament staining by phalloidin in wild-type, scramble and Ahi1-knockdown cells showing a disorganization and decrease in actin filaments in Ahi1-knockdown cells. Scale bars = 2 µm in (A), 0.3 µm in (B) and 5 µm in (C).
Ahi1 is required for localization of Rab8a to the basal body of the primary cilium
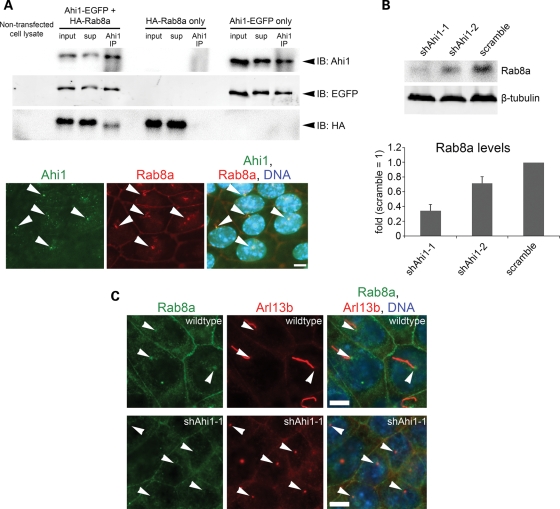
Rab proteins are generally thought to control polarized membrane trafficking in cells (16), and disruptions in Rab function result in significant alterations in protein homeostasis (44). Several members of the Rab GTPase family, particularly Rab8a, have been identified as critical modulators of ciliogenesis (18,45). Given the importance of Rab8a in ciliogenesis, we examined whether Ahi1 and Rab8a were found in similar cellular domains. We observed that both Ahi1 and Rab8a were seen to co-localize to the basal body of the developing and mature cilium (Fig. 5A, bottom). Next, we sought to determine whether Ahi1 interacts with Rab8a in regulating cilium formation. Since our mouse Ahi1 antibody cannot recognize human AHI1 in HEK293 cells, we used this cell line for testing whether Ahi1 and Rab8a form a complex. In HEK293 cells that were co-transfected with Ahi1-EGFP and wild-type HA-tagged Rab8a (HA-Rab8a) constructs, we observed an association of HA-Rab8a with Ahi1-EGFP by co-immunoprecipitation methods (Fig. 5A, top), but not in cells transfected with either Ahi1-EGFP or HA-Rab8a alone (Fig. 5A, top). Similar results were obtained using myc-tagged Ahi1 (myc-Ahi1) and HA-Rab8a constructs (Supplementary Material, Fig. S5A) or IMCD3 cell lysates (Supplementary Material, Fig. S5B), but not using myc-Ahi1 and HA-Rab11 constructs (control experiment; Supplementary Material, Fig. S5C). In addition, bead only controls were devoid of non-specific binding (data not shown). Since these experiments were conducted in sub-confluent HEK293 cells, with relatively few cilia present, these data further suggest that this interaction is apparently not dependent on the presence of a cilium.
Figure 5.
Ahi1 is involved in ciliogenesis through its interactions with the small GTPase, Rab8a. (A, top) Co-immunoprecipitation of Ahi1-EGFP with HA-Rab8a in lysates from HEK293 cells transfected with Ahi1-EGFP and HA-Rab8a, but not in cells transfected with HA-Rab8a alone or Ahi1-EGFP alone. Resin only controls had no labelling (data not shown). (A, bottom) Co-localization of both Ahi1 (green) and Rab8a (red) at the basal body in wild-type cells (denoted by white arrowheads). (B) The protein level of endogenous Rab8a from Ahi1-knockdown (shAhi1-1 and -2) and from scramble shRNAi control cells was analysed by western blotting (top) and graphically displayed (bottom). The level of β-tubulin represents the loading control. The error bars represent the standard error of the mean. (C) Wild-type (top panel) and Ahi1-knockdown IMCD3 cells (lower panel) were stained with antibodies against Rab8a (green) and Arl13b (red; marker for the basal body and the primary cilium). Right panel represents the merged images of Rab8a, Arl13b and DNA (blue) staining. White arrowheads point to the basal body. With this Rab8a antibody, localization is only found at the basal body and does not show any apparent localization to the cilium. Scale bars =5 µm in (A and C).
In order to determine whether the association of Rab8a and Ahi1 was functionally important, we examined the expression and localization of Rab8a in Ahi1-knockdown cells. Significantly lower Rab8a expression was observed, by western blotting, in Ahi1-knockdown cells when compared with the expression in control cells (Fig. 5B) suggesting a role for Ahi1 in stabilizing Rab8a expression. We also observed a significant re-distribution of Rab8a in Ahi1-knockdown cells, relative to the pattern in control cells. In control cells, endogenous Rab8a immunostaining was found mainly at cell junctions and close to the basal body of primary cilium (Fig. 5C). In contrast, in Ahi1-knockdown cells, Rab8a expression was significantly lower, and it could not be detected at the basal body (Fig. 5C). This result suggests that Ahi1 is required for Rab8a localization to the basal body and further indicates that failure of Ahi1 and Rab8a to associate leads to Rab8a destabilization.
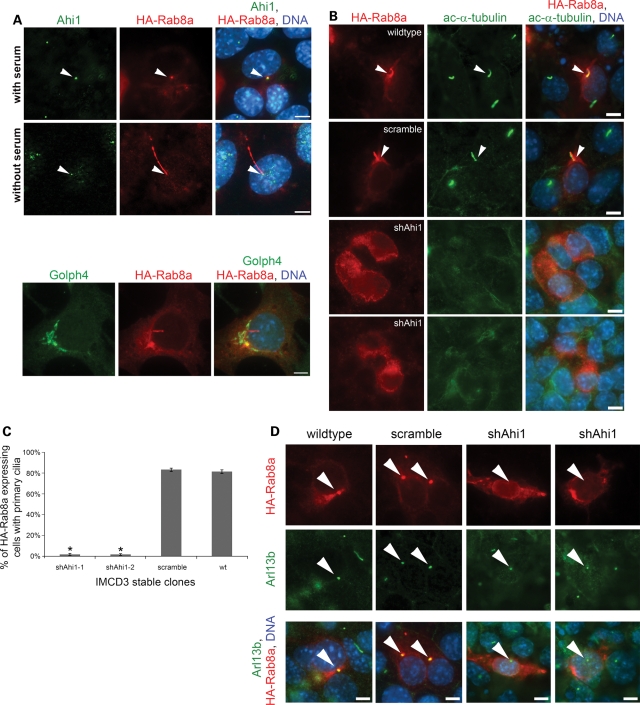
To test this model, HA-Rab8a was overexpressed in control, scramble and Ahi1-knockdown cells. Overexpression of HA-Rab8a was chosen instead of using a Rab8a antibody because our previous data demonstrated a significant reduction in Rab8a levels in Ahi1-knockdown cells to the point that it is not detectable by fluorescent microscopy. In control cells, HA-Rab8a was found mainly in the primary cilium, but variably in the basal body of wild-type cells under cilia growth-optimizing conditions (Fig. 6A, without serum); however, when cells were not in cilia growth-optimizing conditions, HA-Rab8a was found at the basal body (Fig. 6A, with serum). In addition, HA-Rab8a was also found localized throughout the cytoplasm, in the Golgi complex (using the Golgi marker, Golph4), and variably in cytoplasmic vesicles (Fig. 6A, bottom). Overall, these results indicate that Rab8a recruitment to the basal body of the primary cilium precedes ciliogenesis. During ciliary biogenesis, Rab8a transitions from the basal body to the primary cilium itself.
Figure 6.
