Abstract
To equalize X-chromosome dosages between the sexes, the female mammal inactivates one of her two X-chromosomes. X-chromosome inactivation (XCI) is initiated by expression of Xist, a 17-kb noncoding RNA that accumulates on the X in cis. Because interacting factors have not been isolated, the mechanism by which Xist induces silencing remains unknown. Here, we discover a 1.6 kb ncRNA (RepA) within Xist and identify the polycomb complex, PRC2, as its direct target. PRC2 is initially recruited to the X by RepA RNA, with Ezh2 serving as the RNA-binding subunit. The antisense Tsix RNA inhibits this interaction. RepA depletion abolishes full-length Xist induction and H3-K27 trimethylation of the X. Likewise, PRC2 deficiency compromises Xist upregulation. Therefore, RepA/PRC2 is required for the initiation and spread of XCI. We conclude that a ncRNA cofactor recruits polycomb complexes to their target loci.
The mouse X-inactivation center harbors several noncoding genes, including Xist (1, 2) and its antisense repressor, Tsix (3). On the future Xa (active X), Tsix blocks Xist upregulation and prevents the recruitment of silencing factors in cis. On the future Xi (inactive X), Tsix is downregulated, enabling Xist transactivation and spread of Xist RNA along the chromosome (4). The accumulation of Xist transcripts correlates with a cascade of chromatin changes (5), but how Xist directs these changes is unknown. In principle, the act of transcribing Xist could induce structural changes which could alter chromosome-wide function (1). Alternatively, Xist could work as a transcript (1, 2) by recruiting chromatin modifiers or by targeting the X to a specialized compartment (6). Though universally attractive, RNA-based models have remained hypothetical, as Xist-interacting proteins have yet to be identified.
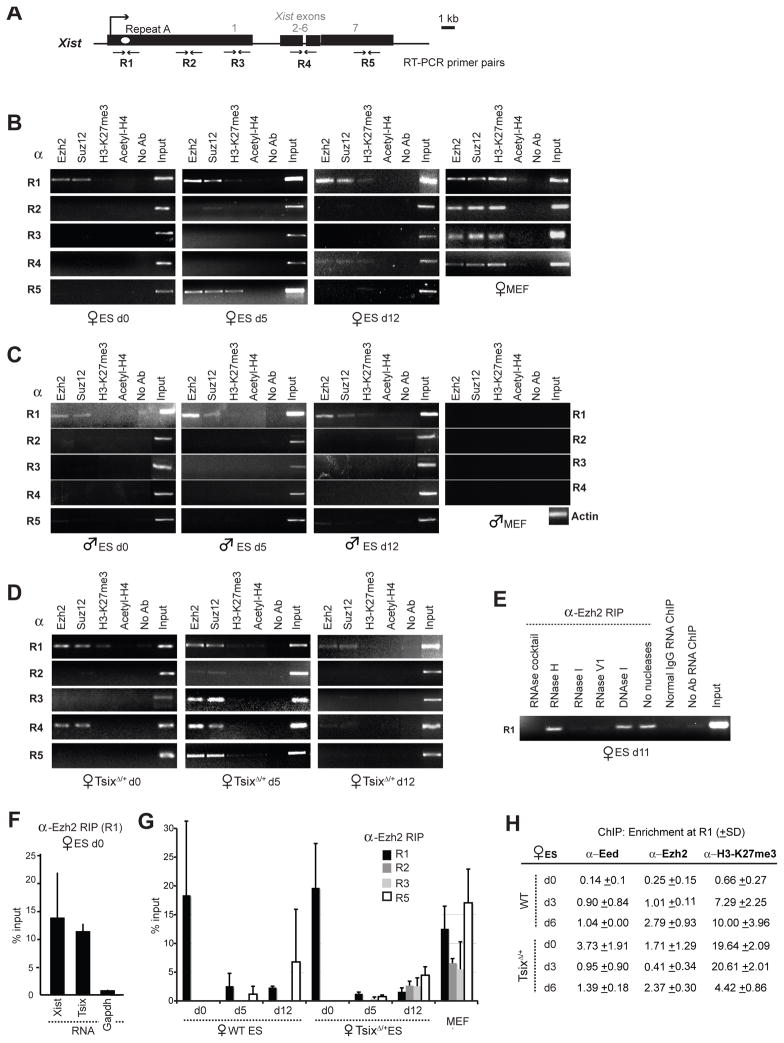
To circumvent conventional difficulties with purifying Xist-interacting proteins, we carried out RNA immunoprecipitations (RIP) and asked if Xist RNA can be found in a specific protein complex. We isolated nuclear RNAs and their binding proteins in the native state to avoid fixation artifacts and tested two cell types -- mouse embryonic stem (ES) cells, which exist in the pre-XCI state but recapitulate XCI when induced to differentiate; and mouse embryonic fibroblasts (MEFs) which faithfully maintains Xi. Because H3-K27 trimethylation (H3-K27me3) closely follows Xist up- and down-regulation (6–9), we asked if Xist RNA binds the H3-K27 methylase, PRC2, the polycomb complex that includes Eed, Suzl2, RbAp48, and the catalytic subunit, Ezh2 (10). Indeed, α-Ezh2 and α-Suz12 antibodies co-immunoprecipitated Xist RNA (Fig. 1A–D). By contrast, Xist sequences were not detected in α-H3-K27me3, α-H4Ac, and no-antibody controls. Pre-treatment with RNases that digest single-stranded (RNase I) and double-stranded (RNase V1) RNA abolished RIP signals, whereas pre-treatment with RNase H (which digests RNA in RNA:DNA hybrids), DNase I, or no nucleases had no effect (Fig. 1E). By inference, the RIP products must be single- or double-stranded RNA.
