Abstract
Killer lymphocytes recognize stress-activated NKG2D ligands on tumors. We examined NKG2D ligand expression in head and neck squamous cell carcinoma (HNSCC) cells and other cell lines. HNSCC cells typically expressed MHC class I chain-related gene A (MICA), MICB, UL16-binding protein (ULBP)2, and ULBP3, but they were uniformly negative for cell surface ULBP1 and ULBP4. We then studied how cancer treatments affected NKG2D ligand expression. NKG2D ligand expression was not changed by most cancer-relevant treatments. However, bortezomib and other proteasome inhibitor drugs with distinct mechanisms of action dramatically and specifically up-regulated HNSCC ULBP1 mRNA and cell surface protein. Proteasome inhibition also increased RNA for ULBP1 and other NKG2D ligands in nontransformed human keratinocytes. Proteasome inhibitor drugs increased ULBP1 transcription by acting at a site in the 522-bp ULBP1 promoter. Although the DNA damage response pathways mediated by ATM (ataxia-telangiectasia, mutated) and ATR (ATM and Rad3-related) signaling had been reported to up-regulate NKG2D ligand expression, we found that ULBP1 up-regulation was not inhibited by caffeine and wortmannin, inhibitors of ATM/ATR signaling. ULBP1 expression in HNSCC cells was not increased by several ATM/ATR activating treatments, including bleomycin, cisplatin, aphidicolin, and hydroxyurea. Ionizing radiation caused ATM activation in HNSCC cells, but high-level ULBP1 expression was not induced by gamma radiation or UV radiation. Thus, ATM/ATR signaling was neither necessary nor sufficient for high-level ULBP1 expression in human HNSCC cell lines and could not account for the proteasome effect. The selective induction of ULBP1 expression by proteasome inhibitor drugs, along with variable NKG2D ligand expression by human tumor cells, indicates that NKG2D ligand genes are independently regulated.
Lymphocyte activation is governed by a balance between activating and inhibitory receptor signaling. This balance is especially critical because appropriately activated killer lymphocytes defend against intracellular pathogens and cancers, but inappropriately activated killer lymphocytes attack healthy cells and cause autoimmune disease. The activating receptor NKG2D is expressed on virtually all human NK lymphocytes, CD8 T lymphocytes, and lymphocytes that express the γδ form of the TCR (1-3). NKG2D engagement may directly activate NK cells and it costimulates T cells (4). The importance of NKG2D is underscored by observations that tumors arise more frequently in NKG2D-deficient mice (5) and that many viruses and tumors have evolved mechanisms to escape NKG2D recognition (6, 7).
NKG2D binds to ligands that are up-regulated by a variety of stimuli that have been collectively termed “cell stress” (1-3, 8). NKG2D is unusual in binding to multiple ligands that have limited sequence homology (6, 9). In fact, NKG2D recognizes at least seven distinct protein ligands in mice (Rae-1 family members, Mult1, and H60) and humans (MHC class I chain-related gene A (MICA),4 MICB, UL16-binding protein (ULBP)1−4, and retinoic acid early transcript (RAET)1G). Among the human NKG2D ligands, MICA, MICB, ULBP4, and RAET1G are type I cell surface transmembrane proteins, whereas ULBP1−3 are GPI linked and contain either two (ULBP1−4, RAET1G) or three (MICA, MICB) extracellular domains (2, 6). All of the NKG2D ligands are distantly related to MHC class I molecules, but they do not associate with β2-microglobulin or bind either peptide or nonpeptide Ags. The distinct NKG2D ligand structures and divergent promoter sequences imply that they are differentially regulated (6, 10, 11).
Killer lymphocyte recognition of cancer cell NKG2D ligands allows rejection of experimental tumors and boosts subsequent adaptive immunity (12-15). These interactions may be relevant in human cancer patients because naturally occurring tumors selectively down-regulate killer lymphocyte NKG2D or cancer cell NKG2D ligand, resulting in defective NKG2D-mediated tumor surveillance (16-19). For example, cell surface NKG2D ligand expression correlated with decreased uveal melanoma metastasis (20), but with worse breast cancer grade and prognosis (21). Therefore, it is relevant to examine NKG2D ligand expression patterns in different types of human tumors. NKG2D ligands are expressed in many cancers, but it is rare for any cancer to express all NKG2D ligands. MICA was expressed on most human melanomas and melanoma cell lines, ULBP2 and ULBP3 were expressed rarely, and MICB and ULBP1 were not expressed; when a given NKG2D ligand expression was observed, it often was heterogeneous (22, 23). Among carcinoma cell lines, MICA expression was usually positive, ULBP2, ULBP3, and ULBP4 expression were variable, MICB expression was often absent, and ULBP1 expression was rare (24-27). In contrast to these solid tumors, T cell leukemias less often expressed MICA and more often expressed ULBP1 (17, 24). Only a few groups have studied NKG2D ligand expression in head and neck squamous cell carcinoma (HNSCC). These tumors account for 3% of cancer in the United States, and overall 5-year survival has not improved significantly in the last 50 years (28). One group found that MICB RNA, but not MICA RNA, was significantly overexpressed in HNSCC compared with noncancer tissues from the same patients, whereas another group found that MICA was overexpressed, but they did not study MICB (29, 30). Because HNSCC appear susceptible to NKG2D-mediated immune recognition (27, 31), we investigated the pattern of NKG2D ligand expression by human HNSCC cell lines and how it is affected by clinically relevant drugs and treatments. We found that bortezomib and other proteasome inhibitor drugs dramatically and specifically up-regulated ULBP1 cell surface protein, mRNA, and gene transcription. We then tested the hypothesis that proteasome inhibitor drugs increase ULBP1 expression via ATM (ataxia-telangiectasia, mutated)/ATR (ATM and Rad3-related) signaling.
Materials and Methods
Cell culture and treatment
Human FaDu, HeLa, CAL27, PCI-15A, PCI-15B, Tu167, DM14, and SKMES-1 cells were cultured in DMEM with additives (Invitrogen) and in 10% iron supplemented bovine serum or FBS (HyClone). Tu167 and DM14 cells were gifts from Dr. J. Meyers (M. D. Anderson, Houston, TX). PCI-4A, PCI-4B, PCI-15A, and PCI-15B cells were gifts from Dr. T. L. Whiteside (University of Pittsburgh, Pittsburgh, PA). Jurkat cells were a gift of Dr. G. Koretzky (University of Iowa). Neonatal human epidermal keratinocytes from Invitrogen were obtained from H. Swanson (University of Kentucky) and grown in EpiLife media plus the growth supplement EDGS (EpiLife Defined Growth Supplement; Invitrogen). Cells were seeded at 50−60% confluency 24 h before treatment. MG132, lactacystin, epoxomicin (Sigma-Aldrich), and bortezomib (Millennium Pharmaceuticals) were resuspended as recommended by the manufacturer and used 1/1000 directly into cell culture at final concentrations of 10 μM, 10 μM, 1 μM, and 1 μg/ml, respectively, except as noted. FaDu cells were treated with cisplatin (10 μg/ml; Calbiochem/EMD Chemicals), bleomycin (0.3 U/L; Ben Venue Laboratories), hydroxyurea and aphidicolin (both 5 mM; Sigma-Aldrich), caffeine, and wortmannin (10 mM and 6 μM, respectively; Sigma-Aldrich). In some cases caffeine and wortmannin were added to cell culture 2 h before the addition of MG132 (10 μM) and cultured for an additional 5 h. FaDu and CAL27 cells (2.5 × 106 cells per 100-mm dish) were cultured overnight before treatment with UV light in a custom-made radiator consisting of UVA and UVB bulbs (GE Healthcare) or with gamma radiation at 15 Gy/min from a 137Cs source (Mark I Model 68; J. L. Shepherd and Associates). Control cells were removed from the incubator for the same time as treated cells. Cells then were cultured before harvesting for RNA quantitation (5 h) or Western blotting (1.5 h).
Expression analysis
Anti-NKG2D ligand mAbs were provided by Amgen. ULBP1 (huULBP1-M295), ULBP2 (huULBP2-M311), and ULBP3 (huULBP3-M551) have been described (32). Anti-ULBP4 mAb (huULBP4-M475) was generated by immunizing mice with an ULBP4-Fc construct (33). Anti-HLA class I mAb (HP-1F7) was a gift of Dr. M. Lopez-Botet (University Pompeu Fabra, Barcelona, Spain). Cells were incubated with unlabeled primary mAb or with nonspecific control mouse IgG Ab (SouthernBiotech), washed, and then stained with PE-conjugated goat anti-mouse IgG (SouthernBiotech). Cell surface staining was analyzed using Summit software (Dako). For DNA content analysis, cells were fixed in 70% ethanol for 2 h on ice, washed, and resuspended in propidium iodide/Triton X-100 solution with RNase A, as described (34). Cell cycle was analyzed using ModFit software (Verity Software). After DNase I treatment and reverse transcription of unselected RNA into cDNA with random hexamer priming, expression was analyzed using quantitative PCR, as described (35). Briefly, NKG2D ligand values were calculated using the slope and y-intercept of a standard curve for each primer set. For each sample, these values were adjusted for RNA integrity using GAPDH values. Then, the GAPDH-corrected values for each test sample were normalized by dividing by values for the DMSO solvent treatment values for the same time point. This is presented as relative RNA in the figures. For each cDNA, PCR amplicons crossed predicted mRNA exon-exon junctions to ensure that genomic DNA was not amplified (illustrated in Fig. 7A). Lack of significant genomic DNA contamination was confirmed by amplifying controls without reverse transcriptase. The forward and reverse primers, respectively, were as follows: MICA, TCCTGCTTCTGGCTGGCAT and GACAGCACCGTGAG GTTA; MICB, CTACATGGAACACAGCGGGA and GCATAGC AGCAGAAACATATGGA; ULBP1 mRNA, TTTCCTTAAAGGGCAAC TGCT and AGGAACTGCCAAGATCCTCT; ULBP2, CATTACTTCT CAATGGGAGACTGT and TGTGCCTGAGGACATGGCGA; ULBP3, CTGATGCACAGGAAGAAGAG and TATGGCTTTGGGTTGAGCT AA; ULBP4, AAGGGAGACTGCGATCA and GAAGACCAGTGGATA TCTGAA. To measure ULBP1 heteronuclear (unspliced) RNA (hnRNA), we used intron-based primers as illustrated in Fig. 7A, CAAGAGGTT GAGGAGCAT and CTGAACTCGAGACAGCTTCCAT. These primers did not produce PCR product in the absence of reverse transcription, showing an absence of genomic DNA contamination of the extracted RNA (data not shown). All primers showed minimal cross-priming of other NKG2D ligand templates, including RAET1G (36). After this study was completed, we learned that RAET1L is expressed in a limited number of cell lines (Rob Eagle and John Trowsdale, unpublished observation). Therefore, we tested primer specificity with RAET1L. RAET1L cDNA was significant amplified by ULBP2 primers, but not other primers (data not shown). Because apparent expression of both ULBP2 RNA and cell surface protein changed little in our experimental conditions, potential cross-priming of RAET1L cDNA with ULBP2 primers did not affect our conclusions.
