Abstract
Adult teleost fish and urodele amphibians possess a spectacular ability to regenerate amputated appendages, based on formation and maintenance of progenitor tissue called a blastema. While injury-induced, or facultative, appendage regeneration has been studied extensively, the extent to which homeostatic regeneration maintains these structures has not been examined. Here, we found that transgenic inhibition of Fgf receptors in uninjured zebrafish caused severe atrophy of all fin types within two months, revealing a requirement for Fgfs to preserve dermal bone, joint structures, and supporting tissues. Appendage maintenance involved low-level expression of markers of blastema-based regeneration, focused in distal structures displaying recurrent cell death and proliferation. Conditional mutations in the ligand Fgf20a and the kinase Mps1, factors critical for regeneration of amputated fins, also caused rapid, progressive loss of fin structures in otherwise uninjured animals. Our experiments reveal that the facultative machinery that regenerates amputated teleost fins also has a surprisingly vigorous role in homeostatic regeneration.
INTRODUCTION
The capacity for organ regeneration is remarkably elevated in certain non-mammalian vertebrates like urodele amphibians and teleost fish. Together, these species regenerate amputated appendages and jaws, resected heart muscle, depleted sensory hair cells, damaged retinae and brain, dissected lenses, transected spinal cord, and portions of intestine. The teleost zebrafish represents a unique example of a highly regenerative vertebrate model system that is amenable to genetic approaches, making it well-suited to illuminate how complex tissue regeneration occurs at the molecular level. One of the most spectacular examples of regeneration in adult zebrafish is their rapid and virtually indelible renewal of amputated fins, complex tissues comprised of segmented bone, connective tissue, blood vessels, nerves, and epidermis. Fin regeneration is divided into three stages – wound healing, blastema formation, and regenerative outgrowth (Akimenko et al., 2003; Poss et al., 2003; Stoick-Cooper et al., 2007a). First, the amputation injury is healed through migration of epidermal cells, covering the wound within an hour of trauma. Formation of the regeneration blastema, a mass of undifferentiated, proliferative mesenchymal cells, is the second and defining stage of regeneration. Here, presumptive blastemal cells are stimulated to disorganize, migrate distally, and accumulate within 1–2 days of injury. It is currently unclear whether the blastema consists of a homogeneous population of multipotent progenitor cells, or heterogeneous subpopulations. During the final stage called regenerative outgrowth, a repeated series of proliferation, patterning, and differentiation events in blastemal tissue leads to deposition of new bone-secreting scleroblasts and distal addition of all lost bone segments within ~2 weeks.
Multiple studies in recent years have identified key molecular regulators of blastemal formation and function. Several different approaches revealed that signaling by Fibroblast growth factors (Fgfs) is critical for regeneration (Lee et al., 2005; Poss et al., 2000; Tawk et al., 2002; Thummel et al., 2006; Whitehead et al., 2005). The ligand fgf20a, found in a genetic screen for temperature-sensitive regeneration mutants, is required for normal morphogenesis of the regeneration epidermis and for mesenchymal proliferation during blastema formation (Whitehead et al., 2005). During regenerative outgrowth, the Fgf receptor (Fgfr) fgfr1, as well as Fgf target genes mkp3, sef, and spry4, are expressed in blastemal mesenchyme and in the surrounding basal epidermal layer. As regeneration proceeds, Fgf signaling tightly controls the amount of blastemal proliferation and the rate of growth, resulting in different regenerative rates dependent on the proximodistal level of amputation (Lee et al., 2005; Poss et al., 2000). Other studies have shown that a suite of signaling molecules such as Sonic hedgehog (Shh), Bone morphogenetic proteins, Activin-betaA, and canonical and noncanonical Wnts, influence blastemal proliferation and patterning during regenerative outgrowth (Jazwinska et al., 2007; Laforest et al., 1998; Quint et al., 2002; Smith et al., 2006; Stoick-Cooper et al., 2007b).
Vertebrate organs generally exhibit two forms of regeneration: facultative and homeostatic. Facultative regeneration describes mechanisms that are activated by stimuli like amputation or chemical injury; following the initial trauma, progenitor and/or structural cells near the injury site proliferate to replace dead or lost tissue. By contrast, homeostatic regeneration refers to regular replacement of cells lost through apoptosis, daily wear, and aging (Jones and Wagers, 2008). Interestingly, surveys of regenerative capacity among mammalian organs have found that the capacity of an organ for facultative regeneration often correlates positively with its baseline level of cell turnover (Rando, 2006). For instance, blood and skin undergo frequent cell loss and replacement through the activity of self-renewing stem cells, and utilize similar processes to quickly regenerate after injury (Blanpain and Fuchs, 2006; Scadden, 2006). Conversely, the mammalian brain and heart possess low levels of cellular turnover, and, despite evidence for resident stem cells (Alvarez-Buylla and Lim, 2004; Beltrami et al., 2003; Laugwitz et al., 2005), there is little or no regeneration after major injury. Furthermore, facultative regenerative capacity in mammalian organs tends to decrease with age, a phenomenon observed in concert with age-dependent reductions in the frequency of homeostatic structural cell or progenitor cell proliferation (Janzen et al., 2006; Krishnamurthy et al., 2006; Molofsky et al., 2006). These correlations indicate that common cellular and molecular mechanisms are responsible for recurrent cell replacement and injury-induced regeneration in many tissues. However, neither the vigor nor mechanisms of homeostatic regeneration have been examined in complex tissues that regenerate through a blastema-based mechanism, such as urodele or teleost appendages.
In this study, we tested the idea that the molecular pathways that control blastema formation and function during regeneration in amputated zebrafish fins have additional homeostatic functions in the absence of injury. We found that long-term inhibition of Fgfrs in uninjured zebrafish led to the progressive loss of distal fin structures, revealing homeostatic maintenance of fins by this pathway. Homeostatic regeneration was characterized by low-level expression of several mediators of facultative regeneration, including shh, msxb, and the Fgf target gene mkp3, in areas of cell proliferation and apoptosis. Using a conditional mutant strain, we found that Fgf-dependent homeostatic regeneration is mediated at least in part by the specific ligand Fgf20a. Taken together, our findings reveal robust new requirements for Fgfs in the day-to-day homeostatic preservation of zebrafish appendages, and have implications for why elevated regenerative capacity has been selectively preserved in certain vertebrate species.