Figure 1. The PRG2 complex contains Xist.
A. Map of Xist.
B,C,D. RIP in indicated cells, α, antibodies.
E.. Effects of RNase pretreatment on RIP signals.
F. Strand-specific RIP at R1 by realtime PCR, normalized to input RNA. Error bar, 1 standard deviation (SD).
G. Quantitative RIP by realtime PCR at positions R1–R5.
H. DNA ChIP using indicated antibodies, shown as a fraction of input and standardized to normal IgG ChIP.
In female cells, RNA could be detected in the complex even in the pre-XCI state (day 0, d0) when there are <10 transcripts/cell (11). On d0, PRC2 bound only Repeat A (R1), a motif required for silencing (12, 13). Interestingly, quantitative strand-specific RIP showed that both sense and antisense strands were highly enriched in the PRC2 complex (Fig. 1F). Not until cell differentiation and Xist upregulation could PRC2 coimmunoprecipitate more 3′ regions of Xist, suggesting that other regions of Xist eventually comes in contact with PRC2, though Repeat A remained the epicenter of binding (Fig. 1G). To determine when PRC2 is loaded onto chromatin, we performed DNA chromatin immunoprecipitation (ChIP) (Fig. 1H). While bound to RNA in d0 wildtype cells, PRC2 was not enriched on DNA until differentiation (d3, d6) when Eed/Ezh2 levels increased ~10-fold. Accordingly, H3-K27me3 levels rose > 10-fold. Together, RIP and ChIP showed that, although PRC2 bound Repeat A in pre-XCI cells, H3-K27me3 of chromatin was not evident until differentiation (Fig. 1B,H). For males, PRC2 coimmunoprecipitated Xist sequences only in ES cells, not in MEFs (Fig. 1C), consistent with the absence of XCI. In TsixδCpG/+ female cells where XCI choice is predetermined and accelerated (3), PRC2 spreading occurred earlier, consistent with preemptive H3-K27me3 (Fig. 1D,H)(11). Thus, PRC2 recruitment by RNA and its activity on chromatin are biochemically separable.
Examination of TsixδCpG/+ cells enabled us to determine when Xist transactivation occurred relative to PRC2 recruitment. In this mutant, XCI always occurs on the mutated X and H3-K27 methylation preempts Xist upregulation, indicating that H3-K27me3 and Xist transactivation are genetically separable (11). Indeed, DNA ChIP showed high Eed/Ezh2 enrichment on Repeat A on d0 with accompanying H3-K27me3 (Fig. 1H). Xist expression remained low until differentiation(11). Therefore, PRC2 is recruited by RNA to Xist’s 5′ end on d0, but PRC2 transfers to chromatin and catalyzes H3-K27me3 only after differentiation is triggered. These events occur prior to Xist transactivation.