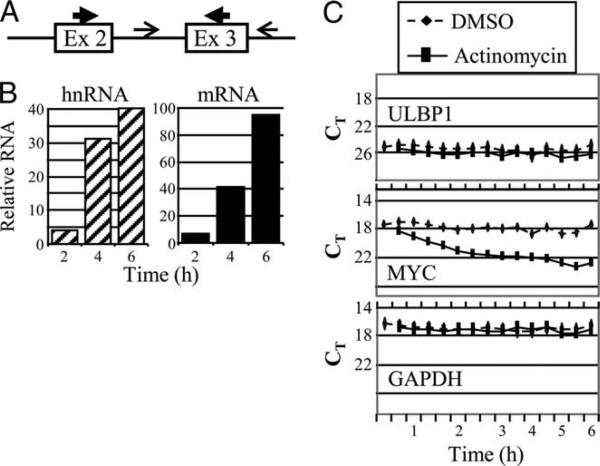
FIGURE 7.
MG132 induces ULBP1 transcription. A, Schematic illustration of the primers used to detect nascent hnRNA (thin arrows flanking exon 3) and mRNA (thick arrows in exon 2 and exon 3). B, FaDu cells were treated with DMSO solvent or MG132 for the times indicated before cell lysis and analysis of relative ULBP1 hnRNA (left panel, hatched bars) and mRNA (right panel, solid bars). PCR amplification of RNA that had not undergone reverse transcription gave uniformly negative results, indicating the absence of genomic DNA. The finding is typical of three independent experiments. C, ULBP1 mRNA is stable. FaDu cells were treated with DMSO or actinomycin D, as indicated. Cells were harvested at the times indicated and ULBP1, c-myc, and GAPDH mRNA was measured by quantitative RT-PCR. The x-axis is not linear; on the y-axis, a lower CT (threshold cycle) value indicates greater mRNA amount. The finding is typical of at least three independent experiments.
Promoter analysis
Three ULBP1 promoter fragments of 2299, 875, and 522 bp were PCR amplified from PAC clone RP11−409C11 (BACPAC Resources Center, Children's Hospital Oakland Research Institute, Oakland, CA) using a common reverse primer (CCGCTCGAGCGGTCGACCTGGAGCTCCG TGGACAGGAG at chr6:150326851) and three forward primers (CG GGGTACCATCCCACTTCACTCCAGCCTGGGCAAC, CGGGTACCT CCACCTCCCTCTGGCTTC, and GTGGAGAGGCAAAAAAGGTTCG CT, located at chr6:150,324,579, chr6:150,326,003, and chr6:150,326,356, respectively, as reported by the University of California Santa Cruz Genome Browser (genome.ucsc.edu/cgi-bin/hgGateway)). Promoter fragments were cloned into the pGL3 luciferase reporter plasmid. FaDu cells were transfected with 1.0 μg of ULBP1 promoter plasmid and either 2.5 ng of CMV-Renilla or 10 ng of SV40-Renilla control plasmids using Lipofectamine and Plus reagents (Invitrogen) following the manufacturer's instructions. After culture for 24 h, cells were treated with 10 μM MG132 or DMSO solvent and lysed 13 h later. Luciferase and Renilla activity was measured in lysates as described (37). In this and other analyses, Microsoft Excel was used to calculate means and 95% confidence limits.
Immunoblotting
FaDu and CAL27 cells were trypsinized, washed twice with PBS, and frozen at −80°C. Whole-cell extracts were prepared by resuspending the cell pellets for 15 min in 60 μl of ice-cold lysis buffer (25 mM Tris-HCl, 25 mM NaCl, 30 mM KCl, 5 mM CaCl2, 5 mM MgCl2, 0.05% Nonidet P-40, 1 mM EDTA, 1 mM NaVO4; Sigma-Aldrich), 1× Roche protease inhibitor cocktail (Roche Diagnostics), 0.5 mM PMSF (Sigma-Aldrich), and 0.55 mg/ml DNase 1 (Worthington Biochemical), pH 7.6. DNA digestion was stopped with addition of 2.0 μl of 0.5 M EDTA, and the debris was pelleted in a cold microcentrifuge for 5 min. Supernatant containing 40 μg of protein, determined as described (38), was denatured in boiling 2× SDS gel-loading buffer (39) for 10 min, electrophoresed on an 8% polyacrylamide-SDS gel, and transferred to polyvinylidene difluoride membrane (Millipore) by electroblotting. Membranes were incubated with control anti-β-actin Ab (Sigma-Aldrich), anti-ATM Ab (Cell Signaling Technology), or anti-phospho-Ser1981 ATM mAb (10H11.E12; Cell Signaling Technology). Membranes were washed and incubated with HRP-coupled secondary goat anti-rabbit Ab (Santa Cruz Biotechnology) using standard methods. Signal was developed using ECL detection reagents (GE Healthcare) and x-ray film (Kodak BioMax light film).
Results
HNSCC express a restricted set of NKG2D ligands
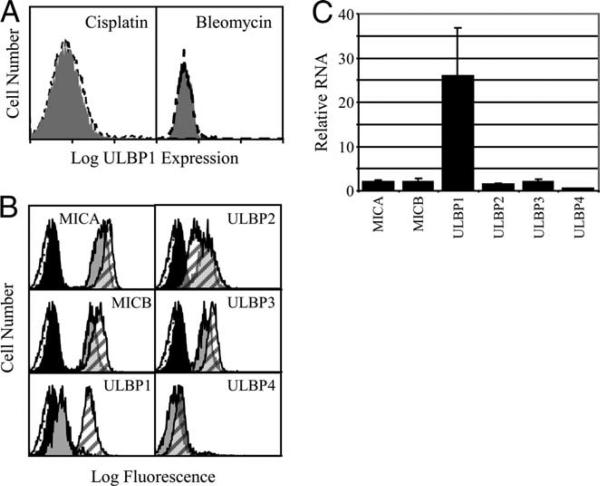
NKG2D ligand expression patterns vary between and within tumor types, so we measured cell surface expression levels on seven HNSCC cell lines by flow cytometry. Similar to the colon cancer cell line, CaCo-2, CAL27, and most other HNSCC cell lines expressed MICA, MICB, ULBP2, and ULBP3, but not ULBP1 or ULBP4 (Table I and Fig. 1). HeLa cells expressed little MICB and little or no cell surface protein for any of the ULBPs. In contrast to these epithelial tumors, Jurkat leukemic T cells expressed considerable cell surface ULBP1 (Table I).
Table I.
Head and neck squamous cell carcinoma cell lines do not express cell surface ULBP1 or ULBP4a
| Cells | Source | MICA | MICB | ULBP1 | ULBP2 | ULBP3 | ULBP4 |
|---|---|---|---|---|---|---|---|
| FaDu | HNSCC | + | + | − | + | + | − |
| CAL27 | HNSCC | + | + | − | + | + | − |
| PCI-15A | HNSCC | + | + | − | + | + | − |
| Tu167 | HNSCC | + | + | − | + | + | − |
| DM14 | HNSCC | + | + | − | − | + | − |
| CaCo-2b | Colon | + | + | − | + | + | − |
| HeLac | Cervical | + | Weak | − | Weak | Weak | − |
| Jurkatd | T cell | + | + | + | + | − | − |
Cell surface NKG2D ligand expression, assessed by mAb staining and flow cytometry.
Colon adenocarcinoma.
Cervical adenocarcinoma.
T cell lymphoma/leukemia.
FIGURE 1.
HNSCC cell lines express NKG2D ligands selectively. CAL27 cells were stained as indicated and analyzed by flow cytometry. The open histogram plot shows staining with nonspecific mouse IgG, and the filled histogram plot shows staining with the mAb indicated. mAb staining is presented on a four-decade fluorescence intensity scale, which was truncated in some cases for compact display. The experiment shown is representative of two independent experiments with CAL27 and is typical of most HNSCC cell lines tested (see Table I).
Clinically relevant stresses selectively induce NKG2D ligand expression
To investigate whether cancer treatments affect NKG2D ligand expression, we incubated FaDu cells with a variety of anticancer drugs. Cisplatin is commonly used to treat HNSCC (40). Radiation therapy is a mainstay of HNSCC treatment, and radiation effects are mimicked by bleomycin (40, 41). Bortezomib has shown promise in preclinical trials. After a 16-h drug treatment, NKG2D ligand cell surface expression was determined by flow cytometry. Neither cisplatin nor bleomycin affected the expression of any of the NKG2D ligands tested, regardless of their initial level of expression (Fig. 2A and data not shown). Bortezomib treatment produced little or no change in MICA, MICB, ULBP2, ULBP3, or ULBP4 levels (Fig. 2B). In contrast, bortezomib selectively and reproducibly increased ULBP1 expression (Fig. 2B). To assess whether the bortezomib effect was the result of RNA up-regulation, FaDu cells were treated for 5 h and steady-state mRNA levels were determined by quantitative RT-PCR. In parallel with cell surface expression, bortezomib dramatically induced ULBP1 mRNA, but had little effect on other NKG2D ligands (Fig. 2C).