Overexpression of HA-Rab8a in Ahi1-knockdown cells results in an inability to rescue ciliogenesis and an inability for HA-Rab8a to localize to the basal body of the primary cilium. (A) Wild-type IMCD3 cells, cultured under non-cilia forming conditions (top) or under serum-deprived conditions so as to promote cilium formation (bottom), and transfected with an HA-Rab8a expression plasmid were immunostained for Ahi1 (green) and for HA (red). Right panel represents the merged images of Ahi1, HA-Rab8a and DNA (blue) staining. White arrowheads point to the basal body. The bottom panel represents HA-Rab8a localization to the Golgi complex (using the Golgi marker Golph4 (green)). (B) Wild-type, scramble and Ahi1-knockdown cells transfected with HA-Rab8a were immunostained with acetylated α-tubulin (green) for primary cilia and HA for Rab8a (red) distribution. Arrowheads indicate the co-localization of HA-Rab8a and acetylated α-tubulin staining. Right panel represents the merged images of HA-Rab8a, acetylated α-tubulin and DNA (blue) staining. (C) Quantification of cilium formation in HA-Rab8a expressing wild-type, scramble and Ahi1-knockdown cells. Cells were co-stained with HA and Arl13b. Fifty HA-positive cells were counted from each slide and the number of primary cilium was determined. Data were collected from three independent experiments. The error bars represent the standard error of the mean. Asterisks indicate significance from scramble and wild-type cells (P < 0.0001) using Chi-square analyses. (D) Wild-type and scramble cells [grown to sub-confluence (resulting in few cilia); left two panels] and Ahi1-knockdown cells (grown under cilia optimized conditions; right two panels) transfected with HA-Rab8a were stained with antibodies against HA (red, for HA-Rab8a) and Arl13b (green, for the basal body). Arrowheads point to Arl13b labelled basal bodies. The lower panel represents the merged images of HA-Rab8a, Arl13b and DNA (blue) staining. This experiment was designed to capture wild-type and scramble cells at a time when few cells had cilia, so as to easily observe the association of HA-Rab8a in the basal body (before the formation of cilia where HA-Rab8a would enter the cilium making it difficult to differentiate basal body from the primary cilium). Scale bars =5 µm in (A, B and D).
Overexpression of HA-Rab8a in Ahi1-knockdown cells resulted in not only a failure to rescue cilium formation (Fig. 6B and C), but also an inability for Rab8a to localize to the basal body [Fig. 6D (using the basal body/cilia marker, Arl13b) and Supplementary Material, Fig. S6 (using the basal body marker, γ-tubulin)]. Diffuse expression of HA-Rab8a was found throughout much of the cytoplasm in control, scramble and Ahi1-knockdown cells; however, unlike control and scramble cells, HA-Rab8a was never found to localize to the basal body in Ahi1-knockdown cells (Fig. 6B and D and Supplementary Material, Fig. S6). Since Rab8a is not found at the basal body in Ahi1-knockdown cells (Fig. 6B and D and Supplementary Material, Fig. S6), this indicates a fundamental role for Ahi1 in the proper targeting of Rab8a to the basal body. Furthermore, since Rab8a has been proposed to mark the cilium membrane domain (11), the proper distribution of Rab8a is likely critical for cilium formation. Since the normal distribution of Rab8a was disrupted in Ahi1-knockdown cells, particularly in the region of the ciliary basal body, a plausible physical explanation to account for the impairment in ciliogenesis seen in Ahi1-knockdown cells is that Ahi1 is required for Rab8a targeting to the appropriate membrane domain in Ahi1 expressing cells.
Ahi1 is required for vesicular trafficking
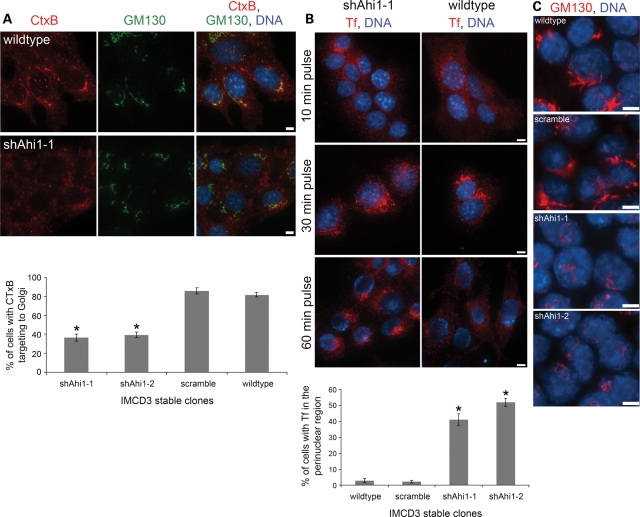
Rab8a regulates the transport of membrane proteins from the Golgi network to the base of the primary cilium and also to the surface of a polarized cell (11,17,44), suggesting a fundamental role of Rab8a in the directional transport of proteins and vesicles to plasma membrane domains. Since the Rab8a distribution was disrupted in Ahi1-knockdown cells, we next examined whether vesicular trafficking was disrupted in these cells. To begin to determine whether clathrin-independent endocytic transport is altered in Ahi1-knockdown cells, we used fluorescently labelled Cholera toxin B (CTxB) to follow endocytic vesicles (46). We found in control cells that CTxB-labelled vesicles were rapidly transported to the Golgi as evidenced by co-localization with the Golgi marker, GM130 (Fig. 7A). In contrast, CTxB-labelled vesicles were not transported to the Golgi in Ahi1-knockdown cells (Fig. 7A). Instead, CTxB vesicles were seen to be randomly distributed in the cytoplasm of these Ahi1-knockdown cells (Fig. 7A). Although longer CTxB exposure resulted in transport of some vesicles to the Golgi, most CTxB vesicles were unable to reach the Golgi. These results suggest that Ahi1 is necessary for trafficking of vesicles, but not for endocytosis itself.
Figure 7.
Membrane trafficking is altered in Ahi1-knockdown cells. (A) Cholera toxin endocytic assay. Wild-type (top row) and Ahi1-knockdown cells (bottom row) were incubated with Alexa Fluor 546-conjugated Cholera toxin subunit B (CTxB; red) for 30 min after cold incubation. GM130 (green) staining delineates the Golgi. The plot shows the percentages of cells showing co-localization of CTxB and GM130 staining. In each case, >200 cells were scored in three independent tests [asterisk indicates a significant effect (P < 0.0001) using Chi-square analysis in comparing Ahi1-knockdown cells with scramble and wild-type cells]. (B) Transferrin endocytic assay. Wild-type (right) and Ahi1-knockdown (left) cells were incubated with Alexa Fluor 546-conjugated transferrin (Tf; red) for 10, 30 or 60 min after cold incubation. DNA is visualized with Hoechst 33258 (blue). Note that following a 60 min pulse, Tf is found throughout the cell membrane in wild-type cells, but in Ahi1-knockdown cells, Tf has accumulated in the perinuclear region of the cell (and not the membrane). The plot shows the percentages of cells with Tf localized to the perinuclear region of the cell after a 60 min pulse. In each case, >200 cells were scored with the asterisk indicating a significant effect (P < 0.0001) using Chi-square analysis in comparing Ahi1-knockdown cells with scramble and wild-type cells. (C) The Golgi structure and position in Ahi1-knockdown cells. Ahi1-knockdown (shAhi1-1 and -2), control scramble and wild-type IMCD3 cells cultured under serum-deprived conditions were stained with the cis-Golgi compartment marker, GM130 (red), to permit examination of the Golgi structure. Similar results were obtained with the trans-Golgi compartment marker, Golph4 (data not shown). Scale bar=5 µm in (A–C).