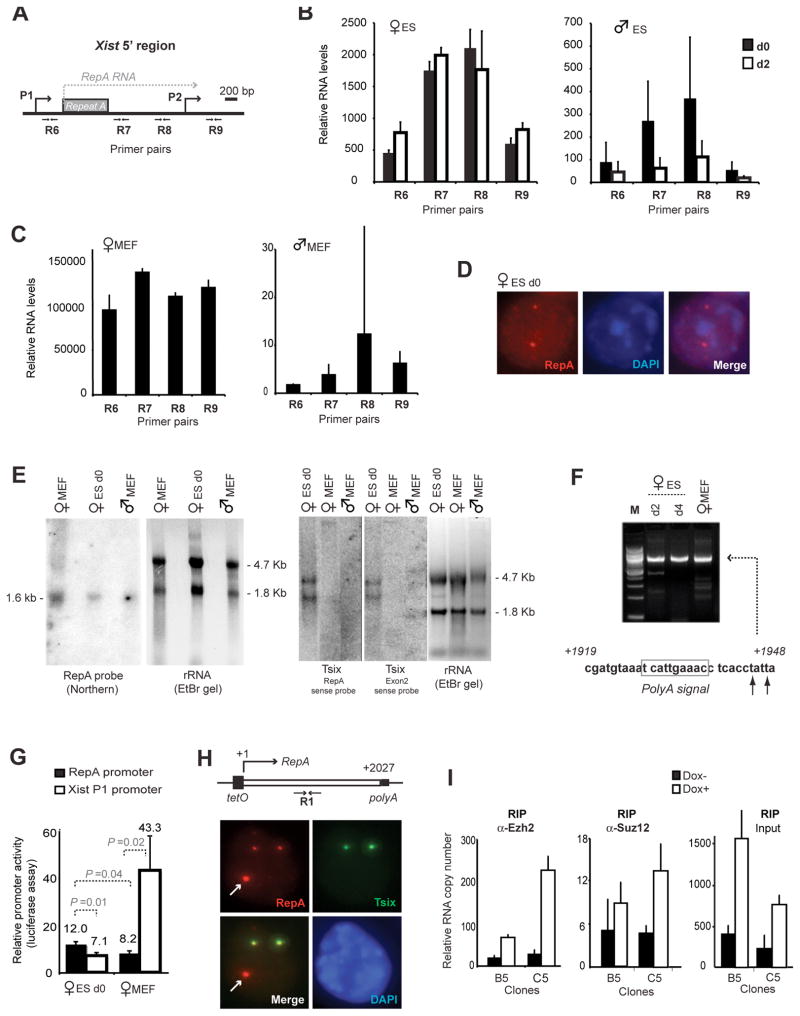
Intriguingly, PRC2 preferentially associates with Repeat A across all timepoints (Fig. 1G), though an intact Xist molecule should theoretically coimmunoprecipitate with PRC2 during native RIP regardless of which RNA domain binds PRC2. To undertake higher resolution analysis, we performed Xist-strand qPCR between Xist promoters P1 and P2 in ES cells and observed 3 to 4 times higher RNA levels at positions R7 and R8 than at R6 and R9 (Fig. 2A,B). During differentiation, Xist upregulated > 100-fold in females but became barely detectable in males (Fig. 2C).
Figure 2. A small RNA within Xist.
A. Map of RepA and the 5′ end of Xist.
B,C. Strand-specific realtime PCR quantitates RNA copies at R6–R9 in ES cells (B) or MEFs (C), normalized to standard curve.
D. RNA FISH using RepA probe.
E. Northern analysis of RepA and Tsix (5′ and 3′ positions).
F. 3′ RACE of RepA.
G. Transient transfection of luciferase reporter constructs comparing RepA (bp79–320) versus Xist P1 promoters, each normalized to vector control. P, student t-tests in indicated pairwise comparisons.
H. DNA FISH of RepA transgenic female ES cells. Xist P1 promoter is not in transgenes. Arrows, transgene. Tsix detected by pSx7.
I. Quantitative RIP in representative clones B5 and C5 +/− Doxycycline induction. No-antibody controls yielded no detectable RNA.