FIGURE 2.
The effect of clinically relevant drugs on NKG2D ligand expression. A, Cisplatin and bleomycin do not induce NKG2D ligand expression. FaDu HNSCC cells were untreated (gray shading) or were treated for 16 h with the indicated drug (dashed line) and stained with anti-ULBP1 mAb. Cisplatin and bleomycin treatment did not change expression of MHC class I, MICA, MICB, or ULBP2−4 (not shown). B, Bortezomib induces cell surface ULBP1 expression. FaDu cells were treated with DMSO or bortezomib for 16 h and stained with nonspecific mouse IgG (speckled and solid histograms representing control and bortezomib-treated cells) and with the indicated anti-NKG2D ligand mAb (gray and hatched histograms representing control and bortezomib-treated cells). For A and B, flow cytometry results are presented on a log scale. C, Bortezomib selectively induces ULBP1 mRNA expression. FaDu cells were treated with DMSO solvent or with bortezomib for 5 h and NKG2D ligand RNA was measured. Shown are means and 95% confidence limits of three independent experiments.
Bortezomib acts via proteasome inhibition
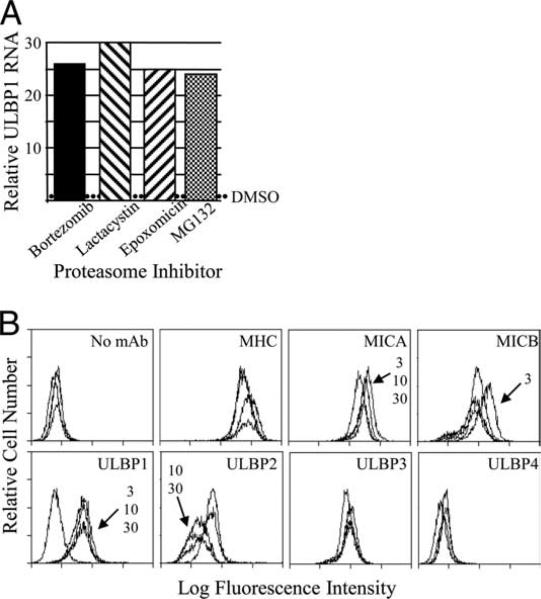
Although bortezomib is a highly specific proteasome inhibitor, it potentially may affect other enzymatic activity. Therefore, we tested additional drugs that inhibit the proteasome via diverse mechanisms (42). FaDu cells were treated with bortezomib, MG132, lactacystin, and epoxomicin, and ULBP1 mRNA was measured 5 h later. Each proteasome inhibitor dramatically increased ULBP1 mRNA levels (Fig. 3A). In separate experiments, the lactacystine effect varied in strength, but it was always strongly positive (data not shown). Thus, four drugs that inhibit proteasome activity by at least three distinct mechanisms all strongly increased ULBP1 mRNA levels. These results demonstrate that proteasome inhibition was responsible for ULBP1 up-regulation. We tested whether MG132 also affected cell surface ULBP1. As with bortezomib, MG132 consistently induced cell surface ULBP1 (Fig. 3B). Only minor shifts were seen in the expression of other NKG2D ligands, which were not consistent (Fig. 3B and data not shown). Because of the cost and limited availability of bortezomib, we routinely performed subsequent experiments using MG132.
FIGURE 3.
Proteasome inhibitor drugs increase ULBP1 expression. A, FaDu cells were treated with DMSO solvent or the proteasome inhibitor as indicated for 5 h before cell lysis and analysis of ULBP1 mRNA. The relative level of mRNA was normalized to that of control DMSO-treated cells in the same experiment, indicated by the dotted line. Each proteasome inhibitor was tested at least twice. B, FaDu cells were treated with DMSO or with 3, 10, or 30 μM MG132 for 24 h and stained with mAb to MHC class I and to the NKG2D ligands, as indicated. Arrows and MG132 concentrations indicate treatment groups with staining intensity different from the DMSO-only control groups. Shown is one experiment out of at least 10 experiments with similar results, except that small shifts in MICA and ULBP2 staining were seen in a minority of experiments.
Proteasome inhibitor drugs induce ULBP1 expression in multiple cell lines
To address whether the induction of ULBP1 by proteasome inhibition was limited to FaDu cells, several other squamous cell carcinomas were tested. MG132 treatment up-regulated ULBP1 cell surface protein on the HNSCC cell lines PCI-15A and CAL27 (Fig. 4A). Based on results with FaDu cells, increased ULBP1 cell surface expression suggested that ULBP1 mRNA levels also would increase. We tested several HNSCC cell lines, the noncancerous HaCaT human keratinocyte cell line, and a non-HNSCC cancer cell line. Following a 5-h MG132 treatment, steady-state ULBP1 mRNA levels were quantified by quantitative RT-PCR. MG132 treatment increased ULBP1 RNA to varying degrees in all cell lines, ranging from ∼4- to 36-fold over DMSO solvent-treated control cells (Fig. 4B). Thus, proteasome inhibitor drug treatment increased ULBP1 mRNA levels in HaCaT nonmalignant keratinocytes, in several HNSCC, and in other carcinoma cell lines.
FIGURE 4.
MG132 induces ULBP1 expression in multiple cell lines. Cells were treated with DMSO or MG132. A, After 16 h of culture, cell surface ULBP1 expression was measured. B, After 5 h culture, ULBP1 mRNA was measured. Solid bars denote HNSCC cell lines, hatched bar represents the nonmalignant HaCaT keratinocyte cell line, and the checkered bar represents the non-HNSCC human cancer cell line, 293T. Each result is typical of at least two independent experiments. C, Human epidermal keratinocytes were treated for 7 or 14 h, as indicated, and RNA was extracted and quantified. Shown are means and 95% confidence limits of three independent experiments.
We also wanted to test the effect of proteasome inhibitor drugs on nontransformed human epidermal keratinocytes. At both 7 and 14 h, MG132 increased ULBP1 RNA by 5- to 6-fold (Fig. 4C). MICA, MICB, ULBP2, and ULBP3 RNA also increased.
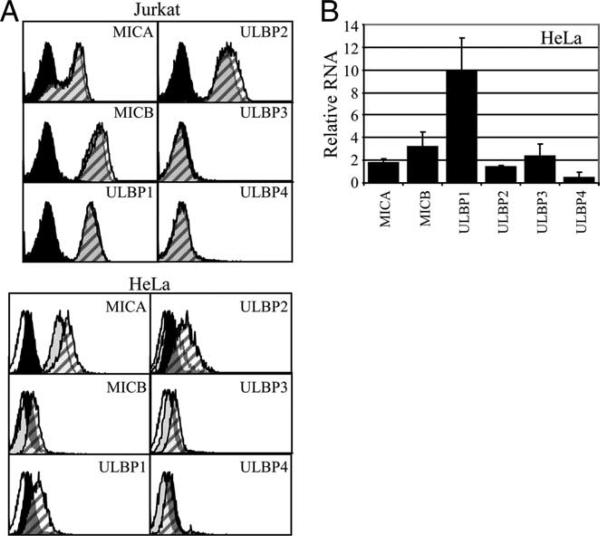
As this manuscript was in preparation, Armeanu et al. reported that low-dose bortezomib and other proteasome inhibitor drugs caused human hepatocellular carcinoma cell lines to produce increased amounts of MICA and MICB, but that ULBP1−3 remained undetectable (43). Additionally, Valés-Gómez et al. reported that proteasome inhibitor drugs up-regulated ULBP2, but not ULBP1, in Jurkat T leukemia cells (44). In the latter study, proteasome inhibitor drugs up-regulated cell surface ULBP2 in several other lines, including HeLa, but ULBP1 was not tested. We analyzed the effect of proteasome inhibitor drugs on NKG2DL expression in Jurkat and HeLa cells. Untreated Jurkat cells expressed considerable MICA, MICB, ULBP1, and ULBP2, but no significant ULBP3 or ULBP4 (Table I and Fig. 5A). As described (44), we treated Jurkat T cells with 0.25 μM MG132; higher doses caused considerable Jurkat cell death (data not shown). ULBP2 was up-regulated in two experiments (Fig. 5A and data not shown) but did not change in two other experiments (data not shown). Jurkat cell surface ULBP1 did not change in any of the experiments (Fig. 5A). The small and variable increase in ULBP2 expression was similar to ULBP2 expression changes that we had observed in HNSCC. In HeLa cells, MG132 treatment consistently up-regulated ULBP1 (Fig. 5A). ULBP2 was variable, ranging from no change (one experiment), to a slight increase (one experiment), and to an increase similar to that of ULBP1 (one experiment, shown in Fig. 5A). Therefore, MG132 caused a small increase in Jurkat and HeLa cell surface ULBP2, but the changes were less than the ULBP1 elevations seen in HNSCC cells. Other NKG2DL showed little or no cell surface change regardless of whether the initial levels were high (MICA) or low (ULBP3 and ULBP4; Fig. 5A). Consistent with elevated HeLa cell surface ULBP1 expression, MG132 increased ULBP1 RNA by 10-fold, with smaller changes in RNA for other NKG2D ligands (Fig. 5B).
FIGURE 5.
Effect of MG132 in the Jurkat and HeLa cell lines. Cells were treated with DMSO or MG132. A, After 16 h of culture, cells were stained with nonspecific mouse IgG (speckled and solid histograms representing control and MG132-treated cells) or with the indicated anti-NKG2D ligand mAb (gray and hatched histograms representing control and MG132-treated cells). Results are typical of at least three to four independent experiments. B, After 5 h of culture, HeLa RNA was measured. Shown are means and 95% confidence limits from three independent experiments.