To determine whether this failure of endocytic vesicle trafficking was a general property of endocytic vesicles in Ahi1-knockdown cells, we examined whether clathrin-dependent endocytosis required Ahi1 by analysing the transferrin and transferrin receptor (Tf/TfR) endocytic pathway. In both control and Ahi1-knockdown cells, fluorescently labelled Tf was found to bind to the cell surface, and to then become incorporated into vesicles and transported toward the perinuclear region (Fig. 7B; 10 and 30 min pulse). This result demonstrates that Ahi1 is not required for general vesicular trafficking, as clathrin-dependent vesicle trafficking to the perinuclear region remains normal, while vesicles derived from clathrin-independent endocytosis do not traffic normally. The Tf/TfR system also allowed us to examine vesicle cycling from the Golgi to the plasma membrane. Whereas control cells recycled the Tf/TfR back to the plasma membrane after 60 min, Ahi1-knockdown cells accumulated Tf/TfR vesicles in the perinuclear region, indicating that vesicle recycling to the plasma membrane is disrupted (Fig. 7B; 60 min pulse). Our results demonstrate that trafficking of proteins/vesicles is altered in the absence of Ahi1, particularly from the Golgi to the plasma membrane, again suggesting that disruptions of the Rab8a-dependent membrane targeting pathway could be responsible for the failure of ciliogenesis and ciliary biogenesis.
Since we observed an alteration in vesicular trafficking in Ahi1-knockdown cells, we hypothesized that Golgi organization was disrupted in these cells. Analysis of Golgi structure in Ahi1-knockdown cells showed abnormalities in Golgi structure, using both a cis-Golgi (GM130) marker (Fig. 7C) and a trans-Golgi (Golph4) marker (data not shown). The Golgi in Ahi1-knockdown cells appeared significantly less-extensive and fragmented in comparison to the perinuclear ribbon-like network observed in control cells (Fig. 7C). In addition, the Golgi in Ahi1-knockdown cells was abnormally positioned, appearing on the apical end of the nucleus, in contrast to the geometry in control cells where the Golgi was positioned laterally with respect to the nucleus (Fig. 7C). Golgi organization was thus apparently altered in Ahi1-knockdown cells, but whether the mechanism for this alteration relates directly to Ahi1 disruption, or whether it is an indirect result of vesicle accumulation remains to be established. Since proper Golgi position, vesicular trafficking and cilium formation are linked to cellular polarity, our results support a role for Ahi1 in establishing polarity or orienting the endocytic system to an existing cellular polarity.
DISCUSSION
Ahi1 is required for ciliogenesis
Many genes have recently been identified in human genetic syndromes that affect the formation or function of the primary cilium. Two of these genetic syndromes, Bardet–Biedl syndrome (BBS) and JBTS, present with different symptoms, but these multigenic syndromes are thought to affect the same organelle, the primary non-motile cilium (47). In this study, we have investigated the role of the JBTS gene, Ahi1, in the function of the primary non-motile cilium. Using well-developed cell culture models of ciliogenesis, we find that Ahi1 is expressed at the mother centriole and basal body in ciliated cells (42). Using multiple shRNAi constructs and mouse embryonic fibroblasts derived from mice with a targeted deletion of Ahi1, we find that depletion of Ahi1 in these cells results in loss of the primary cilium. It still remains to be determined whether the loss of primary cilium in individuals with JBTS and AHI1 mutations is a significant factor in the pathogenesis of JBTS or whether the organ abnormalities in JBTS are related to other molecular mechanisms (i.e. defects in vesicular trafficking). However, our data clearly indicate that the loss of Ahi1 in cells results in defects in the primary non-motile cilium.
Primary cilia have an important role in signalling in the cell, and molecules such as Shh and Wnt, which are critical for development, transduce their signals through cilia (2–5,8). Thus, embryonic defects including embryonic lethality would be expected if a gene involved in the formation of cilia is defective, but the severity of that defect may vary due to differing ciliary-dependent signalling requirements. Genes known to be required for the formation of primary cilia have been found to be embryonic lethal in the mouse. For instance, cells with KIF3A defects (KIF3A is a component of kinesin II) cannot form primary cilium. Mice with targeted deletions of Kif3a show embryonic lethality, which is thought to be due to impairments in Shh signalling (48–50). However, abnormal ciliary function can result in a variety of phenotypes, not including embryonic lethality. For instance, the JBTS-causing genes NPHP1 and CEP290 encode centrosomal/basal body proteins (19,51). However, NPHP1 and CEP290 mutations, in both humans and mice, do not cause embryonic lethality, but manifest other ciliary defects, such as infertility and retinal degeneration (22–24,52–54). Conversely, other JBTS-causing genes, such as RPGRIP1L and ARL13B, which also encode centrosomal/basal body proteins, rarely cause embryonic lethality in humans, but almost always cause embryonic lethality in mice (3,27,28,55). However, little is known about the mechanisms behind these pleiotropic phenotypes.
Our results indicate that significant ciliogenesis defects are observed with either Ahi1-knockdown by shRNAi or in cells from Ahi1 knockout mice, but like mutations in other JS-related genes, no embryonic lethality is observed in human patients with AHI1 mutations (30,31). One possible explanation for the differences in phenotypic expression is likely tissue specificity. Ahi1 is not expressed throughout the body, but is only expressed in a limited number of tissues (34). It is only in the cells in which Ahi1 is expressed, that there appears to be ciliary defects. Therefore, absence of Ahi1 may not impair global ciliogenesis, but only effect the formation of cilia in certain cells and tissues (i.e. those expressing Ahi1). Furthermore, as described earlier, mutations in several ciliopathy genes can result in different phenotype severities in humans. This widely varying phenotype appears to be common in many human ciliopathies, such as BBS, Meckel–Gruber syndrome and JBTS (9).
Ahi1 and Rab8a cooperation in ciliogenesis
Multiple processes are required for the formation of the primary cilium: transport of the mother centriole to the cell membrane, axoneme assembly, IFT and polarized membrane transport to the ciliary membrane domain (1). Rab8a is known to target polarized membrane transport from the trans-Golgi complex to the basolateral membrane of cells (17). Interestingly, Rab8a has also been implicated as playing a crucial role in ciliogenesis (11,56). In BBS, a related ciliopathy, cilium formation is impaired due to improper Rab8a signalling at the cilia (11). Rab8a entry into the ciliary axoneme is regulated by the BBsome, a multiprotein complex located at the basal body (11). Also, CEP290, another gene mutated in JBTS has also been shown to be critical for normal ciliogenesis through a Rab8a mediated pathway (19,20). However, how Rab8a is targeted to the basal body is still largely unknown. We have utilized our Ahi1-knockdown cell lines to determine what role Ahi1 plays in this complex process. We find the localization of Rab8a to the basal body is disrupted in Ahi1-knockdown cells as are the cytosolic levels of Rab8a. Importantly, despite overexpression of Rab8a, proper localization to the basal body and primary cilium formation are not rescued. This indicates that Ahi1 is required for proper Rab8a targeting to the basal body, as well as having a role in the stabilization of the Rab8a protein. These data provide some insight on how Rab8a is localized to the basal body through an Ahi1-dependent mechanism. Whether Cep290 and Ahi1 work cooperatively in mediating the Rab8a-dependent role on ciliogenesis, or whether Cep290 and Ahi1 work through distinctive mechanisms remains unknown. However, if Cep290 and Ahi1 work cooperatively, then it reasons that the common phenotypes observed in JBTS may be through an effect on Rab8a trafficking and localization to the basal body.
Ahi1 is required for proper membrane trafficking and Golgi organization
Defects in vesicular trafficking are also apparent in Ahi1-knockdown cells. In cultured Ahi1-knockdown cells, Tf/TfR and CTxB endocytosis occurs, but only the Tf/TfR endocytic vesicles are able to traffic to the perinuclear region; trafficking of CTxB to the Golgi is retarded in Ahi1-knockdown cells. This suggests that the CTxB vesicles are unable to associate with the transport apparatus. Rab8a is required for the proper transport of CTxB to the Golgi and for transport of Tf/TfR to the perinuclear region of the cell (17,46). Defects in Rab8a function can lead to the disruption of these endocytic pathways (17,46). Furthermore, our data indicate that Ahi1 is likely regulating Rab8a in specific membrane trafficking routes, but not in a universal manner. Remarkably, the Tf/TfR containing vesicles traffic correctly to the perinuclear region, but cannot return to the cellular membrane. Since the Tf/TfR recycling process is regulated by other Rab proteins that are concentrated in the pericentriolar region, and are required for Tf/TfR recycling through the endosomal recycling compartment (57,58), this suggests a role for Ahi1 in other Rab protein-mediated processes. Our data indicate that Ahi1 may be required for coupling of vesicles to the transport apparatus, but in its absence may not provide a specific directional cue. Taken together, Ahi1 affects the function of Rab8a in specific trafficking pathways, as well as potentially affecting other Rab proteins involved in membrane trafficking (58–60).