Quantitative differences at R6-R9 hinted at a novel promoter activity. Indeed, RNA fluorescence in situ hybridization (FISH) detected a pinpoint signal on d0 (Fig. 2D). Northern analysis revealed a ~1.6 kb transcript, with no obvious antisense counterparts other than known processed Tsix transcripts (Fig. 2E)(14). 3′ RACE defined its terminus at bp 1948 downstream of P1 (Fig. 2F), implying a transcription start site at ~bp300. Luciferase reporter assays confirmed promoter activity within bp79–320, appearing equally active in pre- and post-XCI cells, whereas P1 activity increased upon XCI (Fig. 2G). Competitive RT-PCR previously revealed ~10 absolute copies of sense RNA in this region in d0 female ES cells (11). The current stoichiometric data implied that 3–4 copies derive from full-length Xist and 6–7 from Repeat A (Fig. 2B). Upon differentiation, Xist levels increased ~100-fold (Fig. 2C), whereas Repeat A levels increased 1.8-fold (Fig. 2E). Thus, Repeat A produces a small internal transcript, present in both male and female cells before XCI, but restricted to females after XCI. We designate the transcript ‘RepA’ for Repeat A.
To test whether PRC2 is actually recruited by RepA, we generated doxycycline-inducible RepA transgenic female ES cells (Fig. 2H) and asked if RepA could target PRC2 to ectopic autosomal sites independently of Xist. Indeed, for two clones (B5, C5) of low transgene copy number, doxycycline induction resulted in >3-fold increases in RepA and commensurate increases in PRC2 binding (Fig. 21). Thus, RepA is sufficient to recruit PRC2 in vivo without Xist, and recruitment depends on RepA transcription/RNA.
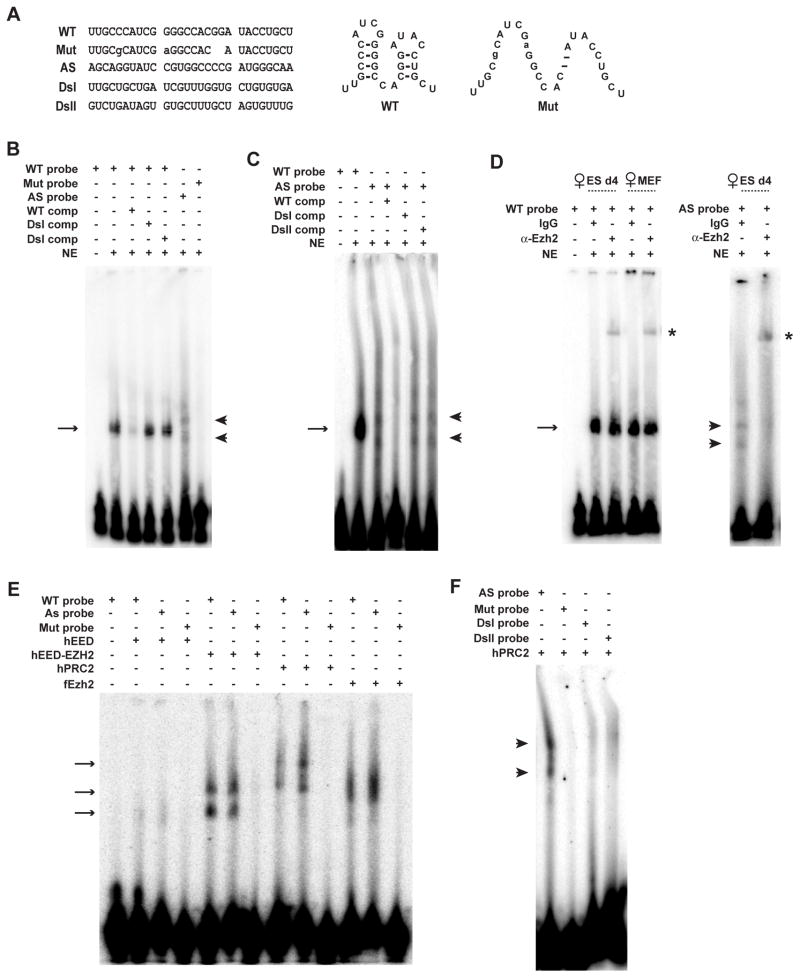
Does RepA RNA directly bind PRC2? To investigate, we tested whether RepA RNA oligos could shift PRC2 in vitro in an electrophorectic mobility shift assay (EMSA). RepA comprises 7.5 tandem repeats of a 28-nt sequence that folds into two conserved stem-loop structures (13) (Fig. 3A). A specific RNA-protein complex was observed when ES nuclear extract was incubated with wildtype sense probe (Fig. 3B,C). Interestingly, a specific complex was also seen with antisense RNA, which harbors complementary stem-loop structures. In both cases, RNA-protein interactions were disrupted by excess cold wildtype but not mutant or random competitors. No shift was observed with a mutant probe lacking the conserved stem-loop structures or with random RNA oligos (DsI, DsII). Therefore, a specific factor in ES nuclei binds RepA and Tsix.