MATERIALS AND METHODS
Zebrafish strains and surgeries
Wildtype, transgenic, and mutant zebrafish of the Ekkwill strain, or hybrids between Ekkwill and the related AB strain, were used in all experiments. Animals were between 4 and 12 months of age. mps1 mutants, fgf20a mutants, and transgenic hsp70:dn-fgfr1 and shh:EGFP fish were described previously (Lee et al., 2005; Poss et al., 2002a; Shkumatava et al., 2004; Whitehead et al., 2005). Heterozygous hsp70:dn-fgfr1 fish and wildtype clutchmates received daily heat shocks using an electric heater to raise the water temperature from 26°C to 38°C, as described previously (Lee et al., 2005). Homozygous mps1 and fgf20a mutants, and wildtype controls, were raised to adulthood at 25°C before transfer to aquaria with recirculating water heated to 33°C. Homozygous mps1 mutants were identified by phenotyping for regenerative defects at 33°C, and then allowed at least one month to regenerate at 25°C before use in homeostasis experiments. Some zebrafish heterozygous for mps1 mutations also showed loss of distal fin structures in our experiments (data not shown). For fin amputations, ~50% of the caudal fin was removed using a razor blade, and animals were allowed to regenerate for 3 days at 33°C, or 3–4 days at 25°C.
Fin length measurement and analysis
Fish were anesthetized in 0.1% Tricaine and the fins were imaged at various timepoints. Imaging software (Openlab) was used to measure the length of the two rays flanking the central-most ray, from the end of the most proximal visible ray segment to the distal tip of the ray. The two values were then averaged to obtain a value for each fin. Each value was divided by the average fin length at day 0 (for the same group) to give a normalized (percentage of starting length) value of fin length. Central rays were chosen because they are the most protected from any potential spontaneous injury, although such injuries rarely occurred in our studies (Fig. S1), and because the degree of tissue loss often varied between the two lobes of the fin, confounding statistical analysis. Tests of statistical significance were performed using the Students t-test, with two-tailed distribution assuming unequal variance. At least 8 fins were assessed at each timepoint for each group.
Spontaneous injury analysis
Fish were separated into groups of 4–5 fish and placed in a 1L tank so that individual fish were recognizable over the course of the experiment. Every 2–3 days, fins were imaged on a dissecting microscope, over a total of 14 to 24 days. A low magnification image of the whole fin and a series of high magnification images of groups of fin rays was acquired for each fin.
Scleroblast visualization
To visualize scleroblasts in tissue sections, fins were fixed in paraformaldehyde (PFA) overnight at 4°C and cryosectioned. Fin sections were then stained with the monoclonal zns-5 antibody as described (Johnson and Weston, 1995; Poss et al., 2002b). For whole-mount visualization of scleroblasts, the monoclonal zn-3 antibody was used, which marks the scleroblast cell membrane (A. A. W. and K. D. P.). Fins were fixed in Carnoy’s solution overnight at 4°C, and stained as described (Newmark and Sanchez Alvarado, 2000; Poss et al., 2002a).
TUNEL staining
For comparison of cell death in the distal and proximal regions of the fin, wildtype fins were fixed and cryosectioned, and slides were dried overnight at room temperature. Slides were then incubated in PBS for 30 minutes at 37°C. DNaseI-treated slides were used as a positive control. Slides were transferred to 0.3% Triton X-100 (Sigma) in PBS for 10 minutes, and then covered with 150 μl of 1X TdT buffer (Invitrogen) for 5 minutes. The buffer was then removed and replaced with 150 μl of 1X TdT buffer containing 0.3U/μl of TdT enzyme (Invitrogen) and 8 μM Biotin-14-dCTP (Invitrogen). Slides were incubated at 37°C for 1 hour, followed by termination in Stop Buffer (300 mM NaCl, 30 mM Na-Citrate) for 15 minutes at room temperature. Slides then rinsed three times in PBS and covered with 20 μg/ml Texas Red Streptavidin (Vector Laboratories) in PBS, pH 8.2, and incubated in the dark at room temperature for 30 minutes. Slides were then washed four times in PBS and coverslipped using Vectashield with DAPI to stain nuclei.
Five different sections were imaged at distal and proximal regions of each fin (n = 10 fins). Distal regions represent approximately 350 μm at the distal end of the fin, and proximal samples represent tissue approximately 700 μm to 1050 μm from the distal end of the fin. This length was chosen because it is the length of a frame at 20X magnification using our imaging equipment. Epidermal and mesenchymal regions were carefully outlined in each image, and the area quantified using Openlab software. Then, TUNEL-positive nuclei, co-stained with DAPI, were counted by hand within these regions. The areas and TUNEL-positive cell counts from these 5 sections were summed, giving each animal 4 indices: distal epidermal, distal mesenchymal, proximal epidermal, and proximal mesenchymal cell death.
BrdU incorporation
For comparison of proliferation in distal and proximal regions of the fin, fish were allowed to swim for 24 hours in a 50 μg/mL solution of bromodeoxyuridine (BrdU) in fish water. After collection, fins were fixed, cryosectioned, and stained as described (Poss et al., 2002b). For each fin, five different sections were analyzed for both distal and proximal samples as described above. A total of 8 fins were analyzed for the number of BrdU-positive cells per area of epidermal or mesenchymal tissue.
BrdU incorporation was also analyzed in wildtype or hsp70:dn-fgfr1 animals given 14 days heat shock followed by 5 days recovery at room temperature. For these experiments, a 2.5 mg/ml solution of BrdU was injected intraperitoneally two hours before fin collection. Fins were fixed in Carnoy’s solution overnight at 4°C, and stained as described (Newmark and Sanchez Alvarado, 2000; Poss et al., 2002a). A total of 10 wildtype and 8 hsp70:dn-fgfr1 fins were analyzed.
In situ hybridization
Whole-mount in situ hybridization was performed as described previously (Poss et al., 2000), using digoxigenin-labeled probes for mkp3, msxb, fgf20a, and mps1 (Akimenko et al., 1995; Lee et al., 2005; Poss et al., 2002a; Whitehead et al., 2005). Wildtype and transgenic fins were hybridized and developed simultaneously. Section in situ experiments were performed as described, using mkp3 and msxb probes (Poss et al., 2002b). The msxb probe was developed overnight, as is standard in our lab (~16 hours); for mkp3 the exposure time was shortened to ~4 hours to limit nonspecific staining.
RT-PCR
RNA samples were prepared from uninjured fins and regenerating fins using TRI Reagent (Sigma) according to the manufacturers protocol. Five μg of total RNA was used for reverse transcription reactions using Superscript III and Oligo-dT20 (Invitrogen) according to the manufacturers protocol. PCR reactions, consisting of 2 minutes at 94°C, 26 to 30 cycles of 94°C for 20 seconds, 52°C for 20 seconds, 72°C for 45 seconds and a final 72°C extension for 5 minutes were performed in an Eppendorf Mastercycler, and samples were run on 2% agarose gels.