Proteasome inhibition induces ULBP1 transcription
To investigate how ULBP1 expression is regulated, we performed a detailed time course experiment following MG132 treatment. Upon proteasome inhibition, ULBP1 mRNA levels increased geometrically between 0 and 4 h of MG132 treatment and slowly increased after 8 h (Fig. 6A). ULBP1 cell surface protein was not detectable above background at 2 or 4 h following MG132 treatment, but expression rapidly increased between 4 and 8 h (Fig. 6B). ULBP1 expression increased slowly after the 8-h time point and was maintained through 24 h of MG132 treatment. The delayed appearance of cell surface ULBP1 is not consistent with a rapid release of ULBP1 protein from intracellular stores or protection from protein degradation. Instead, the early appearance of ULBP1 RNA followed by the subsequent appearance of cell surface protein indicates that proteasome inhibition up-regulated ULBP1 cell surface protein expression by increasing steady-state mRNA levels.
FIGURE 6.
MG132 induces ULBP1 RNA before ULBP1 cell surface protein. A, FaDu cells were treated with DMSO solvent or MG132 for the times indicated before cell lysis and analysis of ULBP1 mRNA. The x-axis is not linear. B, Shown on a logarithmic scale is anti-ULBP1 mAb staining of FaDu cells treated with MG132 for the times indicated. Expression at 2 and 4 h was identical to that of DMSO-treated cells. Results shown are representative of at least two independent experiments.
To determine whether increased ULBP1 mRNA was caused by increased transcription, we measured unspliced nascent hnRNA levels following proteasome inhibition. Fig. 7A schematically illustrates how intron-based PCR primers detect unspliced hnRNA, whereas exon-based primers can detect spliced mRNA. As expected from the very short t½ typical of nascent RNA, the level of hnRNA was much lower than that of mRNA (data not shown). Both spliced and unspliced RNA increased within 2 h of MG132 treatment and continued to rise for 6 h (Fig. 7B). The rapid rise in nascent hnRNA levels indicates that proteasome inhibition induced ULBP1 transcription.
The ULBP1 3′-untranslated region is large (2.3 kb), suggesting that RNA stability might be actively regulated. Therefore, we performed RNA turnover studies using actinomycin D to block de novo mRNA synthesis (Fig. 7C). As noted above, low-level ULBP1 RNA was present in FaDu cells, allowing us to compared ULBP1 t½ with stable GAPDH RNA and unstable c-myc RNA. Upon treatment of FaDu cells with actinomycin D, c-myc mRNA decayed with an ∼40 min t½ . GAPDH mRNA failed to decrease within the 6-h treatment period. These values are consistent with reported t½ of <1 h for c-myc and ∼15 h for GAPDH (45). Similar to GAPDH, ULBP1 mRNA was stable for at least 6 h (Fig. 7C). Although actinomycin-induced cell death precluded collection of later time points and the calculation of a t½ , it is clear that ULBP1 mRNA was stable. As expected, ULBP1 mRNA remained stable after a 5-h treatment with MG132 followed by actinomycin D treatment for an additional 6 h (data not shown), consistent with a long ULBP1 mRNA t½ in the presence or absence of proteasome inhibitor drug-induced transcription. We conclude that any possible MG132-induced increase in mRNA stability beyond its constitutively long t½ in FaDu cells could make only a minor contribution to the rapid ULBP1 mRNA induction that we observed following proteasome inhibition. Thus, proteasome inhibitor drugs increase ULBP1 expression largely by increasing transcription.
Proteasome inhibitor drug treatment activates the ULBP1 promoter
Given that proteasome inhibition induced ULBP1 transcription, we investigated the ULBP1 promoter as a potential target of proteasome signaling. Using the University of California Santa Cruz Genome Browser (genome.ucsc.edu/cgi-bin/hgGateway), we determined that the longest stretch of nonrepetitive DNA extends ∼2.2 kb upstream of the ULBP1 ATG translation start site (data not shown). We cloned promoter fragments into the luciferase reporter vector pGL3 from a site immediately upstream of the ULBP1 ATG translation start site and extending 2299, 875, and 522 bp upstream. All three fragments were active in FaDu cells, demonstrating that the 522-bp proximal promoter contains all of the cis-acting elements required for transcription in HNSCC (Fig. 8A). This result extends prior tests of a 1-kb ULBP1 promoter in HEK293 and HeLa cells (46). To investigate whether proteasome inhibition response elements were located in the ULBP1 promoter, we treated transfected FaDu cells with MG132 during the final 13 h before harvesting. All three ULBP1 promoter constructs, including the short 522-bp fragment, responded to MG132 treatment with increased luciferase activity (Fig. 8B). As corrected for transfection efficiency using cotransfected pRL-SV40 (Renilla luciferase) plasmid, proteasome inhibition increased promoter activity 7- to 8-fold (Fig. 8). In the same experiment the pGL3-control plasmid was apparently stimulated ∼2-fold by MG132 treatment, so we further normalized ULBP1 promoter results using the pGL3-control results. This indicated that MG132 treatment stimulated ULBP1 promoter activity by 3.2- to 3.7-fold. The same relative induction (3- to 4-fold) was observed in experiments with cotransfected pRL-CMV (Renilla luciferase) plasmid or when results were normalized to live cell counts (data not shown). All three ULBP1 promoter fragments showed similar MG132 concentration-dependent activity (data not shown). We conclude that proteasome inhibitor drug signaling increases ULBP1 transcription, at least in part, by activating transcription factors that operate within the proximal 522 bp of the ULBP1 promoter.
FIGURE 8.
The ULBP1 promoter is active in FaDu HNSCC cells and promoter activity is increased by proteasome inhibition. FaDu cells were transfected with the indicated pGL3-luciferase test plasmids as described in Materials and Methods. Promoters 0.5, 0.9, and 2.3 represent pGL3 plasmids containing 522-, 875-, and 2299-bp DNA, respectively, 5′ of the ULBP1 ATG translation start site. Values represent averages from independent tests of at least three different plasmid preparations (each measured in duplicate), with error bars representing 95% confidence limits. A, Cotransfected CMV-Renilla luciferase plasmid was used to control for transfection efficiency. “0” refers to the promoterless pGL3-basic plasmid. B, Cotransfected SV40-Renilla luciferase plasmid was used to control for transfection efficiency. Shown are results with the positive control SV40 promoter and test ULBP1 promoters in pGL3-luciferase. As described in Materials and Methods, 24 h after transfection, cells were cultured with DMSO or with MG132 for an additional 13 h before lysis. The histogram plots show the relative increase in activity over DMSO-stimulated controls.
ATM/ATR signaling is neither necessary nor sufficient for high-level ULBP1 expression in HNSCC
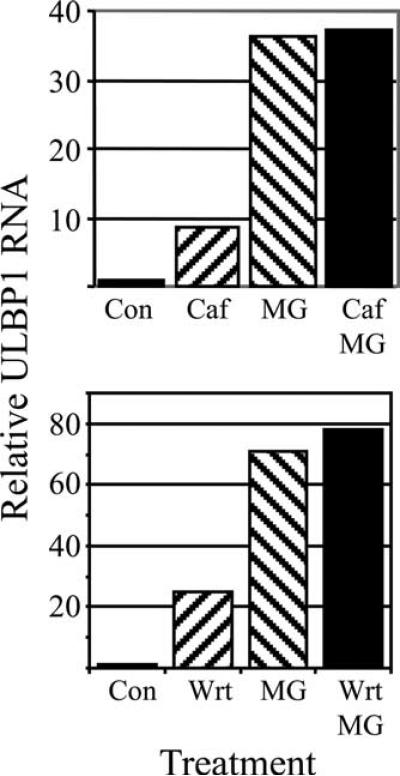
Gasser et al. demonstrated that NKG2D ligand expression was induced by ATM/ATR signaling in the DNA damage response pathway and that induction was prevented by ATM/ATR inhibitors, such as caffeine (47, 48). Therefore, we tested whether caffeine could prevent proteasome inhibitor drug-induced ULBP1 transcription. Surprisingly, caffeine reproducibly increased ULBP1 RNA levels (Fig. 9). Caffeine pretreatment failed to block MG132-induced ULBP1 up-regulation (Fig. 9). We also tested wortmannin at a concentration near its in vivo IC50 for ATM and well above its IC50 for other protein kinases (49). Like caffeine, wortmannin alone stimulated ULBP1 mRNA and did not blunt MG132-induced ULBP1 mRNA or cell surface protein expression (Fig. 9 and data not shown). These studies indicate that ATM/ATR signaling is not necessary for induction of high-level ULBP1 transcription by proteasome inhibition.
FIGURE 9.
The protein kinase inhibitors caffeine and wortmannin do not prevent MG132-induced ULBP1 expression. FaDu cells were treated with caffeine (Caf) or wortmannin (Wrt) for 7 h, with MG132 (MG) for 5 h, or with caffeine or wortmannin for 7 h, with the addition of MG132 during the final 5 h (Caf/MG and Wrt/MG). ULBP1 mRNA was measured and normalized to control DMSO-treated cells (Con). Shown are results that are typical of two independent experiments.