Figure 3. RepA RNA directly binds PRC2 in vitro.
A. One Repeat A unit. WT, wildtype sense. Mut, mutated. AS, antisense. DsI and DsII, randomized Xist sequences.
B. EMSA using female ES nuclear extract (NE). Comp, competitors at 500X molar excess. Arrow, sense shift. Arrowhead, AS shift.
C. Antisense binding competed by sense but not nonspecific RNAs.
D. EMSA supershifts (*) with α-Ezh2.
E. EMSA using recombinant hPRC2 (sub)complexes. fEzh2, Drosophila Ezh2.
F. hPRC2 bound by antisense but not by randomized probes.
To identify the factor, we asked if α-Ezh2 could supershift the complex and found that pre-incubation in nuclear extract (d4 female ES or MEF) produced a supershift, whereas normal IgG did not (Fig. 3D). Therefore, PRC2 directly binds RepA and Tsix, in agreement with RIP results (Fig. 1F). To confirm, we generated recombinant human PRC2 (hPRC2) containing EED, EZH2, SUZ12, and RBAP48 (15) and observed that hPRC2 shifted both sense and antisense RNAs but not mutated or random RNA (Fig. 3E,F; additional bands may indicated subcomplexes). The hEED-hEZH2 subcomplex and the complete hPRC2 complex bound wildtype RNAs equally well. Ezh2 alone could also bind RNA, but hEED alone could not. Thus, Ezh2 must be the RNA-binding subunit of PRC2 (Fig. S1). Given that Tsix also binds PRC2 and is a known Xist antagonist, Tsix could block XCI by titrating away PRC2. Indeed, RepA and Tsix oligos competed with each other for PRC2 in vitro (Fig. 3C) and, in the absence of Tsix in vivo (TsixδCpG/+), H3-K27me3 occurred prematurely on d0 (Fig. 1H). We propose that RepA directly interacts with Ezh2 and that Tsix competitively inhibits this interaction.
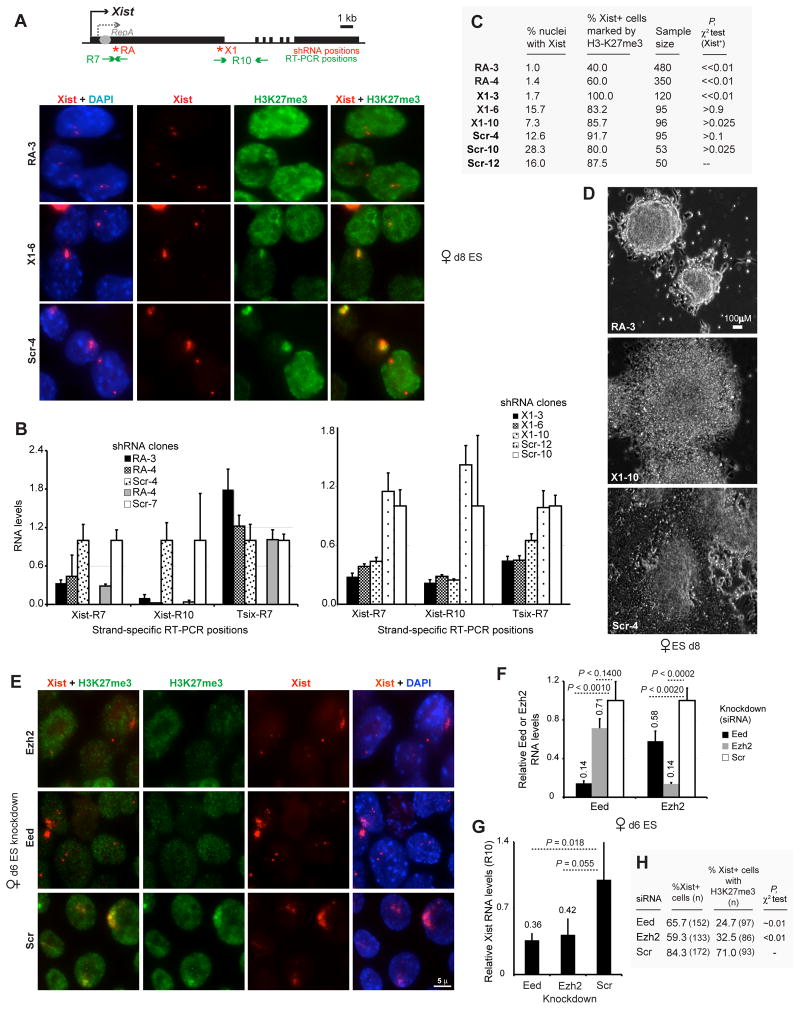
Previously, PRC2 seemed an unlikely direct target of Xist, as one report suggested that PRC2 is recruited without Repeat A (7). However, another report showed that PRC2 recruitment drops 80–90% in Repeat A mutants (9). To test if RepA functions in XCI, we created female ES clones carrying shRNA transgenes directed against RepA (RA; Fig. 4A). Because RepA and Xist overlap, shRNA against RepA could potentially affect Xist. To distinguish RepA from Xist, we created shRNA against the end of Xist exon 1 (X1), which does not overlap RepA. qRT-PCR confirmed knockdown efficacy and specificity (Fig. 4B: Xist contains R7 sequence, so it may be affected by X1 knockdown; residual R7 levels may represent RepA).