RESULTS
Long-term inhibition of Fgf receptors in uninjured zebrafish causes progressive fin atrophy
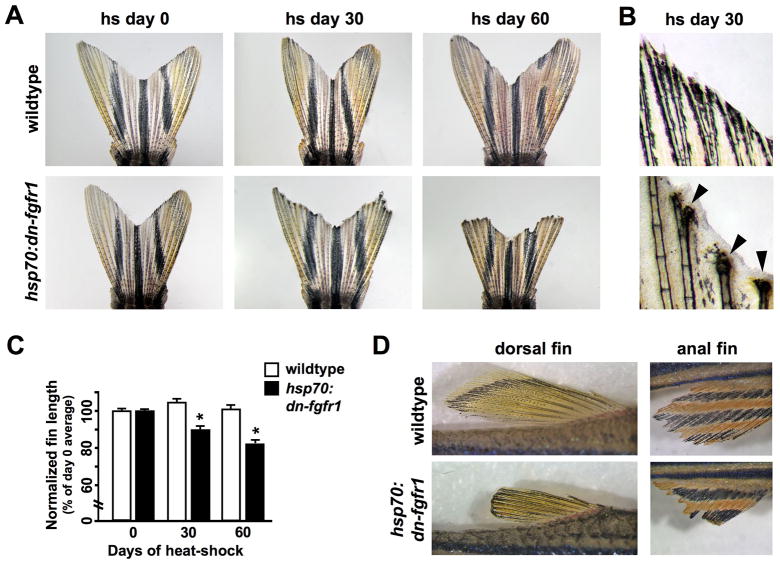
To test the extent to which facultative mechanisms also support homeostatic regeneration of zebrafish appendages, we examined requirements for Fgfs, robust regulators of blastema formation and function in a variety of organisms and systems. We used a transgenic strain in which blastemal proliferation can be inducibly blocked by heat-induced expression of a transgenic dominant-negative Fgfr construct (hsp70:dn-fgfr1), slowing or blocking regeneration of amputated fins (Lee et al., 2005; Yin et al., 2008). Adult transgenic hsp70:dn-fgfr1 fish and wildtype clutchmates were given a strong, daily heat-shock regimen for 30 or 60 days, after which animals were imaged to assess caudal fin length and morphology. Surprisingly, these conditions caused rapid, easily visible loss of distal fin tissue in transgenic animals (Fig. 1A). Transgenic fins often terminated with damaged bone and an excess of epidermal tissue, suggesting recent injury and epidermal repair (Fig. 1B). Wildtype clutchmates occasionally showed similar morphology (without tissue loss), but only in isolated rays rather than the entirety of the fin (data not shown). The degree of damage was variable in hsp70:dn-fgfr1 outer rays but generally consistent within the inner rays; therefore, we quantified the lengths of centrally located rays and found reductions of fin length of ~10% after 30 days, and ~17% after 60 days. Under identical conditions, wildtype clutchmates maintained their fin length (Fig. 1C). Isolated hsp70:dn-fgfr1 animals, removed from possible aggressive interactions with other fish, also displayed progressive loss of fin structures (data not shown). Furthermore, similar tissue loss also occurred in dorsal, anal, pelvic, and pectoral fins, indicating that Fgf signaling is required for homeostasis in all fin types (Fig. 1D and data not shown). If Fgf signaling was restored to these animals for 30 days following long-term Fgfr inhibition, the majority of fin rays were able to recover lost structures (Fig. S1).
Fig. 1. Inhibition of Fgf signaling causes progressive tissue loss from zebrafish fins.
(A) Images of wildtype and hsp70:dn-fgfr1 fins at day 0, day 30, and day 60 of heat-shock. Wildtype fins were unaffected by daily heat-shocks, while transgenic fins showed progressive loss of distal fin tissue. Fins shown are representative, and not from the same animal at each timepoint. (B) High magnification images of distal fin structures after 30 days of heat shock. Many hsp70:dn-fgfr1 rays exhibited severe tissue loss, which was often accompanied by an excess of epidermal tissue (arrowheads). (C) Quantification of fin loss by measurement of centrally located rays (see Materials and methods). hsp70:dn-fgfr1 animals displayed significant reductions in fin length following both 30 and 60 days of heat-shock, whereas wildtype clutchmates showed no changes (mean ± SEM; *Student’s t-test, p ≪ 0.001 at days 30 and 60). (D) Dorsal and anal fins of hsp70:dn-fgfr1 zebrafish showed fin atrophy after 60 days of Fgfr blockade. The fins of wildtype clutchmates retained their length and morphology after 60 days of similar heat treatments.
We examined transgenic fin tissues closely to identify the underlying pathology responsible for atrophy. One explanation was the inability during extended Fgfr inhibition to regenerate tissues lost through minor injuries that occur normally in zebrafish fins. In this model, zebrafish experience regular bone fragmentation and loss, recurrently countered by activation of regenerative mechanisms. To examine this possibility, we observed and imaged wildtype fin rays at high magnification every 2–3 days over a period of 2–3 weeks. We found that events of bone loss and regeneration occur very rarely in our animal facility (2 events in 13 fish over 14 to 24 days; Fig. S2). While not fully excluding a model of recurring injury, these observations indicate that visible injuries to the fin are normally too rare or mild to account for the consistent degeneration we observed during Fgfr inhibition.
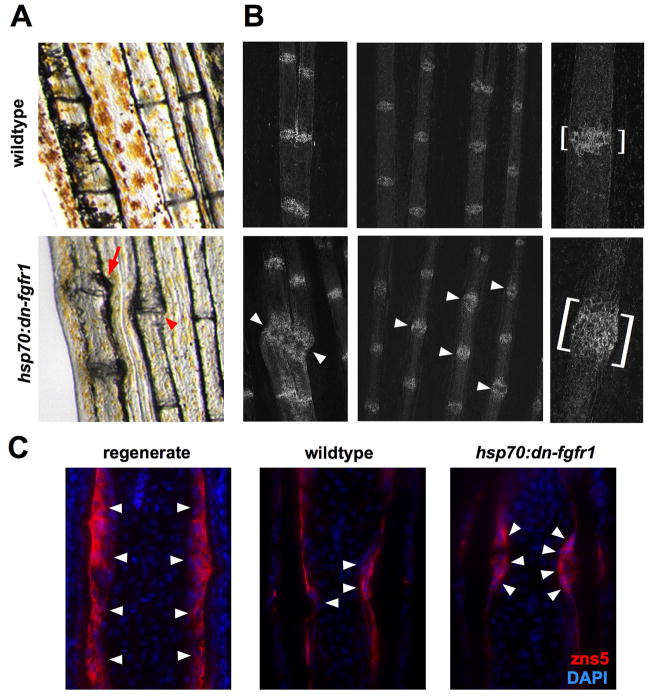
Interestingly, upon examining fins at high magnification after 30 days of Fgf inhibition, we found that over half of the transgenic fins exhibited swelling, discoloration, and/or separation or slipping of rays at the intersegmental joints (16/30 fins, Fig. 2A). Joint swelling or segment separation was only occasionally observed in wildtype clutchmates (2/30 fins), and never with the degree of severity exhibited by transgenics. This suggested that pathology or weakness at segment joints contributes to major tissue loss in hsp70:dn-fgfr1 fins. We examined the joints more closely by confocal microscopy and by histology to identify disturbances in cellular organization. The zebrafish fin lepidotrichia consist of two bony, facing hemirays, which are initially formed by mineralization of bone matrix secreted by the flattened scleroblasts that encase them. At the intersegmental joints between lepidotrichial segments, there exist clusters of scleroblasts with a different, rounded morphology. In projections of confocal slices and in tissue sections, these rounded scleroblasts appear as a small bulge in the intraray mesenchyme around the joints (Fig. 2B,C). Scleroblasts in regenerating fins also have a similar rounded morphology (Fig. 2C). In confocal projections and sections through hsp70:dn-fgfr1 fin rays, the bulge at the intersegmental joints was often enlarged, with large numbers of rounded scleroblasts protruding deep into the surrounding mesenchyme (Fig. 2B,C). We observed this hypertrophy in areas of severe joint dysmorphology (Fig. 2C, bottom left), as well as in transgenic rays that lacked major anatomical changes (Fig. 2B, middle and bottom right). Our observations indicate that one way in which Fgf signaling maintains fin integrity is through control of scleroblast activity and/or patterning, particularly at intersegmental joints.