To investigate whether ATM/ATR activation was sufficient for ULBP1 induction in HNSCC cells, we treated FaDu cells with known ATM/ATR activators. Bleomycin induces DNA damage and activates ATM/ATR signaling, resulting in cell cycle arrest (50). As noted above, bleomycin and cisplatin had no effect on NKG2D ligand expression. To expand on this finding, FaDu cells were treated with bleomycin for 16 h, and the effectiveness of this treatment was tested by flow cytometry. Bleomycin treatment arrested FaDu cells in the G2/M phase of the cell cycle (Fig. 10A). Cell cycle arrest was nearly complete, indicating that FaDu cells were highly susceptible to the dose of bleomycin used. Bleomycin did not induce ULBP1 protein expression in the same experiment, although the cells responded to MG132 (Fig. 10B). We then tested other DNA damaging agents that had been employed previously to induce ATM/ATR signaling and NKG2D ligand up-regulation (47). Hydroxyurea and aphidicolin failed to change ULBP1 mRNA levels (Fig. 10C). Ionizing radiation is a classic DNA damaging treatment (51). Gasser et al. had demonstrated NKG2D ligand induction after 40 Gy of gamma radiation (47). We exposed FaDu cells to gamma radiation and measured ULBP1 RNA 5 h later. A dose of 1 Gy did not change ULBP1 RNA level, a dose of 10 Gy caused a <2-fold increase, and a dose of 40 Gy caused only a 4-fold increase in ULBP1 RNA (Fig. 10D). ULBP1 expression that was observed following high-dose ionizing radiation may have been secondary to non-ATM/ATR signaling that resulted from radiation-induced lipid peroxidation, damage to other cellular components, or cell death (52). Given that bleomycin, cisplatin, hydroxyurea, aphidicolin, and low-level ionizing radiation are known to activate ATM signaling, yet did not increase ULBP1 expression, we conclude that treatments sufficient to activate ATM are not sufficient to induce high-level ULBP1 transcription in FaDu HNSCC cells. ATR signaling also has been shown to induce NKG2D ligand expression (47). Therefore, we irradiated FaDu cells with UV light, a classic ATR activator (53). ULBP1 mRNA was not increased by UV treatment at 40, 60, 80, or 200 J/m2, although these doses have been shown to activate ATR (53). Higher doses (400−800 J/m2) increased ULBP1 mRNA ∼2-fold in one experiment and none in a second experiment (data not shown). As with high-dose gamma radiation, minimal induction of ULBP1 RNA only above 400 J/m2 UV irradiation may have been secondary to non-ATM/ATR signaling pathways. Failure to induce ULBP1 RNA at UV doses sufficient to activate ATR signaling provides evidence that ATR activation is not sufficient to induce high-level ULBP1 transcription in HNSCC cell lines (53). Our results indicate that ATM/ATR signaling is neither necessary nor sufficient for proteasome inhibitor drug induction of ULBP1 expression in FaDu HNSCC cells.
FIGURE 10.
ATM activating agents do not induce high-level ULBP1 expression. A and B, FaDu cells were cultured with media (top panel) or with 0.3 U/L bleomycin (bottom panel) for 16 h. A, Cells were permeabilized and DNA content was measured with propidium iodide staining and flow cytometry. DNA content is shown on a linear scale, and cell cycle stages G0/G1 (open histogram), S (hatched), and G2/M (cross-hatched) are indicated. B, Shown on a log scale is anti-ULBP1 mAb staining of FaDu cells treated with DMSO solvent alone (gray shading), bleomycin (thin line), or MG132 (thick line). C, FaDu cells were treated with aphidicolin (Aph) or hydroxyurea (HU) and cultured for 5 h before measurement of ULBP1 mRNA. Results are shown relative to the DMSO-treated control (Con). Show is a single experiment that is typical of at least two independent experiments. D, FaDu cells were given the indicated doses of gamma radiation, cultured for 5 h, and lysed. ULBP1 RNA was measured relative to GAPDH RNA and values were normalized to control nonirradiated cells. Shown are the means of two independent experiments and 95% confidence limits.
Ionizing radiation activates ATM kinase in HNSCC cells
Because many cancers, including HNSCC, have defective DNA repair mechanisms (54), we investigated whether the ATM kinase was functional in HNSCC. ATM autophosphorylation of Ser1981 is a key early signaling event in the DNA damage response cascade (51, 53, 55). We γ-irradiated FaDu and CAL27 HNSCC cells, lysed the cells 1.5 h later, and measured phospho-Ser1981 ATM in cell lysates by immunoblotting. ATM Ser1981 phosphorylation was induced by 10 and 40 Gy ionizing radiation (Fig. 11) and persisted at least 5 h (data not shown). Given that there was minimal induction of ULBP1 RNA at 10 Gy ionizing radiation, this result demonstrates that ATM activation is not sufficient to induce high-level ULBP1 transcription in FaDu HNSCC cells. Although we have not ruled out other defects in the DNA damage response pathway, this result strengthens the conclusion that ATM activation is neither necessary nor sufficient for the high-level ULBP1 transcription observed following proteasome inhibition in HNSCC cell lines.
FIGURE 11.
HNSCC phosphorylate ATM at Ser1981 following ionizing radiation. FaDu and CAL27 cells were treated with 0, 1, 10, and 40 Gy of gamma radiation and cultured for 1.5 h before lysis and immunoblotting. Top, ATM phospho-Ser1981 is detected. Bottom, The same membrane was stripped and reprobed with control Ab to β-actin. Results are typical of two independent experiments.
Discussion
In both experimental animals and human cancer patients, tumor NKG2D ligand expression is associated with tumor eradication and superior patient survival (5, 12-15, 26, 56). NKG2D recognizes ligands that often are expressed at higher levels on tumor cells than in the surrounding normal tissue and that can be induced further by cancer treatments. Therefore, effective cancer treatments might both directly damage tumor cells and induce NKG2D ligand expression and subsequent attack by killer lymphocytes. We investigated NKG2D ligand regulation in a little-studied tumor system, human HNSCC.
We found a common expression pattern of NKG2D ligands in HNSCC cell lines: MICA, MICB, ULBP2, and ULBP3 proteins were expressed at the cell surface in the absence of ULBP1 and ULBP4. The cell surface protein expression pattern reflected steady-state RNA levels, although ULBP1 mRNA was detected at very low levels. The NKG2D ligand expression pattern among HNSCC lines appeared to be more uniform than the pattern within leukemias, colorectal cancers, or gliomas (14, 24, 26, 57-60), which may indicate that immune editing of HNSCC is more uniform than in other tumors. Although ULBP1 and ULBP4 were poorly expressed by HNSCC, their expression is not inherently “weak”, as they are well expressed by cells from other tissues (26, 57, 60). We tested three drugs relevant to HNSCC treatment, bleomycin, cisplatin, and bortezomib. Only bortezomib had a substantial effect on NKG2D ligands, dramatically and specifically up-regulating ULBP1. Bortezomib is a novel proteasome inhibitor that has been approved for the treatment of plasma cell myeloma and is undergoing clinical trials for the treatment of several epithelial cancers, including HNSCC. To address the possibility that ULBP1 was induced by a proteasome-independent process, we tested a total of four drugs that inhibit proteasome activity via at least three mechanisms (42). All of the proteasome inhibitor drugs induced ULBP1 expression, effectively ruling out “off-target” effects. Although proteasome inhibitor drugs are known to increase the t½ of unstable mRNA (61), ULBP1 mRNA was stable in untreated cells, and any possible increase in ULBP1 mRNA t½ could have made only a minor contribution to the rapid and dramatic increase in ULBP1 mRNA. Drug treatment also induced nascent hnRNA, indicating that proteasome inhibition stimulated ULBP1 transcription. Consistent with this conclusion, proteasome inhibitor drug-sensitive elements were located to the 522-bp ULBP1 proximal promoter.
As this manuscript was being finalized, Armeanu et al. reported that low-dose proteasome inhibitor drugs caused hepatocellular carcinoma cell lines to selectively up-regulate MICA and MICB, but not ULBP1−3 (43). In contrast, Valés-Gómez et al. reported that several proteasome inhibitor drugs increased Jurkat T cell surface ULBP2 levels by <2-fold to <4-fold; cell surface MICA, MICB, ULBP1, ULBP3, or ULBP4 proteins did not change significantly (44). In response to MG132 treatment, Jurkat ULBP2 RNA increased, but it was not quantified. The MG132 effect varied, inducing ULBP2 on some tumor cells lines but not on others (44). The Raulet group recently reported that MG132 treatment of thymocytes increased Mult1 RNA 4-fold, relative to DMSO (62). In our study of nontransformed human keratinocytes, MG132 treatment increased RNA by 2- to 6-fold for all NKG2D ligands except ULBP4. In HNSCC, MG132 and other proteasome inhibitor drugs dramatically increased ULBP1 RNA and surface protein expression, but had minimal effects on MICA, MICB, and the other ULBP, although 2- to 3-fold changes were detected in ULBP2 and ULBP3 cell surface protein in a minority of experiments. To account for variation between cells, we propose that proteasome inhibitor drugs cannot further increase NKG2D ligand expression beyond a maximal range. We further propose that short-term proteasome inhibitor drug treatment cannot induce expression of NKG2D ligand genes that are silenced by DNA methylation and repressive histone proteins.
NKG2D ligands are induced by a wide variety of signals that have collectively been termed “cell stress” (1-3, 8, 47, 63). In many cases, cell stress affects the expression of NKG2D ligands differentially. Oxidative stress increased MICB and ULBP1 RNA, but not MICA or ULBP2−4 RNA, in human bronchial cells (57), but it increased both MICA and MICB in the CaCo-2 colon carcinoma cell line (64). Lytic phase EBV infection selectively up-regulated ULBP1, but not MICA, MICB, ULBP2, or ULBP3 (65). Likewise, Mycobacterium tuberculosis infection of human monocytes up-regulated cell surface ULBP1, but not ULBP2, ULBP3, or MICA/MICB (66). Both influenza A and Sendai virus infection of human macrophages were reported to up-regulate MICB RNA, but not ULBP RNA (67). In mouse macrophages, TLR stimulation or MCMV infection up-regulated Rae-1 family members, but not H60 (68, 69). Myc-driven carcinogenesis induced Rae-1ε, but not Mult1, on B lineage lymphomas (63). In contrast to their findings with other DNA damaging agents, the Raulet group reported that UV treatment decreased mouse fibroblast Mult1 RNA (47, 62). This occurred even while fibroblast Mult1 cell surface protein increased via a posttranslational mechanism (62). UV treatment of thymocytes did not increase Mult1 expression (62). Therefore, cell stresses differentially induce NKG2D ligands, often in a tissue-dependent fashion.