The structure of the Golgi appears altered in Ahi1-knockdown cells; the Golgi is less-extensive, fragmented and has altered polarity with respect to the nucleus. These characteristics are often found in cells with abnormal membrane trafficking (61). However, it is clear that Ahi1 is not required for general Golgi function, since Ahi1 expression is not widespread in all cell types (34,62). This suggests that the defects in Golgi structure might possibly relate to the excess build-up of cytoplasmic vesicles either to or from the Golgi in Ahi1-knockdown cells (63,64). This build-up could itself result in abnormal Golgi. In addition, aberrant Golgi structure may also result from an excess of vesicles in the cytoplasm that then would provide a loss of membrane back to the Golgi or within the Golgi (63,64).
Ahi1 and the formation of polarity
Significant alterations in the cytoskeleton were found in Ahi1-knockdown cells suggesting a role of Ahi1 in cytoskeletal dynamics. However, given the lack of organized actin filaments and acetylated-tubulin in cells lacking Ahi1, the decreases in cilia may be accounted for by a more global effect of Ahi1 in regulating the actin/microtubule cytoskeleton. Microtubule involvement in the formation of cilia has long been recognized as an important contributor to this process, but the role of actin in ciliogenesis has only recently been appreciated through studies demonstrating that apical enrichments of actin are required for the proper docking of the basal body to the cell membrane to initiate axoneme outgrowth in ciliogenesis (65–67).
Given the abnormal localization of the Golgi and the absence of primary cilium in Ahi1-knockdown cells, these findings suggest a potential role of Ahi1 in establishing or responding to cellular polarity, or in polarized membrane trafficking in other cell types. Since Ahi1 is highly expressed in neurons and the major anatomical defects in JBTS involve brain structures (21,30,31,34,68,69), whether Ahi1 is involved in neuronal polarity remains to be determined. Neurons are highly polarized cells, where specific membrane trafficking is required for neurite development (70,71). One of the major neuroanatomical defects in JBTS is abnormalities in axonal decussation in the cerebellar vermis and the brainstem; however, little is known about the mechanisms involved in these processes. The necessity of Ahi1 in regulating Rab8a-dependent polarized membrane trafficking may provide insight into the mechanisms involved in JBTS and in the establishment of polarity in developing neurons.
Conclusion
Overall, our data suggest Ahi1 is a crucial regulator of cilium formation through the targeted localization of Rab8a to the basal body and a regulator of both vesicular trafficking to and from the Golgi. Given the role of Rab8a in membrane trafficking in the primary cilium, our data provide strong evidence that Ahi1 plays an important role in targeting ciliary membrane and/or receptors to the growing cilium. In the absence of Ahi1, cells are incapable of targeting membrane to the ciliary membrane domain and are therefore unable to form a cilium. Lastly, our data also illustrate a role of Ahi1 in vesicle trafficking in the cell.
It has been proposed that the primary cilium functions as a central sensory organelle for the cell (7). In order to serve this function, receptors must localize to this structure and it is likely that this occurs by transport of vesicles to the ciliary membrane domain and subsequent transport to the ciliary tip via IFT (2–5,8). However, very little is known about the regulation of these pathways and how this specificity in receptor recruitment to the primary cilium is achieved. Our present work adds to the growing list of genes involved in either the formation or function of the cilium (1,72–74). In stark contrast to previously identified genes, Ahi1 is expressed in only a subset of all ciliated cells (34). The unexpected role of Ahi1 in vesicular trafficking suggests a fundamental role of Ahi1 in the regulation of cilia bound vesicular traffic, possibly responsible for providing specificity to cilia. If true, this indicates that cells may have adopted differing mechanisms for ciliogenesis and for the specific targeting of only certain receptors to the tips of the primary cilium. A special subset of proteins, to which Ahi1 might belong, may act as adaptors that recruit proteins to the primary cilium in a tissue-specific manner. While there does not appear to be any Ahi1 family members in mammals, most analyses have only been conducted using bioinformatic approaches (31,33,42). Since these analyses rely mostly on protein sequence and structure, proteins that might not fit these criteria, but still serve a similar function as Ahi1, would be missed. Our data begin to suggest that Ahi1 may be a protein that is present only in a subset of ciliated cells that may be involved in the regulation or trafficking of proteins necessary for the formation and/or functioning of different subtypes of cilium with presumed independent functions.
MATERIALS AND METHODS
Cell culture and transfection
Mouse inner medullary collecting duct cells (IMCD3; from ATCC, Manassas, VA, USA) were maintained in DMEM/F12 medium (Cellgro, Manassas, VA, USA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA), non-essential amino acids (BioWhittaker, East Rutherford, NJ, USA) and penicillin (100 U/ml)/streptomycin (100 µg/ml) (GIBCO, Carlsbad, CA, USA) at 37°C in a humidified incubator under 5% CO2. To induce cilium formation, we cultured IMCD3 cells to confluence, followed by an additional 2 days in serum-deprived medium.
Stable and transiently transfected IMCD3 cells, containing either shRNAi sequences directed against Ahi1 or a scramble sequence, were analysed. The RNAi targeting sequences for mouse Ahi1 (RNAi 1: 5′-GATTTCTCACCCAATGGTAAA-3′; RNAi 2: 5′-TGAAATTCCTTCTGGACGTTT-3′; RNAi 3: 5′-GAAACTGTCACAGAGGTGATA-3′) and for a control scramble sequence (5′-GAAACAAGGGTGCCAGTGTCT-3′) were synthesized. The sense and antisense oligonucleotides were cloned into a pSUPER-neo/gfp plasmid according to the manufacturer's instructions (OligoEngine, Seattle, WA, USA). To establish the stable Ahi1-knockdown cells, we linearized the pSUPER-neo/gfp-shRNA plasmids with SacI, and transfected them into IMCD3 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After 24 h, cells were cultured in G418 selecting medium (1 mg/ml G418; Sigma, St Louis, MO, USA) until cell colonies formed. For cells with Ahi1 RNAi knockdown, 10 EGFP-expressing/G418-resistant clones were assayed for Ahi1 protein levels, by western blotting, resulting in four lines (shAhi1-1, -2, -3, -4). Control scramble RNAi-expressing IMCD3 cells were established using the same procedure (scramble-1 and -2). Once the stable clones were established, cells were maintained in normal IMCD3 medium.
For transient transfection, IMCD3 cells were plated on glass coverslips in 24-well culture dishes for 24 h before transfection with Lipofectamine 2000. Cells were fixed for immunostaining 24 h after transfection.
Cell cycle synchronization
Localization of Ahi1 at time points throughout the cell cycle was determined through synchronization of the cells at various stages of the cell cycle. To enrich the population of cells at specific stages of the cell cycle, we cultured IMCD3 cells under the following conditions: for G0/G1 phase (serum-depriving medium for 24 h); for G1 phase (100 µM mimosine (Sigma) for 16 h); for G1/S phase (1 µg/ml aphidicolin (Sigma) for 16 h); for M phase (100 nM paclitaxel (Sigma) for 16 h). Low cell-density IMCD3 cultures, in normal medium without any treatments, were also assessed for cell cycle stage on the basis of DNA and cell morphological-based criteria.
Immunostaining and imaging
Overexpressed hemagglutinin (HA)-tagged Rab8a was transfected into the stably integrated IMCD3 cells using Lipofectamine 2000, with fixation of cells 48 h later. In addition, untransfected cell lines were also analysed throughout our studies.