Fig. 2. Fgf receptor inhibition causes pathology at intersegmental joints.
(A) Many hsp70:dn-fgfr1 fins exhibited swelling (arrow) or dislocation (arrowhead) of the ray segments at the intersegmental joints (bottom). A representative image of a wildtype fin is provided for comparison (top). (B) Confocal images of whole-mount hsp70:dn-fgfr1 and wildtype fins after 30 days of heat-shock, stained with zn3 antibody to visualize scleroblasts. (Left) Example of scleroblast expansion at hsp70:dn-fgfr1 segment joints (bottom) in a case of visible joint pathology (arrowheads). (Middle) Joint hypertrophy was also observed in regions of hsp70:dn-fgfr1 fins without obvious structural damage (arrowheads). (Right) Segmental joints viewed at high magnification, with expansion and disorganization in an hsp70:dn-fgfr1 joint (brackets). (C) Sections of hsp70:dn-fgfr1 and wildtype fins after 30 days of heat-shock, stained to visualize scleroblasts with zns-5 antibody. There is an expanded zone of rounded scleroblasts (red) surrounding the segment joints of transgenic hemirays. (Left) Regenerates at 4 days post-amputation also have rounded scleroblast morphology.
Homeostasis mechanisms include distally focused areas of cell turnover and developmental signaling
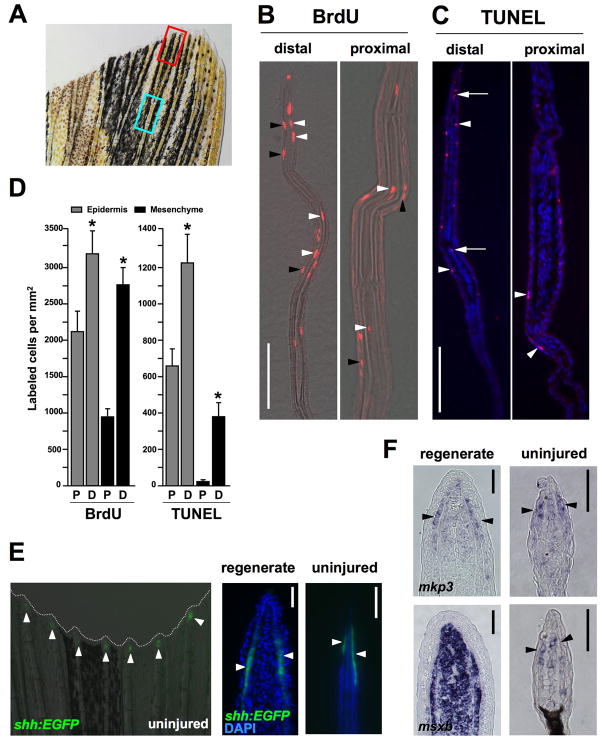
The fin atrophy exposed by Fgfr blockade indicated vigorous homeostatic regeneration; thus, we searched for regions of cell turnover and activated developmental programs in uninjured fins. Previous studies have described cell proliferation in the uninjured zebrafish caudal fin, reporting qualitatively higher levels of proliferation at the distal fin structures and lower levels in more proximal regions. This graded proliferation is purported to reflect growth; specifically, coordinated bursts or saltations of proliferation that periodically add new ray segments as part of adult indeterminate growth (Goldsmith et al., 2003; Iovine and Johnson, 2000; Nechiporuk and Keating, 2002). We quantified cell proliferation in distal and medial regions of uninjured fins using assays for BrdU incorporation (Fig. 3A). Proliferation was low compared to that seen during regeneration of amputated structures. Nevertheless, our data confirmed significantly higher BrdU incorporation indices in distal structures. Epidermal BrdU incorporation was ~43% higher in the most distal 350 μm of the fin versus an identical length of medial structures, while BrdU incorporation in mesenchymal tissue, which includes connective tissue cells and scleroblasts, was ~197% higher in distal structures (Fig. 3B,D). Although proliferation was highest at the distal ray tip, proliferating cells within both the epidermal and mesenchymal compartments were not restricted to a specific cell type or a concentrated area as prominent as the regeneration blastema.
Fig. 3. Homeostatic regeneration programs active in zebrafish fins.
(A) Ventral lobe of an uninjured caudal fin. The red box indicates areas taken for distal measurements; the blue box is for proximal measures. Each box is 350 μm, separated by 350 μm, a length chosen because it is the length of a frame at 20X magnification using our imaging equipment. (B) Distal and proximal sections of the caudal fin stained for BrdU incorporation, after a 24-hour labeling period. BrdU-labeled cells (red) are observed in epidermal (black arrowheads) and mesenchymal (white arrowheads) compartments of both distal and proximal regions. Distal regions are at the top of each image. (C) TUNEL stains of distal and proximal regions, labeling apoptotic cells. TUNEL-positive cells (red) are observed in the epidermal (arrowheads) and mesenchymal (arrows) compartments. Nuclei are labeled with DAPI (blue). (D) Quantification of BrdU and TUNEL labeled cells in epidermal and mesenchymal compartments of proximal (P) and distal (D) fin tissue. (mean ± SEM; *Student’s t-test, p < 0.05). (E) Analysis of a shh:EGFP transgenic reporter strain. (Left) Whole-mount detection of EGFP fluorescence at the distal tips of each ray of an uninjured shh:EGFP transgenic zebrafish. (Right) shh is expressed in the epidermis adjacent to the blastema in the regenerating fin (arrowheads), and in a similarly restricted, epidermal domain at the distal tips of the rays in the uninjured fin (arrowheads). Nuclei are labeled with DAPI (blue). (F) In situ hybridization of tissue sections for mkp3 and msxb. (Top) mkp3 is expressed in the distal lateral epidermis and distal-most mesenchyme of uninjured fins (arrowheads indicate epidermal expression, right), a pattern similar to its expression in the basal epidermal layer and blastema during regeneration (left). (Bottom) msxb expression in the uninjured fin is predominant in distal mesenchyme (right), reminiscent of blastemal expression of msxb after amputation (left). Scale bars = 50μm.