To account for the panoply of NKG2D ligand-inducing cell stresses, Gasser et al. proposed the unifying hypothesis that NKG2D ligand expression is induced by ATM/ATR signaling (47). ATM/ATR signaling is exquisitely sensitive to DNA damage and altered chromatin configuration (55). Therefore, we explored whether ULBP1 induction by proteasome inhibitor drug treatment of HNSCC cells could be explained by the ATM/ATR hypothesis. In contrast to the dramatic effect of proteasome inhibitor drugs in HNSCC cell lines, several DNA damaging treatments had little or no effect on ULBP1 expression. For example, bleomycin, which induces DNA damage and activates ATM (50), caused nearly complete cell cycle arrest, but did not change NKG2D ligand expression. Several doses of ionizing radiation and UV radiation that are sufficient to activate ATM or ATR (51-53) had little or no effect on ULBP1 RNA levels. This is consistent with the recent findings that DNA damage did not induce NKG2D ligand expression in Myc-driven mouse prelymphoma cells (63) and that UV treatment decreased Mult1 RNA in fibroblasts (62). Therefore, classic ATM/ATR activating agents did not increase NKG2D ligand expression. Because DNA damage response pathways are defective in many cancers (54), we investigated ATM activation. ATM Ser1981 autophosphorylation is a key early signaling event in the DNA damage response cascade (51, 53, 55). We found that ATM was phosphorylated at Ser1981 in two HNSCC cell lines in response to ionizing radiation, including a dose that produced little change in ULBP1 expression. We also blocked ATM and ATR activity with wortmannin and caffeine. Pretreatment of cells with either kinase inhibitor failed to block ULBP1 induction by proteasome inhibitor drugs. Indeed, the kinase inhibitors alone caused a small increase in ULBP1 mRNA. Collectively, these results indicate that ATM/ATR signaling was neither necessary nor sufficient to induce high-level NKG2D ligand expression in HNSCC cells. This result is not surprising, because proteasome inhibitor drugs lead to ATM degradation via a caspase-mediated process (70). Although the ATM/ATR hypothesis explains how seemingly unrelated treatments increase NKG2D ligand expression, this hypothesis cannot explain the extensive heterogeneity of NKG2D ligand expression within and between tumor types (14, 24, 26, 57-60). The relatively low homology between transcription control regions of the seven human NKG2D ligand genes makes it probable that multiple signaling mechanisms differentially control NKG2D ligand gene expression (10).
How can we reconcile the current findings with the ATM hypothesis? ATM directly or indirectly leads to the phosphorylation of a myriad of downstream targets (50). It also may be relevant that ATM activation is necessary, but not sufficient, to respond to DNA damage (55). We postulate that NKG2D ligand gene transcription is induced by a combination of one or more of the ATM/ATR downstream transcription factors acting in concert with transcription factors that are activated by non-ATM/ATR signaling. This proposed multiplicity of transcription factors plus the known sequence diversity in NKG2D ligand gene promoters can explain why NKG2D ligand gene expression is heterogeneous in cancer cells and why many cell stresses differentially induce NKG2D ligand expression. It was surprising that increased NKG2D ligand expression in fibroblasts and in oncogene-transformed p53-deficient cell lines apparently required 40 Gy of ionizing radiation (47), because ATM is activated in fibroblasts by as little as 0.5 Gy of gamma radiation (51). However, high-dose ionizing radiation induces non-ATM signaling that is secondary to lipid peroxidation, damage to other cellular components, and cell death (52). The apparent requirement for high-dose ionizing radiation to induce NKG2D ligand expression (47) may be explained by an additional requirement for ATM/ATR-independent signaling.
Only three transcription factors are known to be consistently induced by clinically relevant doses of ionizing radiation (≤2 Gy): NF-κB, Sp1 family members, and p53 (52). ATM activates NF-κB, which has been implicated in MICA expression in human T lymphocytes (48, 71). Although NF-κB may be relevant for NKG2D ligand induction in T lymphocytes, it does not readily account for the ability of proteasome inhibitor drugs to induce ULBP1 transcription in HNSCC. This is because proteasome inhibitor drugs block both the classical and alternative NF-κB signaling pathways (72).
Sp1 family members (also referred to as the SP1-related retinoblastoma control proteins) are up-regulated by low-dose ionizing radiation (52). Sp1 family members have been implicated in ULBP1, MICA, and MICB transcription (46, 73). López-Soto et al. proposed that transcription is controlled by competition between stimulatory Sp3 and repressive AP-2α transcription factors at a cAMP response element (CRE)-like site in the ULBP1 promoter (46). However, this proposal cannot explain our observations. Together with other findings, the López-Soto et al. study implies that low-dose ionizing radiation is predicted to up-regulate Sp1 family members and compete with repressive AP-2α (46, 50). Contrary to this prediction, low-dose ionizing radiation induced little or no ULBP1 in HNSCC cells. AP-2α undergoes proteasome-mediated degradation, and proteasome inhibitor drugs would be predicted to increase repressive AP-2α stability and inhibit NKG2D ligand expression (46, 74). Contrary to this prediction, we found that proteasome inhibitor drugs increased ULBP1 transcription in HNSCC cells.
Heat shock induces MICA and MICB expression in proliferating and quiescent carcinoma cells, acting via HSF1 transcription factor binding to MICA and MICB promoters (59, 73). Heat shock did not induce ULBP1 expression in HNSCC cells (data not shown). However, several potential heat shock elements can be predicted from the ULBP1 promoter sequence, including in the 522-bp proximal fragment (data not shown). Proteasome inhibitor drugs increase protein stability of HSF2, a transcription factor that recognizes the same sequence motifs as HSF1 (75). Therefore, it is possible that proteasome inhibitor drugs induce ULBP1 transcription via a heat shock-like response.
The p53 transcription factor is well known to be up-regulated by low-dose ionizing radiation (50, 52). Gasser et al. found that NKG2D ligands could be up-regulated in p53 knockout cells, but they did not rule out a role for p53 family members, p63 and p73, that also are targets of ATM/ATR signaling and MDM2 regulation (47, 76). p63 and p73 play prominent roles in HNSCC carcinogenesis (77). In HNSCC cell lines, ULBP1 was up-regulated both by proteasome inhibition (this paper) and by MDM2 inhibition (data not shown). Taken together, these observations support the hypothesis that p53 family members are important in proteasome inhibitor drug-induced ULBP1 up-regulation in HNSCC.
A major contribution of the ATM/ATR hypothesis was that it suggested how diverse stimuli could up-regulate NKG2D ligands in infection, inflammation, and cancer (47). However, it would be simplistic to conclude that a single signaling pathway accounts for the complex regulation of eight MIC, ULBP, and RAET genes with diverse promoter sequences (10). The number and diversity of NKG2D ligands in both mice and humans suggest that these molecules play distinct roles. ULBP1 appears to be selectively down-regulated in many epithelial malignancies studied, especially in HNSCC, suggesting a potential unique role for this NKG2D ligand. ULBP1 transcription control also may be unique, given that potential BNC, MZF1, and ATF6 motifs that are found in most other ULBP/RAET promoters are not shared by ULBP1 (10). Distinct biological roles require differential regulation of gene expression. This is a worthy subject of future investigation.
Acknowledgments
We thank Jeffrey Meyers, Teresa Whiteside, Jane McCutcheon, Miguel Lopez-Botet, Rob Eagle, John Trowsdale, Alexander Steinle, Lewis Lanier, Cam Dingle, and Hollie Swanson for providing cells, mAbs, and plasmids. Rob Eagle and John Trowsdale shared unpublished results. We thank Maria Bruno, Charlotte Kaetzel, Alan Kaplan, William St. Clair, and Daret St. Clair for scientific discussions. Fred P. Rawlings, Xiuqin Li, Tiffany Griggs, Maria Bruno, Charlotte Kaetzel, Brett Spear, John D'Orazio, and Jeffrey Ebersole provided technical advice and assistance, and Diana Howard provided bortezomib. We thank the Immunex/Amgen Hybridoma group for their help in generating the anti-NKG2D ligand mAb.
This work was supported by National Institutes of Health Grants DE11139 and AI050656 and the Kentucky Lung Cancer Research Program. J.E.B. was supported by National Institutes of Health Grant T32-CA09509.
Footnotes
Abbreviations used in this paper: MICA, MHC class I chain-related gene A; ATM, ataxia-telangiectasia, mutated; ATR, ATM and Rad3-related; hnRNA, heteronuclear (unspliced) RNA; HNSCC, head and neck squamous cell carcinoma; MICB, MHC class I chain-related gene B; RAET, retinoic acid early transcript; ULBP, UL16-binding protein.
Disclosures
N. Jan Chalupny is employed by Amgen, which provided mAb used in this research. Amgen does not market the mAb used. The other authors have no financial conflicts of interest.