Cells were either fixed with ice-cold methanol at −20°C for 15 min or with 4% paraformaldehyde, and were then permeabilized in 0.04% Triton X-100/PBS (PBSTx) for 10 min at room temperature. Fixed cells were incubated in 10% normal donkey serum (for Odf2 antibodies) or 10% normal goat serum (all other antibodies) in PBSTx for 1 h at room temperature. Primary antibodies were diluted in 1% donkey serum (for Odf2 antibodies) or 1% normal goat serum (all other antibodies) in PBSTx, and were then incubated with cells at 4°C overnight. The following antibodies were used for immuno-labelling: rabbit polyclonal anti-Ahi1 (1:1000; (34)), mouse monoclonal anti-γ-tubulin (1:1000; Sigma), mouse monoclonal anti-acetylated α-tubulin (1:1000; Sigma), mouse monoclonal anti-GM130 (1:1000; BD Biosciences, San Jose, CA, USA), rabbit polyclonal anti-Golph4 (1:500; Abcam, Cambridge, MA, USA), mouse monoclonal anti-Rab8a (1:50; Novus Biologicals, Littleton, CO, USA), rabbit polyclonal anti-ninein (1:100; Millipore, Billerica, MA, USA), goat polyclonal anti-Odf2 (1:100; Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse monoclonal anti-HA (1:1000; Cell Signalling Technology, Danvers, MA, USA), and rabbit polyclonal anti-HA (1:100; Clontech, Mountain View, CA, USA). Cells were then incubated with fluorescently conjugated secondary antibodies (1:500, Invitrogen (Molecular Probes) or Jackson Immunoresearch Laboratories, West Grove, PA, USA) for 1 h at room temperature, followed by Hoechst DNA staining (1 µg/ml). After staining, coverslips were mounted on slides with Fluoromount-G antifade solution (Southern Biotechnology, Birmingham, AL, USA).
For actin staining, IMCD3 (shAhi1-1, shAhi1-2, scramble and wild-type) cell lines were grown for 2 days on glass coverslips and then fixed in 4% paraformaldehyde for 15 min. Cells were permeabilized in PBSTx and incubated in rhodamine-conjugated phalloidin (Sigma, 100 µM) for 2 h. The cells were washed and then incubated in Hoechst dye for 5 min. Coverslips were washed again and mounted for imaging.
All immunofluorescent images were visualized with a Zeiss AxioImager-Z1 microscope, and were collected with a Zeiss AxioCam MRm monochrome camera with AxioVision Rel 4.6 software. The objective lenses were a Zeiss EX Plan-NeoFluar 40X/0.75, ∞/0.17 lens and a Zeiss Plan-Apochromat 63X/1.4 oil DIC, ∞/0.17 lens. The immersion medium for the 63X lens was Zeiss Immersol 518F immersion oil. All images were taken at room temperature. Fluorochromes were Alexa Fluor 488 (Invitrogen); Alexa Fluor 546 (Invitrogen); Cy5 (Jackson Immunoresearch). Contrast and brightness of images were adjusted through linear level adjustments, as needed, to optimize the intensity ranges of the images using Adobe Photoshop CS2 (version 9.0.2; Adobe Systems Inc., San Jose, CA, USA).
Generation of mice with a targeted deletion of Ahi1
The mouse genomic region coding Ahi1 was identified through DNA database searches. A BAC (RPCI23-225H6) or genomic DNA, from C57BL/6J mice, which contained the complete genomic structure of Ahi1, was used as a template for generating the 5′- and 3′-arms of the targeting construct. Our targeting strategy involved deleting coding exons 2–5 (genomic exons 3–6), producing a downstream frameshift and premature stop codon (Supplementary Material, Fig. S4A). The targeting construct was a modified pSA-BGAL-PGK-NEO plasmid (courtesy Sheila Thomas, Beth Israel Deaconess Medical Center, Boston, MA, USA) containing a phosphoglycerate kinase (PGK) promoter-neomycin resistance (Neo) gene flanked by short polylinker sites, and a PGK-promoter driving a diphtheria toxin cassette. A 5 kb fragment of intronic region surrounding and containing exon 2 (containing the ATG start-site) of Ahi1 was PCR amplified (using primers containing AscI/PacI sites) and subcloned into the AscI/PacI sites to form the 5′-arm of the targeting vector. A 7.5 kb fragment of intronic region surrounding and containing exons 7–9 (containing coding exons 6–8) of Ahi1 was PCR amplified (using primers containing NheI/SalI sites) and subcloned into the NheI/SalI sites to form the 3′-arm of the targeting vector. The vector was linearized with AscI and electroporated into C57BL/6-derived Bruce-4 embryonic stem cells resulting in positive recombinant heterozygous clones. Targeting of Ahi1 was determined by Southern blotting of EcoRV digested genomic DNA with a PCR-amplified 1.1 kb probe derived from a genomic region upstream of the 5′-arm of the targeting vector (Supplementary Material, Fig. S4A). Southern blotting confirmed homologous recombination (Supplementary Material, Fig. S4B) and two independent cell lines were injected into blastocysts. Correctly targeted embryonic stem cells were injected into recipient C57BL/6 -Tyrc-2J blastocysts (Jackson Laboratories, Bar Harbor, ME, USA). Resulting chimeric males were crossed to wild-type C57BL/6J females to obtain germline transmission of the mutant allele. Genotyping was performed by PCR using primers specific to a region in exon 4 and the ensuing intronic region for detecting the wild-type allele (Ahi1, For-WT: 5′-CAGTTCATCCCAAGTGCTTGCTGGATGATG-3′; Rev-WT: 5′-CCACGAGGGGCAGCAGAGAGGATTTCTAGT-3′; 228 bp product), and primers specific for lacZ (Ahi1, For-KO: 5′-GTTGCAGTGCACGGCAGATACACTTGCTGA-3′; Rev-KO: 5′-GCCACTGGTGTGGGCCATAATTCAATTCGC-3′; 389 bp product) for detecting the mutant allele. Heterozygous Ahi1 mice were maintained by breeding to wild-type C57BL/6J mice. Ahi1+/− mice were bred to produce Ahi1+/+ and Ahi1−/− mice for the analysis. Mice were maintained on a normal 12 h light–dark cycle (06:00 to 18:00) with unlimited access to food and water. Western blot analysis and immunohistochemistry on brain tissue further confirmed that Ahi1−/− mice had no detectable Ahi1 protein (Supplementary Material, Fig. S4C; data not shown). Mice from both ES cell lines had identical phenotypes (data not shown). All mouse procedures were performed under approval from the Institutional Animal Care and Use Committees of both Rensselaer Polytechnic Institute and the Wadsworth Center (NY State Department of Health), in accordance with The National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Mouse embryonic fibroblast isolation from Ahi1+/+ and Ahi1−/− embryos
Mouse embryonic fibroblasts (MEFs) were prepared (69) from mouse embryos (E14.5–E16.5) derived from intercrossing Ahi1+/− mice. In brief, embryos were removed from the uterus and placed in sterile PBS containing 10% fetal bovine serum. Skin tissue was minced in 0.05% trypsin/EDTA followed by a 10 min incubation at 37°C. The tissue was then gently dissociated to release the cells. The cell suspension was collected and then cultured in DMEM with 10% fetal bovine serum and 1% penicillin/streptomycin. For immunostaining, MEFs were cultured on gelatin-coated glass coverslips to ∼90% confluency and then starved for 3 days in media containing 0.1% fetal bovine serum. The cells were fixed and processed for immunostaining as described previously.
Scanning electron microscopy
IMCD3 cells (shAhi1, scramble and wild-type) were grown on coverslips in six-well dishes. They were fixed with cold 6.5% glutaraldehyde in PBS (pH 7.2) for 2 h, washed in PBS and then in 0.1 m cacodylate buffer (pH 7.2) and post-fixed with 2% osmium in cacodylate buffer for 1 h. After washing in double-distilled water, the fixed cells on coverslips were dehydrated in a graded ethanol series and were stored in 100% ethanol overnight at 4°C. The coverslips were then critical-point dried in a Samdri-795 critical point dryer (Tousimis, Rockville, MD, USA) and coated with gold using a SPI sputter coater (West Chester, PA, USA). Cells were observed and photographed on a LEO 1550 VP field emission scanning electron microscope (Carl Zeiss SMT, Peabody, MA, USA).