We postulated that these graded differences in proliferation indices in uninjured fins might reflect graded cell turnover, in addition to contributions to distal organ growth. To test this idea, we assessed apoptosis by TUNEL staining. Both mesenchymal and epidermal compartments all along the proximodistal axis of the fin contained low numbers of apoptotic cells. However, each of the mesenchymal and epidermal TUNEL indices was significantly higher in distal fin tissue than in more medial tissue (Fig. 3C,D). That rates of TUNEL-detectable cell apoptosis were higher in more proliferative areas of the fin suggests that the ongoing cell proliferation in distal fin structures represents, at least in part, a homeostatic response to counter higher levels of cell death.
Next, we searched for evidence of developmental programs associated with these turnover events, paying particular attention to programs known to regulate regeneration of amputated fins. We first took advantage of an transgenic EGFP reporter strain that marks expression of shh, a reported blastemal mitogen and patterning factor synthesized in regeneration epidermis adjacent to the blastema (Laforest et al., 1998; Quint et al., 2002; Shkumatava et al., 2004). We found that this strain visualizes shh expression during regeneration in a manner consistent with published in situ hybridization results. Moreover, shh was consistently detectable in similar epidermal expression domains in the distal-most ~100 μm of uninjured fins (Fig. 3E). We similarly examined two other markers: 1) msxb marks cells throughout the regeneration blastema as well as regenerating scleroblasts, and was recently shown using electroporation of antisense morpholinos to be essential for regenerative growth (Akimenko et al., 1995; Thummel et al., 2006); and 2) mkp3 is an Fgf target gene that is induced in the blastema as well as cells of the basal epidermal layer encasing the blastema, with expression levels that correlate with the rate of regenerative outgrowth (Lee et al., 2005). Semi-quantitative RT-PCR demonstrated that both of these markers are expressed in the uninjured fin, albeit at much lower levels than that seen after amputation (Fig. S3). By in situ hybridization of tissue sections, we detected faint expression of both of these molecules at the distal fin tip. Interestingly, detectable expression was restricted to domains similar to those occupied by these molecules during regenerative outgrowth: mkp3 was expressed in cells of the distal epidermis and mesenchyme, while msxb expression was restricted to mesenchymal cells, suggesting that these molecules have similar functions in amputated and uninjured fins (Fig. 3F). Specific localization of these regeneration mediators suggested homeostatic involvement in the cell turnover and maintenance functions we had identified.
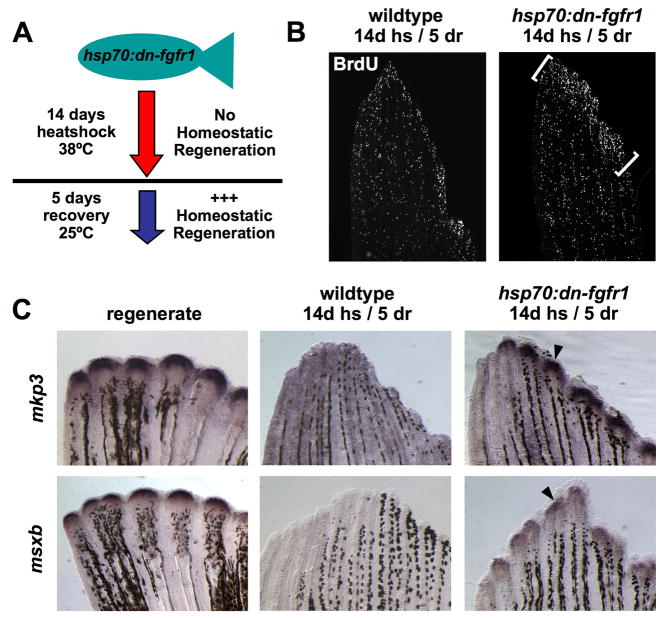
Do cell proliferation and low-level shh, msxb, and Fgf signaling/mkp3 expression in uninjured fins reflect ongoing homeostatic events? To test whether these events rise in the face of a greater homeostatic obligation, we gave transient periods of Fgfr inhibition with the intention that it might “prime” fins for a burst of proliferation and marker expression after Fgf signaling is restored. In these experiments, a 7- or 14-day protocol of daily heat-shocks was given to hsp70:dn-fgfr1 and wildtype clutchmates, followed by a 5-day recovery period at room temperature (Fig. 4A). We saw no significant difference in TUNEL-positive cells in transgenic fins versus wildtype fins during the heat-shock period, suggesting little effects of Fgfr inhibition on apoptosis, as well as no significant differences in mesenchymal BrdU incorporation (data not shown). The latter observation might reflect a role for Fgfs in proliferation of just a subset of cell types within the uninjured fin. Alternatively, our assay may be limited by variability caused by very low baseline numbers of proliferating cells or the possibility of missing saltatory bursts of proliferation.
Fig. 4. Homeostatic responses of cell proliferation and gene expression in zebrafish fins.
(A) A model for “priming” homeostatic regeneration through manipulation of Fgf signaling. If developmental gene expression and cell proliferation are homeostatic events that rely on Fgf signaling, then these events should increase in intensity as a response after fins recover from a period of Fgfr inhibition. (B) BrdU incorporation in uninjured fins after a priming protocol of 14 days of daily heat-shocks, and 5 days at room temperature (14d hs/5 dr). This protocol was predicted to repress, and then release and increase, homeostatic proliferation in hsp70:dn-fgfr1 fins. Transgenic fins display a burst of BrdU incorporation in distal fin tissue during recovery (brackets) that is not detectable in wildtype fins. One lobe of the caudal fin is shown. (C) Expression of regeneration marker genes increases during recovery of Fgf signaling. mkp3 and msxb are robustly expressed in regenerating fins (4 dpa, arrowheads), but expression in the uninjured fin or primed wildtype fin is undetectable by whole mount in situ hybridization. However, mkp3 and msxb levels increase visibly following homeostatic priming of the fin by transient Fgfr inhibition (arrowheads).
By contrast, following the recovery period from heat-shock, proliferation was markedly increased in distal fin regions of transgenic, but not wildtype animals, indicative of an enhanced homeostatic response (Fig. 4B). Furthermore, expression of mkp3 and msxb could be easily visualized by whole-mount in situ hybridization in transgenic fins, but not wildtype fins, during the recovery period of restored Fgf signaling (Fig. 4C). These changes would not be expected if marker expression and cell proliferation were a purely ontogenetic or saltatory growth signature of adult fins. Together, these data reveal molecular and cellular indicators of a responsive program of homeostatic regeneration in uninjured zebrafish fins.