References
- 1.Long EO, Rajagopalan S. Stress signals activate natural killer cells. J. Exp. Med. 2002;196:1399–1402. doi: 10.1084/jem.20021747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat. Rev. Immunol. 2003;3:781–790. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 3.Gleimer M, Parham P. Stress management: MHC class I and class I-like molecules as reporters of cellular stress. Immunity. 2003;19:469–477. doi: 10.1016/s1074-7613(03)00272-3. [DOI] [PubMed] [Google Scholar]
- 4.Upshaw JL, Leibson PJ. NKG2D-mediated activation of cytotoxic lymphocytes: unique signaling pathways and distinct functional outcomes. Semin. Immunol. 2006;18:167–175. doi: 10.1016/j.smim.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Guerra N, Tan YX, Joncker NT, Choy A, Gallardo F, Xiong N, Knoblaugh S, Cado D, Greenberg NR, Raulet DH. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 2008;28:571–580. doi: 10.1016/j.immuni.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eagle RA, Trowsdale J. Promiscuity and the single receptor: NKG2D. Nat. Rev. Immunol. 2007;7:737–744. doi: 10.1038/nri2144. [DOI] [PubMed] [Google Scholar]
- 7.Mistry AR, O'Callaghan CA. Regulation of ligands for the activating receptor NKG2D. Immunology. 2007;121:439–447. doi: 10.1111/j.1365-2567.2007.02652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schrambach S, Ardizzone M, Leymarie V, Sibilia J, Bahram S. In vivo expression pattern of MICA and MICB and its relevance to auto-immunity and cancer. PLoS ONE. 2007;2:e518. doi: 10.1371/journal.pone.0000518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McFarland BJ, Kortemme T, Yu SF, Baker D, Strong RK. Symmetry recognizing asymmetry: analysis of the interactions between the C-type lectin-like immunoreceptor NKG2D and MHC class I-like ligands. Structure. 2003;11:411–422. doi: 10.1016/s0969-2126(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 10.Eagle RA, Traherne JA, Ashiru O, Wills MR, Trowsdale J. Regulation of NKG2D ligand gene expression. Hum. Immunol. 2006;67:159–169. doi: 10.1016/j.humimm.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 11.Kriegeskorte AK, Gebhardt FE, Porcellini S, Schiemann M, Stemberger C, Franz TJ, Huster KM, Carayannopoulos LN, Yokoyama WM, Colonna M, et al. NKG2D-independent suppression of T cell proliferation by H60 and MICA. Proc. Natl. Acad. Sci. USA. 2005;102:11805–11810. doi: 10.1073/pnas.0502026102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerwenka A, Baron JL, Lanier LL. Ectopic expression of retinoic acid early inducible-1 gene (RAE-1) permits natural killer cell-mediated rejection of a MHC class I-bearing tumor in vivo. Proc. Natl. Acad. Sci. USA. 2001;98:11521–11526. doi: 10.1073/pnas.201238598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diefenbach A, Jensen ER, Jamieson AM, Raulet DH. Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature. 2001;413:165–171. doi: 10.1038/35093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friese MA, Platten M, Lutz SZ, Naumann U, Aulwurm S, Bischof F, Buhring HJ, Dichgans J, Rammensee HG, Steinle A, Weller M. MICA/NKG2D-mediated immunogene therapy of experimental gliomas. Cancer Res. 2003;63:8996–9006. [PubMed] [Google Scholar]
- 15.Smyth MJ, Swann J, Kelly JM, Cretney E, Yokoyama WM, Diefenbach A, Sayers TJ, Hayakawa Y. NKG2D recognition and perforin effector function mediate effective cytokine immunotherapy of cancer. J. Exp. Med. 2004;200:1325–1335. doi: 10.1084/jem.20041522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. 2002;419:734–738. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- 17.Salih HR, Antropius H, Gieseke F, Lutz SZ, Kanz L, Rammensee HG, Steinle A. Functional expression and release of ligands for the activating immunoreceptor NKG2D in leukemia. Blood. 2003;102:1389–1396. doi: 10.1182/blood-2003-01-0019. [DOI] [PubMed] [Google Scholar]
- 18.Lee J-C, Lee K-M, Kim D-W, Heo DS. Elevated TGF-β1 secretion and down-modulation of NKG2D underlies impaired NK cytotoxicity in cancer patients. J. Immunol. 2004;172:7335–7340. doi: 10.4049/jimmunol.172.12.7335. [DOI] [PubMed] [Google Scholar]
- 19.Oppenheim DE, Roberts SJ, Clarke SL, Filler R, Lewis JM, Tigelaar RE, Girardi M, Hayday AC. Sustained localized expression of ligand for the activating NKG2D receptor impairs natural cytotoxicity in vivo and reduces tumor immunosurveillance. Nat. Immunol. 2005;6:928–937. doi: 10.1038/ni1239. [DOI] [PubMed] [Google Scholar]
- 20.Vetter CS, Lieb W, Brocker E-B, Becker JC. Loss of nonclassical MHC molecules MIC-A/B expression during progression of uveal melanoma. Br. J. Cancer. 2004;91:1495–1499. doi: 10.1038/sj.bjc.6602123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madjd Z, Spendlove I, Moss R, Bevin S, Pinder SE, Watson NFS, Ellis I, Durrant LG. Upregulation of MICA on high-grade invasive operable breast carcinoma. Cancer Immun. 2007;7:17. [PMC free article] [PubMed] [Google Scholar]
- 22.Pende D, Cantoni C, Rivera P, Vitale M, Castriconi R, Marcenaro S, Nanni M, Biassoni R, Bottino C, Moretta A, Moretta L. Role of NKG2D in tumor cell lysis mediated by human NK cells: cooperation with natural cytotoxicity receptors and capability of recognizing tumors of nonepithelial origin. Eur. J. Immunol. 2001;31:1076–1086. doi: 10.1002/1521-4141(200104)31:4<1076::aid-immu1076>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 23.Vetter CS, Groh V, Straten PT, Spies T, Bröcker E-B, Becker JC. Expression of stress-induced MHC class I related chain molecules on human melanoma. J. Invest. Dermatol. 2002;118:600–605. doi: 10.1046/j.1523-1747.2002.01700.x. [DOI] [PubMed] [Google Scholar]
- 24.Pende D, Rivera P, Marcenaro S, Chang CC, Biassoni R, Conte R, Kubin M, Cosman D, Ferrone S, Moretta L, Moretta A. Major histocompatibility complex class I-related chain A and UL16-binding protein expression on tumor cell lines of different histotypes: analysis of tumor susceptibility to NKG2D-dependent natural killer cell cytotoxicity. Cancer Res. 2002;62:6178–6186. [PubMed] [Google Scholar]
- 25.Maccalli C, Pende D, Castelli C, Mingari MC, Robbins PF, Parmiani G. NKG2D engagement of colorectal cancer-specific T cells strengthens TCR-mediated antigen stimulation and elicits TCR independent anti-tumor activity. Eur. J. Immunol. 2003;33:2033–2043. doi: 10.1002/eji.200323909. [DOI] [PubMed] [Google Scholar]
- 26.Conejo-Garcia JR, Benencia F, Courreges MC, Gimotty PA, Khang E, Buckanovich RJ, Frauwirth KA, Zhang L, Katsaros D, Thompson CB, et al. Ovarian carcinoma expresses the NKG2D ligand Letal and promotes the survival and expansion of CD28− antitumor T cells. Cancer Res. 2004;64:2175–2182. doi: 10.1158/0008-5472.can-03-2194. [DOI] [PubMed] [Google Scholar]
- 27.Wrobel P, Shojaei H, Schittek B, Gieseler F, Wollenberg B, Kalthoff H, Kabelitz D, Wesch D. Lysis of a broad range of epithelial tumour cells by human γδ T cells: involvement of NKG2D ligands and T-cell receptor-versus NKG2D-dependent recognition. Scand. J. Immunol. 2007;66:320–328. doi: 10.1111/j.1365-3083.2007.01963.x. [DOI] [PubMed] [Google Scholar]
- 28.Gustin DM. Chemoprevention of head and neck cancer. Semin. Oncol. 2004;31:769–777. doi: 10.1053/j.seminoncol.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Liu C-J, Lui M-T, Chen H-L, Lin S-C, Chang K-W. MICA and MICB overexpression in oral squamous cell carcinoma. J. Oral Pathol. Med. 2007;36:43–47. doi: 10.1111/j.1600-0714.2006.00471.x. [DOI] [PubMed] [Google Scholar]
- 30.Reinders J, Rozemuller EH, van der Weide P, Oka A, Slootweg PJ, Inoko H, Tilanus MGJ. Genes in the HLA region indicative for head and neck squamous cell carcinoma. Mol. Immunol. 2007;44:848–855. doi: 10.1016/j.molimm.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Li D, Ronson B, Guo M, Liu S, Bishop JS, Van Echo DA, O'Malley BW., Jr. Interleukin 2 gene transfer prevents NKG2D suppression and enhances antitumor efficacy in combination with cisplatin for head and neck squamous cell cancer. Cancer Res. 2002;62:4023–4028. [PubMed] [Google Scholar]
- 32.Cosman D, Müllberg J, Sutherland CL, Chin W, Armitage R, Fanslow W, Kubin M, Chalupny NJ. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14:123–133. doi: 10.1016/s1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- 33.Chalupny NJ, Sutherland CL, Lawrence WA, Rein Weston A, Cosman D. ULBP4 is a novel ligand for human NKG2D. Biochem. Biophys. Res. Commun. 2003;305:129–135. doi: 10.1016/s0006-291x(03)00714-9. [DOI] [PubMed] [Google Scholar]
- 34.Darzynkiewicz Z, Huang X. Analysis of cellular DNA content by flow cytometry. In: Coligan JE, Kruisbeek AM, Margulies DH, Shevach EM, Strober W, editors. Current Protocols in Immunology. Wiley; New York: 2004. pp. 5.7.1–5.7.18. [Google Scholar]
- 35.Chan HW, Miller JS, Moore MB, Lutz CT. Epigenetic control of highly homologous killer Ig-like receptor gene alleles. J. Immunol. 2005;175:5966–5974. doi: 10.4049/jimmunol.175.9.5966. [DOI] [PubMed] [Google Scholar]
- 36.Bacon L, Eagle RA, Meyer M, Easom N, Young NT, Trowsdale J. Two human ULBP/RAET1 molecules with transmembrane regions are ligands for NKG2D. J. Immunol. 2004;173:1078–1084. doi: 10.4049/jimmunol.173.2.1078. [DOI] [PubMed] [Google Scholar]
- 37.Presnell SR, Zhang L, Ramilo CA, Chan H-W, Lutz CT. Functional redundancy of transcription factor-binding sites in the killer cell Ig-like receptor (KIR) gene promoter. Int. Immunol. 2006;18:1221–1232. doi: 10.1093/intimm/dxl043. [DOI] [PubMed] [Google Scholar]