Western blot analysis and co-immunoprecipitation
Cells were lysed or mouse brains were homogenized in RIPA buffer [50 mm Tris (pH 8), 150 mm NaCl, 1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride and a 1X protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN, USA)], incubated on ice for 30 min and centrifuged at 10 000g for 30 min at 4°C. The supernatant was collected, and the protein concentration was determined with the Advanced Protein Assay Reagent kit (Cytoskeleton Inc, Denver, CO, USA). Five micrograms of protein lysate was resolved on an SDS/PAGE gel and transferred onto PVDF membrane (Millipore). After blocking in 5% skim milk/TBSTx [100 mm Tris (pH 7.4), 150 mm NaCl and 0.01% Triton-X100] for 1 h at room temperature, the membrane was incubated with primary antibody [loading controls consisted of using chicken anti-βIII tubulin antibodies (Millipore)] diluted in blocking solution at 4°C overnight. Primary antibodies were detected with either the SuperSignal West Femto Maximum Sensitivity Substrate Chemiluminescence kit (Pierce, Rockford, IL, USA) or with fluorescent secondary antibodies (Alexa Fluor 488, Alexa Fluor 546 or Cy5). Signals were analysed with a Typhoon Trio+ scanner and ImageQuant analysis software (GE Healthcare, Piscataway, NJ, USA). In order to quantify and compare the signal intensities of each sample, the detected signals were unsaturated and in the linear range of detection.
For co-immunoprecipitation in IMCD3 cell lysates, Protein-A magnetic beads (Invitrogen) were used according to the manufacturer's manual. In brief, the lysate was incubated with Ahi1 antibodies (1 µg) at 4°C overnight, followed by a 15 min incubation with pre-washed Protein-A beads at room temperature. Following five washes with 0.1 m NaPO4 buffer (pH 8.0), the precipitated protein was eluted by boiling of the beads for 10 min in 1× SDS/PAGE sample buffer. The supernatant was collected after centrifugation and resolved on an SDS/PAGE gel for western blotting. Controls included a resin only condition to demonstrate that the co-immunoprecipitated proteins are not non-specifically binding to the resin.
For co-immunoprecipitation in HEK293 cells, cells were plated in 6-well plates and transfected with (i) Ahi1-EGFP (34) and HA-tagged Rab8a, (ii) myc-tagged Ahi1 and HA-tagged Rab8a or (iii) myc-tagged Ahi1 and HA-tagged Rab11 using Lipofectamine 2000. Twenty-four hours after transfection, cell lysates were collected as described previously. Ahi1 antibodies (1 µg) were incubated with 100 µl of cell lysate for 1 h at room temperature, and then mixed with 30 µl of Protein-A Dynabeads and incubated at room temperature for 15 min. After three washes with PBS, proteins precipitated by antibodies were eluted by boiling the beads in 20 µl of 1× SDS/PAGE protein sample buffer and were resolved on a 12% SDS/PAGE gel for western blot analysis.
Cholera toxin and transferrin uptake assays
Cholera toxin B subunit and transferrin labelling experiments were carried out as previously described (46). Briefly, IMCD3 cells were rinsed with serum-free medium three times before incubation with 1 µg/ml of Alexa Fluor 546 conjugated cholera toxin B (Invitrogen) in serum-free medium for 30 min on ice, washed, and then were incubated at 37°C for 30 or 60 min before fixing. For quantitative analysis, GM130 was used to mark the Golgi, and the cells with co-localization of cholera toxin B and GM130 were counted and compared with cells that did not have this co-localization. Analysis was conducted from five random image fields under a 40X objective from three independent experiments in Ahi1-knockdown, scramble and wild-type cells.
For the transferrin uptake assay, IMCD3 cells were washed three times with serum-free medium and incubated in the same medium for 2 h at 37°C. Alexa Fluor 546-conjugated-human transferrin (100 µg/ml; Invitrogen) was added and incubated for 30 min on ice, followed by three washes in serum-free medium. Cells were then incubated in serum-free medium for various times (10, 30 or 60 min) at 37° C. Cell surface-bound transferrin was removed by an acid wash (0.05% acetate, 0.5 m NaCl, pH 3) for 45 s. Cells were then rinsed with PBS three times before fixing. For quantification, the number of cells with transferrin in the perinuclear region were counted and compared with number of cells where the transferrin labelling was back on the plasma membrane. Analysis was conducted from six random image fields for Ahi1-knockdown cells, scramble and wild-type cells.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported in part by the National Institutes of Health (MH71801 to R.J.F.) and the March of Dimes Foundation (5-FY09-29 to R.J.F.).
ACKNOWLEDGEMENTS
The authors wish to thank Dr Tamara Caspary (Emory) for generously supplying the Arl13b antibody and Drs Adriana Verschoor (Wadsworth Center) and Fern Finger (RPI) for their critical reading of our manuscript. The authors would also like to thank Dr. Judy Liu (Beth Israel Deaconess Medical Center, Harvard Medical School) for assistance with Southern blotting. The electron microscopy portion of this work was done at the Electron Microscopy Core Facility at the Wadsworth Center.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Satir P., Christensen S.T. Overview of structure and function of mammalian cilia. Annu. Rev. Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- 2.Murcia N.S., Richards W.G., Yoder B.K., Mucenski M.L., Dunlap J.R., Woychik R.P. The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left-right axis determination. Development. 2000;127:2347–2355. doi: 10.1242/dev.127.11.2347. [DOI] [PubMed] [Google Scholar]
- 3.Caspary T., Larkins C.E., Anderson K.V. The graded response to Sonic Hedgehog depends on cilia architecture. Dev. Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Gerdes J.M., Liu Y., Zaghloul N.A., Leitch C.C., Lawson S.S., Kato M., Beachy P.A., Beales P.L., DeMartino G.N., Fisher S., et al. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat. Genet. 2007;39:1350–1360. doi: 10.1038/ng.2007.12. [DOI] [PubMed] [Google Scholar]
- 5.Huangfu D., Liu A., Rakeman A.S., Murcia N.S., Niswander L., Anderson K.V. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 6.Zariwala M.A., Knowles M.R., Omran H. Genetic defects in ciliary structure and function. Annu. Rev. Physiol. 2007;69:423–450. doi: 10.1146/annurev.physiol.69.040705.141301. [DOI] [PubMed] [Google Scholar]
- 7.Pazour G.J., Witman G.B. The vertebrate primary cilium is a sensory organelle. Curr. Opin. Cell Biol. 2003;15:105–110. doi: 10.1016/s0955-0674(02)00012-1. [DOI] [PubMed] [Google Scholar]
- 8.Singla V., Reiter J.F. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 9.Badano J.L., Mitsuma N., Beales P.L., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 10.Takai Y., Sasaki T., Matozaki T. Small GTP-binding proteins. Physiol. Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 11.Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peranen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 12.Segev N. Ypt and Rab GTPases: insight into functions through novel interactions. Curr. Opin. Cell Biol. 2001;13:500–511. doi: 10.1016/s0955-0674(00)00242-8. [DOI] [PubMed] [Google Scholar]
- 13.Pfeffer S. Filling the Rab GAP. Nat. Cell Biol. 2005;7:856–857. doi: 10.1038/ncb0905-856. [DOI] [PubMed] [Google Scholar]