fgf20a is expressed in the intact fin and required for homeostasis
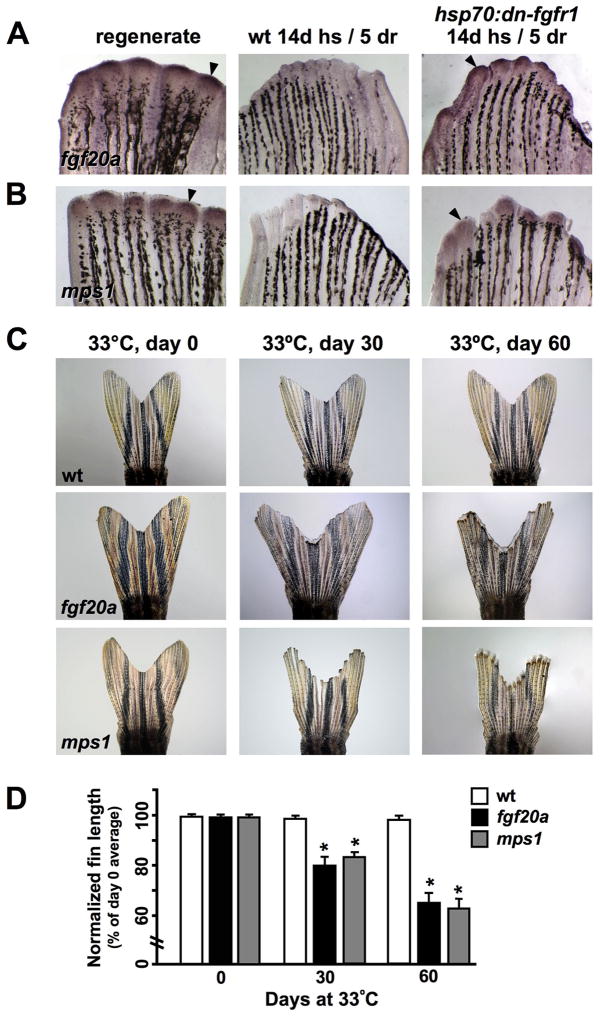
While the above experiments demonstrate that a long-term blockade of Fgfrs causes fin regression, it remained possible that such a strong, animal-wide manipulation disrupts fins indirectly through effects on other organs. For example, extrinsic, blood-borne factors that change with age are known to regulate the regenerative capacity of skeletal muscle satellite cells (Conboy et al., 2005). To address this possibility and also refine our analysis of Fgf signaling during homeostasis, we focused on fgf20a, an Fgf ligand with functions believed to be specific for regeneration of amputated adult fins. Incubation of developing fgf20a embryos at the restrictive temperature of 33°C has little effect on ontogenetic development, while incubation at this temperature arrests adult fin regeneration during blastema formation (Whitehead et al., 2005). Semi-quantitative RT-PCR revealed that fgf20a is expressed at low levels in the uninjured fin (Fig. S2), and in situ hybridization of fins from primed hsp70:dn-fgfr1 animals indicated that fgf20a is a member of a homeostatic response program that includes msxb and mkp3 (Fig. 5A). These expression data suggested that fgf20a might mediate Fgf-dependent homeostatic regeneration.
Fig. 5. Fgf20a and Mps1 are required for homeostatic regeneration in zebrafish fins.
(A, B) In situ hybridization for fgf20a and mps1 during regeneration and priming. fgf20a and mps1 increase expression is increased upon recovery of Fgf signaling (arrowheads), as described for mkp3 and msxb. Expression is undetectable through this method in wildtype fins. (C) Images of wildtype, fgf20a, and mps1 mutant fins at day 0, day 30, and day 60 at the restrictive temperature (33°C). Both mutants exhibited a significant loss in distal tissue that was not seen in wildtype controls maintained at the restrictive temperature. (D) Quantification of length changes in centrally located rays of fgf20a and mps1 mutants. Both mutant strains showed a significant reduction in fin length after 30 and 60 days at the restrictive temperature, while wildtype controls maintained fin length (mean ± SEM, *Student’s t-test, p ≪ 0.001 at days 30 and 60).
To test whether fgf20a is essential for appendage homeostasis, we placed adult fgf20a mutants at 33°C for 30–60 days and assessed fin size and integrity. While wildtype controls maintained fin length at 33°C in these experiments, fgf20a zebrafish displayed a progressive loss of distal fin structures that was quantitatively very similar to results observed with hsp70:dn-fgfr1 animals (Fig. 5C). Quantification of central ray length revealed that fgf20a mutants lost ~19% of their fin tissue after 30 days at the restrictive temperature, and roughly a third of their fin length (~35%) after 60 days (Fig. 5D). Unlike hsp70:dn-fgfr1 zebrafish, joint morphology appeared normal by gross visual inspection, and tissue loss at outer fin rays was less severe. This may be related to different strengths of genetic interventions, different conditions for removing Fgf functions, genetic redundancy, and/or functions specific to Fgf20a apart from other Fgf ligands. In any case, our data indicate that signaling by Fgf20a is essential to maintain zebrafish appendages, accounting for at least part of the Fgf-dependency of homeostatic regeneration.
To test whether other known regulators of the blastema show similar expression characteristics and requirements during homeostasis, we examined mps1, a dual-specificity kinase essential for the spindle checkpoint (Fisk and Winey, 2004). mps1 expression is induced in the newly formed blastema and colocalizes with highly proliferative blastemal cells during regenerative outgrowth. Like fgf20a, mps1 was shown to be essential for regeneration by positional cloning of a temperature-sensitive mutation (Poss et al., 2002a). RT-PCR assays indicated low-level mps1 expression in the uninjured fin (Fig. S2), and, like fgf20a, mps1 expression was induced by a 14-day block of Fgf signaling and 5 days of recovery in hsp70:dn-fgfr1 animals (Fig. 5B). When mps1 mutant zebrafish were placed at the restrictive temperature of 33°C for 30 or 60 days, the animals showed severe distal tissue loss. Measurement of the central rays revealed a ~17% loss of tissue by 30 days at 33°C, and ~36% after 60 days (Fig. 5C,D). Thus, Mps1, a kinase essential for proliferation of blastemal cells after fin amputation, also is necessary to maintain tissue in intact fins. Taken together, our data suggest that factors that enable facultative regeneration are generally required for homeostatic regeneration within uninjured fins.
DISCUSSION
Homeostatic regeneration in zebrafish fins
We conclude that the Fgf-dependent developmental machinery responsible for blastema-based, facultative regeneration of zebrafish fins also mediates homeostatic events that maintain existing fin ray structures. Most notably, rapid loss of distal appendage tissue occurred after multiple genetic disruptions of programs that form and maintain the regeneration blastema. Two independent methods to block Fgf signaling had striking effects on tissue maintenance, pinpointing a requirement for Fgf20a in preserving complex appendage tissue. The kinase Mps1 is also critical for fin homeostasis. Our phenotypic data suggest that Fgf20a functions in maintenance of distal fin regions where cell turnover and expression of msxb, shh, and mkp3 are most visible. We also observed effects of Fgfr blockade on joint anatomy that indicate additional functions of this signaling pathway.