Figure 4. RepA/PRC2 knockdowns compromise XCI initiation.
A. Xist RNA-H3K27me3 immunoFISH in knockdown clones. shRNA: RA, RepA. X1, Xist exon 1. Scr, scrambled control.
B. Xist and Tsix levels at indicated positions in knockdown clones.
C. ImmunoFISH: Frequency of Xist upregulation (Xist+) and H3-K27me3 foci. P, pairwise comparison against Scr-12 control for Xist+ frequencies.
D. EB growth in shRNA clones.
E. Xist-H3K27me3 immunoFISH after Eed, Ezh2, or control knockdown.
F. Eed and Ezh2 mRNA levels after knockdown in TsixΔ/+ES cells. P, student t-test.
G. qRT-PCR of Xist RNA after indicated knockdowns. P, student t-test.
H. ImmunoFISH: Frequency of Xist upregulation and H3-K27 trimethylation after indicated knockdowns. P compares Xist foci numbers in controls (‘expected’) versus Eed/Ezh2 knockdowns.
Xist induction was severely compromised when RepA is depleted in clones RA-3 and RA-4, as few Xist foci were seen on d8 when compared to X1 and scrambled (Scr) controls (Fig. 4A–C). Thus, RepA RNA is required for Xist upregulation. In 100% of RA-3 and RA-4 cells lacking Xist foci, H3-K27me3 was absent on the X (Fig. 4C). In a very small minority of RA-3 and RA-4 cells which upregulated Xist, H3-K27me3 was also compromised, indicating PRC2 recruitment defects - high Xist levels notwithstanding. Consistent with the failure of XCI, RepA-shRNA clones showed extremely poor EB differentiation in contrast to controls (Fig. 4D). X1 clones showed an intermediate phenotype, consistent with intermediate expression of Xist. Although the X1 region is dispensable for silencing and localization (13), its knockdown could affect overall Xist stability and explain the intermediate phenotype. We conclude that RepA RNA functions not only in Xist transactivation but also in H3-K27 methylation and XCI.