- 14.Corbeel L., Freson K. Rab proteins and Rab-associated proteins: major actors in the mechanism of protein-trafficking disorders. Eur. J. Pediatr. 2008;167:723–729. doi: 10.1007/s00431-008-0740-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novick P., Zerial M. The diversity of Rab proteins in vesicle transport. Curr. Opin. Cell Biol. 1997;9:496–504. doi: 10.1016/s0955-0674(97)80025-7. [DOI] [PubMed] [Google Scholar]
- 16.Zerial M., McBride H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 17.Huber L.A., Pimplikar S., Parton R.G., Virta H., Zerial M., Simons K. Rab8, a small GTPase involved in vesicular traffic between the TGN and the basolateral plasma membrane. J. Cell Biol. 1993;123:35–45. doi: 10.1083/jcb.123.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshimura S., Egerer J., Fuchs E., Haas A.K., Barr F.A. Functional dissection of Rab GTPases involved in primary cilium formation. J. Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J., Krishnaswami S.R., Gleeson J.G. CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum. Mol. Genet. 2008;17:3796–3805. doi: 10.1093/hmg/ddn277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsang W.Y., Bossard C., Khanna H., Peranen J., Swaroop A., Malhotra V., Dynlacht B.D. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev. Cell. 2008;15:187–197. doi: 10.1016/j.devcel.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parisi M.A., Doherty D., Chance P.F., Glass I.A. Joubert syndrome (and related disorders) (OMIM 213300) Eur. J. Hum. Genet. 2007;15:511–521. doi: 10.1038/sj.ejhg.5201648. [DOI] [PubMed] [Google Scholar]
- 22.Parisi M.A., Bennett C.L., Eckert M.L., Dobyns W.B., Gleeson J.G., Shaw D.W., McDonald R., Eddy A., Chance P.F., Glass I.A. The NPHP1 gene deletion associated with juvenile nephronophthisis is present in a subset of individuals with Joubert syndrome. Am. J. Hum. Genet. 2004;75:82–91. doi: 10.1086/421846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sayer J.A., Otto E.A., O'Toole J.F., Nurnberg G., Kennedy M.A., Becker C., Hennies H.C., Helou J., Attanasio M., Fausett B.V., et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat. Genet. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 24.Valente E.M., Silhavy J.L., Brancati F., Barrano G., Krishnaswami S.R., Castori M., Lancaster M.A., Boltshauser E., Boccone L., Al-Gazali L., et al. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat. Genet. 2006;38:623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- 25.Baala L., Romano S., Khaddour R., Saunier S., Smith U.M., Audollent S., Ozilou C., Faivre L., Laurent N., Foliguet B., et al. The Meckel-Gruber syndrome gene, MKS3, is mutated in Joubert syndrome. Am. J. Hum. Genet. 2007;80:186–194. doi: 10.1086/510499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arts H.H., Doherty D., van Beersum S.E., Parisi M.A., Letteboer S.J., Gorden N.T., Peters T.A., Marker T., Voesenek K., Kartono A., et al. Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat. Genet. 2007;39:882–888. doi: 10.1038/ng2069. [DOI] [PubMed] [Google Scholar]
- 27.Delous M., Baala L., Salomon R., Laclef C., Vierkotten J., Tory K., Golzio C., Lacoste T., Besse L., Ozilou C., et al. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat. Genet. 2007;39:875–881. doi: 10.1038/ng2039. [DOI] [PubMed] [Google Scholar]
- 28.Cantagrel V., Silhavy J.L., Bielas S.L., Swistun D., Marsh S.E., Bertrand J.Y., Audollent S., Attie-Bitach T., Holden K.R., Dobyns W.B., et al. Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am. J. Hum. Genet. 2008;83:170–179. doi: 10.1016/j.ajhg.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorden N.T., Arts H.H., Parisi M.A., Coene K.L., Letteboer S.J., van Beersum S.E., Mans D.A., Hikida A., Eckert M., Knutzen D., et al. CC2D2A is mutated in Joubert syndrome and interacts with the ciliopathy-associated basal body protein CEP290. Am. J. Hum. Genet. 2008;83:559–571. doi: 10.1016/j.ajhg.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dixon-Salazar T., Silhavy J.L., Marsh S.E., Louie C.M., Scott L.C., Gururaj A., Al-Gazali L., Al-Tawari A.A., Kayserili H., Sztriha L., et al. Mutations in the AHI1 gene, encoding jouberin, cause Joubert syndrome with cortical polymicrogyria. Am. J. Hum. Genet. 2004;75:979–987. doi: 10.1086/425985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferland R.J., Eyaid W., Collura R.V., Tully L.D., Hill R.S., Al-Nouri D., Al-Rumayyan A., Topcu M., Gascon G., Bodell A., et al. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat. Genet. 2004;36:1008–1013. doi: 10.1038/ng1419. [DOI] [PubMed] [Google Scholar]
- 32.Parisi M.A., Doherty D., Eckert M.L., Shaw D.W., Ozyurek H., Aysun S., Giray O., Al Swaid A., Al Shahwan S., Dohayan N., et al. AHI1 mutations cause both retinal dystrophy and renal cystic disease in Joubert syndrome. J. Med. Genet. 2006;43:334–339. doi: 10.1136/jmg.2005.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang X., Hanna Z., Kaouass M., Girard L., Jolicoeur P. Ahi-1, a novel gene encoding a modular protein with WD40-repeat and SH3 domains, is targeted by the Ahi-1 and Mis-2 provirus integrations. J. Virol. 2002;76:9046–9059. doi: 10.1128/JVI.76.18.9046-9059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doering J.E., Kane K., Hsiao Y.C., Yao C., Shi B., Slowik A.D., Dhagat B., Scott D.D., Ault J.G., Page-McCaw P.S., Ferland R.J. Species differences in the expression of Ahi1, a protein implicated in the neurodevelopmental disorder Joubert syndrome, with preferential accumulation to stigmoid bodies. J. Comp. Neurol. 2008;511:238–256. doi: 10.1002/cne.21824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hildebrandt F., Zhou W. Nephronophthisis-associated ciliopathies. J. Am. Soc. Nephrol. 2007;18:1855–1871. doi: 10.1681/ASN.2006121344. [DOI] [PubMed] [Google Scholar]
- 36.Bettencourt-Dias M., Glover D.M. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 37.Mogensen M.M., Malik A., Piel M., Bouckson-Castaing V., Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J. Cell Sci. 2000;113:3013–3023. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- 38.Ishikawa H., Kubo A., Tsukita S., Tsukita S. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 2005;7:517–524. doi: 10.1038/ncb1251. [DOI] [PubMed] [Google Scholar]
- 39.Sorokin S. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J. Cell Biol. 1962;15:363–377. doi: 10.1083/jcb.15.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graser S., Stierhof Y.D., Lavoie S.B., Gassner O.S., Lamla S., Le Clech M., Nigg E.A. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tucker R.W., Pardee A.B., Fujiwara K. Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell. 1979;17:527–535. doi: 10.1016/0092-8674(79)90261-7. [DOI] [PubMed] [Google Scholar]
- 42.Eley L., Gabrielides C., Adams M., Johnson C.A., Hildebrandt F., Sayer J.A. Jouberin localizes to collecting ducts and interacts with nephrocystin-1. Kidney Int. 2008;74:1139–1149. doi: 10.1038/ki.2008.377. [DOI] [PubMed] [Google Scholar]
- 43.Small J., Rottner K., Hahne P., Anderson K.I. Visualising the actin cytoskeleton. Microsc. Res. Tech. 1999;47:3–17. doi: 10.1002/(SICI)1097-0029(19991001)47:1<3::AID-JEMT2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 44.Sato T., Mushiake S., Kato Y., Sato K., Sato M., Takeda N., Ozono K., Miki K., Kubo Y., Tsuji A., et al. The Rab8 GTPase regulates apical protein localization in intestinal cells. Nature. 2007;448:366–369. doi: 10.1038/nature05929. [DOI] [PubMed] [Google Scholar]