While facultative and homeostatic regeneration appear to have many similarities, there are also interesting differences. For instance, fgf20a mutants have a mostly penetrant defect in injury-induced fin regeneration at 25°C (Whitehead et al., 2005), despite developing normally to adulthood and maintaining fins grossly normally at this temperature. This suggests that there is a requirement for some amount or function of Fgf20a that is met during homeostatic regeneration but not facultative regeneration. Similarly, redundancies may be present in one type of regeneration and absent in another. Other differences may relate to restrictive programs with greater presence in intact fins. For instance, recent work has found that signaling by epidermally synthesized Wnt5b has an inhibitory role on blastemal proliferation (Stoick-Cooper et al., 2007b). Candidates for analogous factors in intact fins include several miRNAs that were recently shown to have higher levels in uninjured fins than in fins regenerating after amputation. At least one of these miRNAs, miR-133, also displayed functional properties of a regenerative brake (Yin et al., 2008). Understanding the balance between permissive and restrictive factors in injured and uninjured appendages is likely to be important in unraveling seminal issues in regeneration.
Fgfs, homeostatic regeneration, and positional memory
One of the most fascinating aspects of appendage regeneration is positional memory, the ability of the limb or fin stump to recognize and restore only those structures lost by injury. Positional memory is thought to be based on a gradient of some determinant(s) existing in uninjured tissue or quickly established after amputation. Recently, we found that the amount of Fgf signaling established after amputation is graded along the proximodistal axis, with higher amounts in more proximal tissue and lower amounts distally (Lee et al., 2005). Greater Fgf signaling positively impacts blastemal size and regenerative rate, leading to more rapid outgrowth in proximal regenerates. Similarly, as regeneration proceeds gradually to completion, amounts of Fgf signaling gradually wane.
Our findings here indicate that developmental signaling, including Fgf signaling, is not inactivated after restoration of lost structures; rather, a modicum is maintained at low levels in the distal tips of intact fins. When the capacity to maintain this Fgf signaling is experimentally blocked, tissue loss occurs, revealing an essential role in homeostatic equilibrium. Based on these earlier and current observations, we postulate that weak regenerative presence at the distal tips of intact fins may in fact be a marker or determinant of far-distal positional identity. That is, uninjured fins maintain size in part due to a level of Fgf signaling that precisely opposes ongoing cell death. By contrast, an amputated fin will initiate a position-dependent boost of Fgf signaling for structure-restoring growth. Regeneration then culminates when Fgf signaling decreases to an amount that no longer procures a net gain in growth vis a vis ongoing cell death. While this is an attractive model, what requires characterization is the signal(s) that determines position and sets the appropriate amount of Fgf signaling.
Evolutionary significance of homeostatic regeneration and regenerative capacity
Why is the capacity for appendage regeneration, or organ regeneration in general, distributed unequally among vertebrate species? The selective advantages for non-mammalian regenerative events are not uniformly obvious. For instance, the capacity for tail regeneration in lizards facilitates repeated use of the anti-predatory tactic of autotomy, and also is thought in some species to reduce potential reproductive, social, and locomotor costs (Clause and Capaldi, 2006). By contrast, there is no immediate explanation for the capacity of adult non-mammalian vertebrates to regenerate resected cardiac muscle (Poss et al., 2002b).
Our current study, supported by previous experiments (Nechiporuk and Keating, 2002), suggests that one contributing factor behind the high regenerative capacity of adult zebrafish fins is the particularly dynamic nature by which they are actively maintained. That is, the preservation of capacity to regenerate patterned fin structures after major injury might be the evolutionary consequence of a more critical role for regenerative mechanisms to regularly balance day-to-day cell loss and maintain existing tissue. Accordingly, it is interesting to speculate that teleosts and urodeles possess a wider array of regenerative tissues than mammals in part because of greater cell turnover among those organs. This might also be related to the capacity for indeterminate growth in many of these species, although the newt does not grow throughout its adult life. Supporting this idea, we have found that the adult zebrafish heart, unlike the more static mammalian heart, actively adds cardiac cells during adult animal growth and size maintenance (Wills et al., 2008). Similarly, adult teleost CNS structures like the highly regenerative retina and the brain show unusually high basal rates of neurogenesis (Grandel et al., 2006; Otteson and Hitchcock, 2003).
Notably, the severe fin regression phenotypes we observed after genetic manipulations in zebrafish are highly reminiscent of those seen recently after various gene knockdown experiments in the classic invertebrate model system for blastema-based regeneration, freshwater planarians. Rapid facultative and homeostatic regeneration in planarians are based on stem cell-like neoblasts, the sole proliferative cell type responsible for renewal all structural cells (Birnbaum and Sanchez Alvarado, 2008). In those studies, long-term RNA interference of individual genes frequently had similar effects on facultative and homeostatic regeneration; that is, regeneration could be blocked after animal bisection, while fatal “curling” or progressive changes in adult pattern occurred within weeks of gene perturbation in intact animals (Cebria et al., 2007; Cebria et al., 2002; Cebria and Newmark, 2005; Reddien et al., 2007; Reddien et al., 2005). It will be interesting to compare among different vertebrate species and organs, the extent to which the capacity for injury-induced regeneration correlates with the activity of ongoing homeostatic regeneration maintained by Fgf ligands and other signaling pathways.
Supplementary Material
Acknowledgments
We thank G. Whitehead and E. Lien for sharing fgf20a mutants, J. Moss for shh:EGFP fish, J. Bumgardner, O. Ighile, and C. Wheeler for excellent zebrafish care, J. Holdway for help with imaging, the Zebrafish International Research Center for antibodies, and K. Iovine, Y. Lee, P. Newmark, E. Tanaka, V. Yin, and Poss lab members for helpful comments on the manuscript. This work was supported by grants to K.D.P. from NIGMS, NHLBI, Whitehead Foundation, March of Dimes, and Pew Charitable Trusts.