We next examined whether knocking down PRC2 subunits might have similar effects. Indeed, Eed and Ezh2 knockdown in d6 female EB led to significant reductions in Xist and H3-K27me3 foci (Fig. 4E–H). Therefore, PRC2 also plays a role in Xist upregulation and XCI. Consistent with previous studies (16, 17), among Xist+ cells, PRC2 deficiency did not abrogate genie silencing (Fig. S2), possibly due to functional redundancy of PRC2 and PRC1 (17). By our data (Fig. 1H, 4), the primary effect of the RepA/PRC2 knockdowns may be abrogation of preemptive H3-K27me3 on Xist, an event hypothesized to be necessary for Xist induction (11). Therefore, RepA/PRC2 complex may act during XCI firstly by inducing H3-K27me3 at Xist for its transactivation and secondly by enabling spread of H3-K27me3 along the Xi.
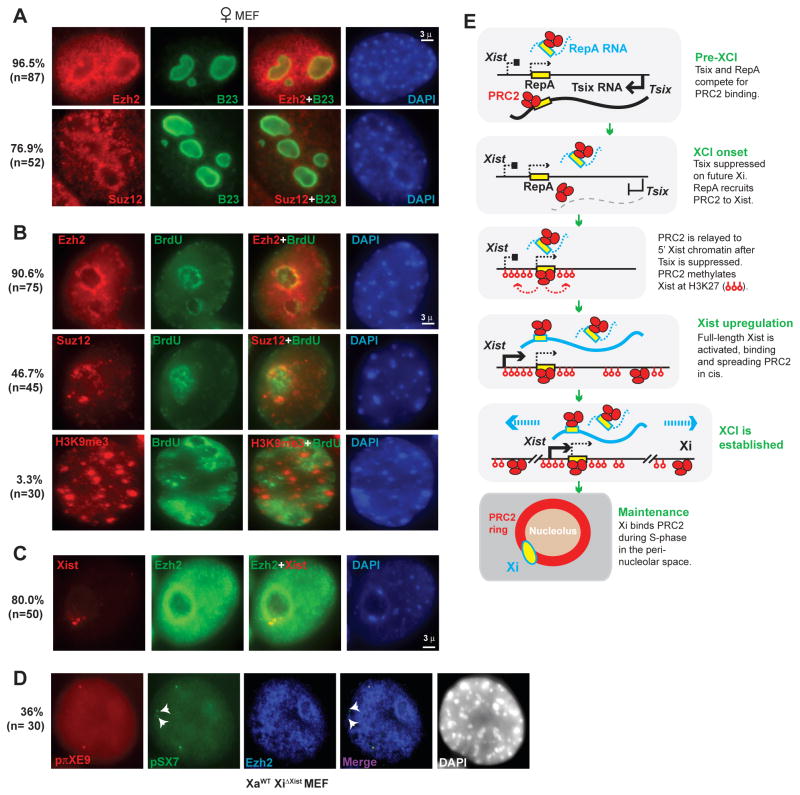
Given the importance of PRC2, it is odd that Xi is decorated by PRC2 only during initiation of XCI, though it stably retains H3-K27me3 thereafter (7, 8). Given the hypothesis that Xi’s epigenetic state is maintained by visiting a perinucleolar compartment during S-phase (6), we wondered if PRC2 association during the maintenancy phase may be likewise compartmentalized and transient. Indeed, we observed high levels of Ezh2, Suz12, and H3-K27me3 in this late-replicating perinucleolar compartment (Fig. 5A,B), to which ~80% of Xi is associated in MEFs (Fig. 5C). When Xi that is deleted for Xist after XCI occurs (XaWTXiΔXist), the chromosome fails to relocalize to this compartment (6). In such cells, we observed that PRC2 localization and H3-K27me3 were abolished (Fig. 5D)(6), supporting the idea that Xi in post-XCI cells associates with PRC2 and maintains H3-K27me3 by visiting the perinucleolar compartment during DNA replication.
Figure 5. PRC2 and Xi associate in the perinucleolar compartment after XCI.