- 45.Qin H., Wang Z., Diener D., Rosenbaum J. Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr. Biol. 2007;17:193–202. doi: 10.1016/j.cub.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hattula K., Furuhjelm J., Tikkanen J., Tanhuanpaa K., Laakkonen P., Peranen J. Characterization of the Rab8-specific membrane traffic route linked to protrusion formation. J. Cell Sci. 2006;119:4866–4877. doi: 10.1242/jcs.03275. [DOI] [PubMed] [Google Scholar]
- 47.Louie C.M., Gleeson J.G. Genetic basis of Joubert syndrome and related disorders of cerebellar development. Hum. Mol. Genet. 2005;14:R235–R242. doi: 10.1093/hmg/ddi264. [DOI] [PubMed] [Google Scholar]
- 48.Lin F., Hiesberger T., Cordes K., Sinclair A.M., Goldstein L.S., Somlo S., Igarashi P. Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc. Natl Acad. Sci. USA. 2003;100:5286–5291. doi: 10.1073/pnas.0836980100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marszalek J.R., Ruiz-Lozano P., Roberts E., Chien K.R., Goldstein L.S. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc. Natl Acad. Sci. USA. 1999;96:5043–5048. doi: 10.1073/pnas.96.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeda S., Yonekawa Y., Tanaka Y., Okada Y., Nonaka S., Hirokawa N. Left-right asymmetry and kinesin superfamily protein KIF3A: new insights in determination of laterality and mesoderm induction by kif3A-/- mice analysis. J. Cell Biol. 1999;145:825–836. doi: 10.1083/jcb.145.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fliegauf M., Horvath J., von Schnakenburg C., Olbrich H., Muller D., Thumfart J., Schermer B., Pazour G.J., Neumann H.P., Zentgraf H., et al. Nephrocystin specifically localizes to the transition zone of renal and respiratory cilia and photoreceptor connecting cilia. J. Am. Soc. Nephrol. 2006;17:2424–2433. doi: 10.1681/ASN.2005121351. [DOI] [PubMed] [Google Scholar]
- 52.Chang B., Khanna H., Hawes N., Jimeno D., He S., Lillo C., Parapuram S.K., Cheng H., Scott A., Hurd R.E., et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol. Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang S.T., Chiou Y.Y., Wang E., Lin H.K., Lee S.P., Lu H.Y., Wang C.K., Tang M.J., Li H. Targeted disruption of Nphp1 causes male infertility due to defects in the later steps of sperm morphogenesis in mice. Hum. Mol. Genet. 2008;17:3368–3379. doi: 10.1093/hmg/ddn231. [DOI] [PubMed] [Google Scholar]
- 54.Jiang S.T., Chiou Y.Y., Wang E., Chien Y.L., Ho H.H., Tsai F.J., Lin C.Y., Tsai S.P., Li H. Essential role of nephrocystin in photoreceptor intraflagellar transport in mouse. Hum. Mol. Genet. 2009;18:1566–1577. doi: 10.1093/hmg/ddp068. [DOI] [PubMed] [Google Scholar]
- 55.Vierkotten J., Dildrop R., Peters T., Wang B., Ruther U. Ftm is a novel basal body protein of cilia involved in Shh signalling. Development. 2007;134:2569–2577. doi: 10.1242/dev.003715. [DOI] [PubMed] [Google Scholar]
- 56.Nachury M.V. Tandem affinity purification of the BBSome, a critical regulator of Rab8 in ciliogenesis. Methods Enzymol. 2008;439:501–513. doi: 10.1016/S0076-6879(07)00434-X. [DOI] [PubMed] [Google Scholar]
- 57.Ren M., Xu G., Zeng J., De Lemos-Chiarandini C., Adesnik M., Sabatini D.D. Hydrolysis of GTP on rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc. Natl Acad. Sci. USA. 1998;95:6187–6192. doi: 10.1073/pnas.95.11.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trischler M., Stoorvogel W., Ullrich O. Biochemical analysis of distinct Rab5- and Rab11-positive endosomes along the transferrin pathway. J. Cell Sci. 1999;112:4773–4783. doi: 10.1242/jcs.112.24.4773. [DOI] [PubMed] [Google Scholar]
- 59.Duclos S., Corsini R., Desjardins M. Remodeling of endosomes during lysosome biogenesis involves ‘kiss and run’ fusion events regulated by rab5. J. Cell Sci. 2003;116:907–918. doi: 10.1242/jcs.00259. [DOI] [PubMed] [Google Scholar]
- 60.Wilcke M., Johannes L., Galli T., Mayau V., Goud B., Salamero J. Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-Golgi network. J. Cell Biol. 2000;151:1207–1220. doi: 10.1083/jcb.151.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zolov S.N., Lupashin V.V. Cog3p depletion blocks vesicle-mediated Golgi retrograde trafficking in HeLa cells. J. Cell Biol. 2005;168:747–759. doi: 10.1083/jcb.200412003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sheng G., Xu X., Lin Y.F., Wang C.E., Rong J., Cheng D., Peng J., Jiang X., Li S.H., Li X.J. Huntingtin-associated protein 1 interacts with Ahi1 to regulate cerebellar and brainstem development in mice. J. Clin. Invest. 2008;118:2785–2795. doi: 10.1172/JCI35339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park B.C., Shen X., Samaraweera M., Yue B.Y. Studies of optineurin, a glaucoma gene: golgi fragmentation and cell death from overexpression of wild-type and mutant optineurin in two ocular cell types. Am. J. Pathol. 2006;169:1976–1989. doi: 10.2353/ajpath.2006.060400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stauber T., Simpson J.C., Pepperkok R., Vernos I. A role for kinesin-2 in COPI-dependent recycling between the ER and the Golgi complex. Curr. Biol. 2006;16:2245–2251. doi: 10.1016/j.cub.2006.09.060. [DOI] [PubMed] [Google Scholar]
- 65.Lanzetti L. Actin in membrane trafficking. Curr. Opin. Cell Biol. 2007;19:453–458. doi: 10.1016/j.ceb.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 66.Pan J., You Y., Huang T., Brody S.L. RhoA-mediated apical actin enrichment is required for ciliogenesis and promoted by Foxj1. J. Cell Sci. 2007;120:1868–1876. doi: 10.1242/jcs.005306. [DOI] [PubMed] [Google Scholar]
- 67.Park T.J., Mitchell B.J., Abitua P.B., Kintner C., Wallingford J.B. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat. Genet. 2008;40:871–879. doi: 10.1038/ng.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yachnis A.T., Rorke L.B. Neuropathology of Joubert syndrome. J. Child Neurol. 1999;14:655–659. doi: 10.1177/088307389901401006. [DOI] [PubMed] [Google Scholar]
- 69.Yachnis A.T., Rorke L.B. Cerebellar and brainstem development: an overview in relation to Joubert syndrome. J. Child Neurol. 1999;14:570–573. doi: 10.1177/088307389901400904. [DOI] [PubMed] [Google Scholar]
- 70.Huber L.A., Dupree P., Dotti C.G. A deficiency of the small GTPase rab8 inhibits membrane traffic in developing neurons. Mol. Cell. Biol. 1995;15:918–924. doi: 10.1128/mcb.15.2.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kobayashi N. Mechanism of the process formation; podocytes vs. neurons. Microsc. Res. Tech. 2002;57:217–223. doi: 10.1002/jemt.10077. [DOI] [PubMed] [Google Scholar]
- 72.Adams M., Smith U.M., Logan C.V., Johnson C.A. Recent advances in the molecular pathology, cell biology and genetics of ciliopathies. J. Med. Genet. 2008;45:257–267. doi: 10.1136/jmg.2007.054999. [DOI] [PubMed] [Google Scholar]
- 73.Pedersen L.B., Veland I.R., Schroder J.M., Christensen S.T. Assembly of primary cilia. Dev. Dyn. 2008;237:1993–2006. doi: 10.1002/dvdy.21521. [DOI] [PubMed] [Google Scholar]
- 74.Santos N., Reiter J.F. Building it up and taking it down: the regulation of vertebrate ciliogenesis. Dev. Dyn. 2008;237:1972–1981. doi: 10.1002/dvdy.21540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.