References
- Akimenko MA, Johnson SL, Westerfield M, Ekker M. Differential induction of four msx homeobox genes during fin development and regeneration in zebrafish. Development. 1995;121:347–57. doi: 10.1242/dev.121.2.347. [DOI] [PubMed] [Google Scholar]
- Akimenko MA, Mari-Beffa M, Becerra J, Geraudie J. Old questions, new tools, and some answers to the mystery of fin regeneration. Dev Dyn. 2003;226:190–201. doi: 10.1002/dvdy.10248. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla A, Lim DA. For the long run: maintaining germinal niches in the adult brain. Neuron. 2004;41:683–6. doi: 10.1016/s0896-6273(04)00111-4. [DOI] [PubMed] [Google Scholar]
- Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–76. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- Birnbaum KD, Sanchez Alvarado A. Slicing across kingdoms: regeneration in plants and animals. Cell. 2008;132:697–710. doi: 10.1016/j.cell.2008.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. Epidermal stem cells of the skin. Annu Rev Cell Dev Biol. 2006;22:339–73. doi: 10.1146/annurev.cellbio.22.010305.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebria F, Guo T, Jopek J, Newmark PA. Regeneration and maintenance of the planarian midline is regulated by a slit orthologue. Dev Biol. 2007;307:394–406. doi: 10.1016/j.ydbio.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebria F, Kobayashi C, Umesono Y, Nakazawa M, Mineta K, Ikeo K, Gojobori T, Itoh M, Taira M, Sanchez Alvarado A, et al. FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature. 2002;419:620–4. doi: 10.1038/nature01042. [DOI] [PubMed] [Google Scholar]
- Cebria F, Newmark PA. Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development. 2005;132:3691–703. doi: 10.1242/dev.01941. [DOI] [PubMed] [Google Scholar]
- Clause AR, Capaldi EA. Caudal autotomy and regeneration in lizards. J Exp Zoolog A Comp Exp Biol. 2006;305:965–73. doi: 10.1002/jez.a.346. [DOI] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weissman IL, Rando TA. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–4. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- Fisk HA, Winey M. Spindle regulation: Mps1 flies into new areas. Curr Biol. 2004;14:R1058–60. doi: 10.1016/j.cub.2004.11.047. [DOI] [PubMed] [Google Scholar]
- Goldsmith MI, Fisher S, Waterman R, Johnson SL. Saltatory control of isometric growth in the zebrafish caudal fin is disrupted in long fin and rapunzel mutants. Dev Biol. 2003;259:303–17. doi: 10.1016/s0012-1606(03)00186-6. [DOI] [PubMed] [Google Scholar]
- Grandel H, Kaslin J, Ganz J, Wenzel I, Brand M. Neural stem cells and neurogenesis in the adult zebrafish brain: origin, proliferation dynamics, migration and cell fate. Dev Biol. 2006;295:263–77. doi: 10.1016/j.ydbio.2006.03.040. [DOI] [PubMed] [Google Scholar]
- Iovine MK, Johnson SL. Genetic analysis of isometric growth control mechanisms in the zebrafish caudal Fin. Genetics. 2000;155:1321–9. doi: 10.1093/genetics/155.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, DePinho RA, Sharpless NE, Scadden DT. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–6. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- Jazwinska A, Badakov R, Keating MT. Activin-betaA signaling is required for zebrafish fin regeneration. Curr Biol. 2007;17:1390–5. doi: 10.1016/j.cub.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Johnson SL, Weston JA. Temperature-sensitive mutations that cause stage-specific defects in Zebrafish fin regeneration. Genetics. 1995;141:1583–95. doi: 10.1093/genetics/141.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DL, Wagers AJ. No place like home: anatomy and function of the stem cell niche. Nat Rev Mol Cell Biol. 2008;9:11–21. doi: 10.1038/nrm2319. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Bonner-Weir S, Sharpless NE. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–7. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- Laforest L, Brown CW, Poleo G, Geraudie J, Tada M, Ekker M, Akimenko MA. Involvement of the sonic hedgehog, patched 1 and bmp2 genes in patterning of the zebrafish dermal fin rays. Development. 1998;125:4175–84. doi: 10.1242/dev.125.21.4175. [DOI] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–53. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132:5173–83. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–52. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechiporuk A, Keating MT. A proliferation gradient between proximal and msxb-expressing distal blastema directs zebrafish fin regeneration. Development. 2002;129:2607–17. doi: 10.1242/dev.129.11.2607. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Sanchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol. 2000;220:142–53. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- Otteson DC, Hitchcock PF. Stem cells in the teleost retina: persistent neurogenesis and injury-induced regeneration. Vision Res. 2003;43:927–36. doi: 10.1016/s0042-6989(02)00400-5. [DOI] [PubMed] [Google Scholar]
- Poss KD, Keating MT, Nechiporuk A. Tales of regeneration in zebrafish. Dev Dyn. 2003;226:202–10. doi: 10.1002/dvdy.10220. [DOI] [PubMed] [Google Scholar]
- Poss KD, Nechiporuk A, Hillam AM, Johnson SL, Keating MT. Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development. 2002a;129:5141–9. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- Poss KD, Shen J, Nechiporuk A, McMahon G, Thisse B, Thisse C, Keating MT. Roles for Fgf signaling during zebrafish fin regeneration. Dev Biol. 2000;222:347–58. doi: 10.1006/dbio.2000.9722. [DOI] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002b;298:2188–90. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Quint E, Smith A, Avaron F, Laforest L, Miles J, Gaffield W, Akimenko MA. Bone patterning is altered in the regenerating zebrafish caudal fin after ectopic expression of sonic hedgehog and bmp2b or exposure to cyclopamine. Proc Natl Acad Sci U S A. 2002;99:8713–8. doi: 10.1073/pnas.122571799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando TA. Stem cells, ageing and the quest for immortality. Nature. 2006;441:1080–6. doi: 10.1038/nature04958. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Kicza AM, Sanchez Alvarado A. BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development. 2007;134:4043–51. doi: 10.1242/dev.007138. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sanchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell. 2005;8:635–49. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–9. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- Shkumatava A, Fischer S, Muller F, Strahle U, Neumann CJ. Sonic hedgehog, secreted by amacrine cells, acts as a short-range signal to direct differentiation and lamination in the zebrafish retina. Development. 2004;131:3849–58. doi: 10.1242/dev.01247. [DOI] [PubMed] [Google Scholar]
- Smith A, Avaron F, Guay D, Padhi BK, Akimenko MA. Inhibition of BMP signaling during zebrafish fin regeneration disrupts fin growth and scleroblasts differentiation and function. Dev Biol. 2006;299:438–54. doi: 10.1016/j.ydbio.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Stoick-Cooper CL, Moon RT, Weidinger G. Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine. Genes Dev. 2007a;21:1292–315. doi: 10.1101/gad.1540507. [DOI] [PubMed] [Google Scholar]
- Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, Moon RT. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007b;134:479–89. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- Tawk M, Tuil D, Torrente Y, Vriz S, Paulin D. High-efficiency gene transfer into adult fish: a new tool to study fin regeneration. Genesis. 2002;32:27–31. doi: 10.1002/gene.10025. [DOI] [PubMed] [Google Scholar]
- Thummel R, Bai S, Sarras MP, Jr, Song P, McDermott J, Brewer J, Perry M, Zhang X, Hyde DR, Godwin AR. Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev Dyn. 2006;235:336–46. doi: 10.1002/dvdy.20630. [DOI] [PubMed] [Google Scholar]
- Whitehead GG, Makino S, Lien CL, Keating MT. fgf20 is essential for initiating zebrafish fin regeneration. Science. 2005;310:1957–60. doi: 10.1126/science.1117637. [DOI] [PubMed] [Google Scholar]
- Wills AA, Holdway JE, Major RJ, Poss KD. Regulated addition of new myocardial and epicardial cells fosters homeostatic cardiac growth and maintenance in adult zebrafish. Development. 2008;135:183–192. doi: 10.1242/dev.010363. [DOI] [PubMed] [Google Scholar]
- Yin VP, Thomson JM, Thummel R, Hyde DR, Hammond SM, Poss KD. Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. Genes Dev. 2008;22:728–33. doi: 10.1101/gad.1641808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.