- 38.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Lab. Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 40.Forastiere AA, Goepfert H, Maor M, Pajak TF, Weber R, Morrison W, Glisson B, Trotti A, Ridge JA, Chao C, et al. Concurrent chemotherapy and radiotherapy for organ preservation in advanced laryngeal cancer. N. Engl. J. Med. 2003;349:2091–2098. doi: 10.1056/NEJMoa031317. [DOI] [PubMed] [Google Scholar]
- 41.Adema AD, Cloos J, Verheijen RH, Braakhuis BJ, Bryant PE. Comparison of bleomycin and radiation in the G2 assay of chromatid breaks. Int. J. Radiat. Biol. 2003;79:655–661. doi: 10.1080/09553000310001596968. [DOI] [PubMed] [Google Scholar]
- 42.Myung J, Kim KB, Crews CM. The ubiquitin-proteasome pathway and proteasome inhibitors. Med. Res. Rev. 2001;21:245–273. doi: 10.1002/med.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Armeanu S, Krusch M, Baltz KM, Weiss TS, Smirnow I, Steinle A, Lauer UM, Bitzer M, Salih HR. Direct and natural killer cell-mediated antitumor effects of low-dose bortezomib in hepatocellular carcinoma. Clin. Cancer Res. 2008;14:3520–3528. doi: 10.1158/1078-0432.CCR-07-4744. [DOI] [PubMed] [Google Scholar]
- 44.Valés-Gómez M, Chisholm SE, Cassady-Cain RL, Roda-Navarro P, Reyburn HT. Selective induction of expression of a ligand for the NKG2D receptor by proteasome inhibitors. Cancer Res. 2008;68:1546–1554. doi: 10.1158/0008-5472.CAN-07-2973. [DOI] [PubMed] [Google Scholar]
- 45.Bruno MEC, Kaetzel CS. Long-term exposure of the HT-29 human intestinal epithelial cell line to TNF causes sustained up-regulation of the polymeric Ig receptor and proinflammatory genes through transcriptional and posttranscriptional mechanisms. J. Immunol. 2005;174:7278–7284. doi: 10.4049/jimmunol.174.11.7278. [DOI] [PubMed] [Google Scholar]
- 46.López-Soto A, Quinones-Lombrana A, López-Arbesú R, López-Larrea C, González S. Transcriptional regulation of ULBP1, a human ligand of the NKG2D receptor. J. Biol. Chem. 2006;281:30419–30430. doi: 10.1074/jbc.M604868200. [DOI] [PubMed] [Google Scholar]
- 47.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cerboni C, Zingoni A, Cippitelli M, Piccoli M, Frati L, Santoni A. Antigen-activated human T lymphocytes express cell-surface NKG2D ligands via an ATM/ATR-dependent mechanism and become susceptible to autologous NK-cell lysis. Blood. 2007;110:606–615. doi: 10.1182/blood-2006-10-052720. [DOI] [PubMed] [Google Scholar]
- 49.Sarkaria JN, Tibbetts RS, Busby EC, Kennedy AP, Hill DE, Abraham RT. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 1998;58:4375–4382. [PubMed] [Google Scholar]
- 50.Kurz EU, Lees-Miller SP. DNA damage-induced activation of ATM and ATM-dependent signaling pathways. DNA Repair (Amst.) 2004;3:889–900. doi: 10.1016/j.dnarep.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 51.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 52.Criswell T, Leskov K, Miyamoto S, Luo G, Boothman DA. Transcription factors activated in mammalian cells after clinically relevant doses of ionizing radiation. Oncogene. 2003;22:5813–5827. doi: 10.1038/sj.onc.1206680. [DOI] [PubMed] [Google Scholar]
- 53.Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh S-Y, Taya Y, Prives C, Abraham RT. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bolt J, Vo QN, Kim W-J, McWhorter AJ, Thomson J, Hagensee ME, Friedlander P, Brown KD, Gilbert J. The ATM/p53 pathway is commonly targeted for inactivation in squamous cell carcinoma of the head and neck (SCCHN) by multiple molecular mechanisms. Oral Oncol. 2005;41:1013–1020. doi: 10.1016/j.oraloncology.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 55.McGowan CH, Russell P. The DNA damage response: sensing and signaling. Curr. Opin. Cell Biol. 2004;16:629–633. doi: 10.1016/j.ceb.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 56.Friese MA, Wischhusen J, Wick W, Weiler M, Eisele G, Steinle A, Weller M. RNA interference targeting transforming growth factor-β enhances NKG2D-mediated antiglioma immune response, inhibits glioma cell migration and invasiveness, and abrogates tumorigenicity in vivo. Cancer Res. 2004;64:7596–7603. doi: 10.1158/0008-5472.CAN-04-1627. [DOI] [PubMed] [Google Scholar]
- 57.Borchers MT, Harris NL, Wesselkamper SC, Vitucci M, Cosman D. NKG2D ligands are expressed on stressed human airway epithelial cells. Am. J. Physiol. 2006;291:L222–L231. doi: 10.1152/ajplung.00327.2005. [DOI] [PubMed] [Google Scholar]
- 58.Groh V, Rhinehart R, Secrist H, Bauer S, Grabstein KH, Spies T. Broad tumor-associated expression and recognition by tumor-derived γδ T cells of MICA and MICB. Proc. Natl. Acad. Sci. USA. 1999;96:6879–6884. doi: 10.1073/pnas.96.12.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Groh V, Steinle A, Bauer S, Spies T. Recognition of stress-induced MHC molecules by intestinal epithelial γδ T cells. Science. 1998;279:1737–1740. doi: 10.1126/science.279.5357.1737. [DOI] [PubMed] [Google Scholar]
- 60.Nowbakht P, Ionescu M-CS, Rohner A, Kalberer CP, Rossy E, Mori L, Cosman D, De Libero G, Wodnar-Filipowicz A. Ligands for natural killer cell-activating receptors are expressed upon the maturation of normal myelomonocytic cells but at low levels in acute myeloid leukemias. Blood. 2005;105:3615–3622. doi: 10.1182/blood-2004-07-2585. [DOI] [PubMed] [Google Scholar]
- 61.Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 62.Nice TJ, Coscoy L, Raulet DH. Posttranslational regulation of the NKG2D ligand Mult1 in response to cell stress. J. Exp. Med. 2009;206:287–298. doi: 10.1084/jem.20081335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Unni AM, Bondar T, Medzhitov R. Intrinsic sensor of oncogenic transformation induces a signal for innate immunosurveillance. Proc. Natl. Acad. Sci. USA. 2008;105:1686–1691. doi: 10.1073/pnas.0701675105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamamoto K, Fujiyama Y, Andoh A, Bamba T, Okabe H. Oxidative stress increases MICA and MICB gene expression in the human colon carcinoma cell line (CaCo-2). Biochim. Biophys. Acta. 2001;1526:10–12. doi: 10.1016/s0304-4165(01)00099-x. [DOI] [PubMed] [Google Scholar]
- 65.Pappworth IY, Wang EC, Rowe M. The switch from latent to productive infection in Epstein-Barr virus-infected B cells is associated with sensitization to NK cell killing. J. Virol. 2007;81:474–482. doi: 10.1128/JVI.01777-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vankayalapati R, Garg A, Porgador A, Griffith DE, Klucar P, Safi H, Girard WM, Cosman D, Spies T, Barnes PF. Role of NK cell-activating receptors and their ligands in the lysis of mononuclear phagocytes infected with an intracellular bacterium. J. Immunol. 2005;175:4611–4617. doi: 10.4049/jimmunol.175.7.4611. [DOI] [PubMed] [Google Scholar]
- 67.Sirén J, Sareneva T, Pirhonen J, Strengell M, Veckman V, Julkunen I, Matikainen S. Cytokine and contact-dependent activation of natural killer cells by influenza A or Sendai virus-infected macrophages. J. Gen. Virol. 2004;85:2357–2364. doi: 10.1099/vir.0.80105-0. [DOI] [PubMed] [Google Scholar]
- 68.Lodoen M, Ogasawara K, Hamerman JA, Arase H, Houchins JP, Mocarski ES, Lanier LL. NKG2D-mediated natural killer cell protection against cytomegalovirus is impaired by viral gp40 modulation of retinoic acid early inducible 1 gene molecules. J. Exp. Med. 2003;197:1245–1253. doi: 10.1084/jem.20021973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hamerman JA, Ogasawara K, Lanier LL. Cutting edge: Toll-like receptor signaling in macrophages induces ligands for the NKG2D receptor. J. Immunol. 2004;172:2001–2005. doi: 10.4049/jimmunol.172.4.2001. [DOI] [PubMed] [Google Scholar]
- 70.Hideshima T, Mitsiades C, Akiyama M, Hayashi T, Chauhan D, Richardson P, Schlossman R, Podar K, Munshi NC, Mitsiades N, Anderson KC. Molecular mechanisms mediating antimyeloma activity of proteasome inhibitor PS-341. Blood. 2003;101:1530–1534. doi: 10.1182/blood-2002-08-2543. [DOI] [PubMed] [Google Scholar]
- 71.Molinero LL, Fuertes MB, Girart MV, Fainboim L, Rabinovich GA, Costas MA, Zwirner NW. NF-κB regulates expression of the MHC class I-related chain A gene in activated T lymphocytes. J. Immunol. 2004;173:5583–5590. doi: 10.4049/jimmunol.173.9.5583. [DOI] [PubMed] [Google Scholar]
- 72.Perkins ND. Integrating cell-signalling pathways with NF-κB and IKK function. Nat. Rev. Mol. Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 73.Venkataraman GM, Suciu D, Groh V, Boss JM, Spies T. Promoter region architecture and transcriptional regulation of the genes for the MHC class I-related chain A and B ligands of NKG2D. J. Immunol. 2007;178:961–969. doi: 10.4049/jimmunol.178.2.961. [DOI] [PubMed] [Google Scholar]
- 74.Li M, Wang Y, Hung M-C, Kannan P. Inefficient proteasomal-degradation pathway stabilizes AP-2α and activates HER-2/neu gene in breast cancer. Int. J. Cancer. 2006;118:802–811. doi: 10.1002/ijc.21426. [DOI] [PubMed] [Google Scholar]
- 75.Mathew A, Mathur SK, Morimoto RI. Heat shock response and protein degradation: regulation of HSF2 by the ubiquitin-proteasome pathway. Mol. Cell. Biol. 1998;18:5091–5098. doi: 10.1128/mcb.18.9.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murray-Zmijewski F, Lane DP, Bourdon J-C. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13:962–972. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- 77.DeYoung MP, Johannessen CM, Leong C-O, Faquin W, Rocco JW, Ellisen LW. Tumor-specific p73 up-regulation mediates p63 dependence in squamous cell carcinoma. Cancer Res. 2006;66:9362–9368. doi: 10.1158/0008-5472.CAN-06-1619. [DOI] [PubMed] [Google Scholar]