A. Immunostain: Ezh2 and Suz12 concentrate around the nucleolus (B23+).
B. Ezh2 and Suz12, but not H3K9me3, showed perinucleolar enrichment.
C. Xist RNA-Ezh2 immunoFISH.
D. Xist DNA-Ezh2 immunoFISH in XaWTXiΔXist MEFs (6).
E. Summary and model.
In summary, we have discovered a small ncRNA that is required to target PRC2 to a specific locus. Long suspected (18), an RNA cofactor may explain why no DNA-binding subunit for mammalian polycomb has emerged so far. Ezh2 is apparently the RNA-binding PRC2 subunit. For XCI, the data provide new insight into how silencing is initiated on Xi (Fig. 5E). Given Tsix’s established role as Xist antagonist (3), ability to bind PRC2 and compete with RepA (Fig. 3), and molar excess over Xist, we propose that Tsix prevents RepA-PRC2 action in pre-XCI cells by titrating RepA away from PRC2, by blocking RepA-PRC2 transfer to chromatin, or by preventing PRC2 catalysis. The latter two possibilities may explain why RepA-PRC2 interactions in males does not induce H3-K27me3 (Fig. 1D). In our model, when Tsix is downregulated on the future Xi, RepA productively engages PRC2, methylates the Xist promoter in cis, and enables Xist transactivation. In support of this, abolishing Tsix (TsixΔCpG/+) results in premature H3-K27 trimethylation (Fig. 1C) and elevated Xist levels (11). Full-length Xist also binds PRC2 (Fig. 1), so the spread of Xist RNA along Xi could distribute PRC2 and H3-K27me3 throughout the chromosome. As ectopic Xist transgenes are known to spread autosomal silencing (13), our data suggest that Xist - possibly RepA itself (Fig. 1) - serves as a nucleation center. After XCI, Xi maintains its association with PRC2 via the perinucleolar compartment in a RepA/Xist-dependent manner. With evidence that RNAi is required to localize Xist and target H3-K27me3 (19), involvement of small RNAs and RNAi proteins may also be possible. Because another ncRNA (“HOTAIR”) was recently identified in connection with PRC2 at a human HOX locus (20), RNA cofactors may emerge as universal requirements for polycomb targeting.
Supplementary Material
References
- 1.Brown CJ, et al. Cell. 1992;71:527. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 2.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Nature. 1996;379:131. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 3.Lee JT, Lu N. Cell. 1999;99:47. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- 4.Clemson CM, McNeil JA, Willard H, Lawrence JB. J Cell Biol. 1996;132:259. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lucchesi JC, Kelly WG, Panning B. Annu Rev Genet. 2005;39:615. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- 6.Zhang LF, Huynh KD, Lee JT. Cell. 2007;129:693. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- 7.Plath K, et al. Science. 2003;300:131. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 8.Silva J, et al. Dev Cell. 2003;4:481. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 9.Kohlmaier A, et al. PLoS Biol. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ringrose L, Paro R. Annu Rev Genet. 2004;38:413. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 11.Sun BK, Deaton AM, Lee JT. Mol Cell. 2006;2l:617. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 12.Hendrich BD, Brown CJ, Willard HF. Hum Mol Genet. 1993;2:663. doi: 10.1093/hmg/2.6.663. [DOI] [PubMed] [Google Scholar]
- 13.Wutz A, Rasmussen TP, Jaenisch R. Nat Genet. 2002;30:167. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 14.Sado T, Wang Z, Sasaki H, Li E. Development. 2001;128:1275. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- 15.Francis NJ, Saurin AJ, Shao Z, Kingston RE. Mol Cell. 2001;8:545. doi: 10.1016/s1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- 16.Kalantry S, Magnuson T. PLoS Genet. 2006;2:e66. doi: 10.1371/journal.pgen.0020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schoeftner S, et al. Embo J. 2006;25:3110. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitt S, Paro R. Genome Biol. 2006;7:218. doi: 10.1186/gb-2006-7-5-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogawa Y, Sun BK, Lee JT. Science. 2008;320:1336. doi: 10.1126/science.1157676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rinn JL, et al. Cell. 2007;129:1311. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.We thank R. Spencer and Y. Jeon for unpublished data and technical advice and Y. Ogawa for critical reading of the manuscript. Grant support: MGH Fund for Medical Discovery (JZ), NSF predoctoral award (JAE), Jane Coffin Childs Fellowship (JJS), and NIH-RO1GM58839 (JTL). JTL is an Investigator of the HHMI.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.