Genetic polymorphisms of the Cytochrome P450 isoform 2C19 (CYP2C19) gene -known to cause poor metabolism of important prescription drugs (e.g., anticonvulsants such as phenytoin, antidepressant, cancer chemotherapy, anti-malaria drugs, anti-ulcer proton-pump inhibitors, and methadone)- were examined in a Puerto Rican newborn population. At least seven single nucleotide polymorphisms (SNPs) in the CYP2C19 gene have been shown to influence enzyme activity. The CYP2C19*2 and CYP2C19*3 variants are most common and best characterized clinically. They both are considered non-functional alleles associated with lack of enzymatic activity. Patients with these variants will require significantly lower doses in order to avoid overexposure and increased risk of adverse effects to standard doses of CYP2C19 substrates. The frequencies of variant alleles for the CYP2C19 marker have been reported to differ among ethnic groups. 2C19*2 is 30% in general population; 85% in Asians and about 15% in Caucasians. 2C19*3 is less common, about 6-10% Asians; but rare (<1%) in Caucasians and African Americans (1-3). The allele frequencies of these two variants in the Puerto Rican population remain to be determined.
The objectives of this study were to determine the prevalence of clinically relevant allele variants (CYP2C19*2, splicing defect G681A SNP and CYP2C19*3, stop codon G636A SNP) in a Puerto Rican study population and finally to determine whether the allele distributions for gene mutations meet Hardy-Weinberg equilibrium (HWE).
Genomic DNA were extracted from leukocytes in 122 dried blood filter samples from different regions of Puerto Rico, which were kindly provided by the Puerto Rico Newborn Screening Program. Samples were analyzed by the PCR-RFLP technique at the Molecular Genetic Core facilities of the School of Medicine, UPR as well as by the Physiogenomic-array with the Illumina BeadArray™ platform, at Laboratory of Personalized Health (LPH), Hartford, CT (4-6).
We were able to obtain preliminary results for the CYP2C19*2 allele from 122 individuals (figures 1 and 2). The results for the CYP2C19*2 allele have showed that 90 of these DNA samples (73.7%) were found to be homozygous for the wild-type allele; and 30 DNA samples (24.6%) were single carrier for the allelic variant*2 (heterozygous); whereas, two DNA sample (1.64%) were double-carrier for this clinically relevant mutation. Genotype analysis also revealed that the allele frequency for CYP2C19*2 was 0.139. Notably, these findings were confirmed by Illumina BeadArrayTM technology genotyping platform using highly multiplexed ligation assays (i.e., minor allele frequency of 0.1406 was detected using a Genomas proprietary PG-array consisting of 384 SNPs from 222 genes). It is known that the prevalence of the poor metabolizer (PM) phenotype in Caucasians ranges from 2 to 5% and that the most common gene defect is the CYP2C19*2 variant, which accounts for 75-83% of the defective alleles in PMs. Single CYP2C19*2 carrier frequency in the Puerto Rican newborn tested was 24.6%. Although the HW goodness-of-fit χ2-test was the only test applied, no major deviations from HWE were claimed (χ2 = 0.28; p = 0.87). Statistical analysis showed that no significant differences in genotypic frequencies were found between our sample and those reported for Caucasians where 2% are PMs and 0.75% harboring the *2 allele (χ2 = 0.723; p = 0.70). We also examined 72 of these samples for the CYP2C19*3 mutant allele (null metabolizer), but we could not detect any carriers or individual homozygous for this mutation, which suggests that the frequency for the CYP2C19*3 allele is lower than that for the CYP2C19*2 allele in the Puerto Rican population. Be advised that this mutation is usually considered rare in non-Asian descendant population (7-8). A larger number of samples need to be genotyped for these two allelic variants in order to validate our findings in Puerto Ricans.
Figure 1.
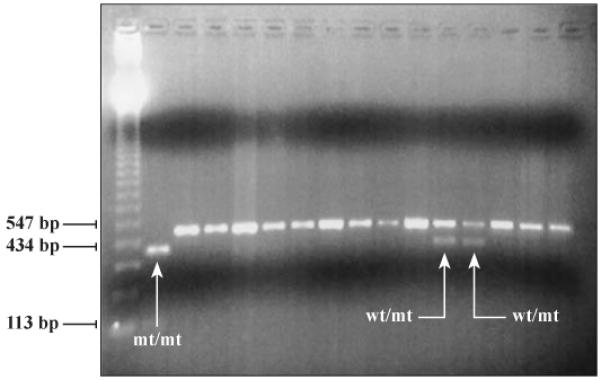
(left-hand side). The agarose gel electrophoresis analysis of CYP2C19*2 allele (G681→A mutation) by PCR-RFLP with BstNI restriction enzyme. Lane 1, 123 bp ladder molecular weight standard, lanes 2-16 are different DNA samples. The sample in lane 2 is homozygous for the CYP2C19*2 mutation and this individual has a reduced capacity to metabolize CYP2C19 substrates and is a PM, lanes 3-11 and 14-16 have samples that are homozygous for the normal allele, and lanes 12 and 13 contain samples that are heterozygous for the CYP2C19*2 allele. Genomic DNA was purified from dried blood filters and eluted in 100 ml. An aliquot of 2 ml was used as template for PCR amplification of exon 5 of the CYP2C19 gene, followed by restriction digestion with 10 units of BstNI enzyme at 60°C, overnight. The PCR-RFLP products were separated in a 2.0% Agarose gel in 0.5X TBE.
Figure 2.

Pie chart for the CYP2C19*2 genotypes in the Puerto Rican population sample (n=122).
Little pharmacogenetic-guided recommendations of clinical relevance are available due in part to limited information in target populations. Accordingly, there is a need to start translating pharmacogenetic data into specific clinical recommendations for labelling and optimal dosage calculation in each individual. Considering the observed prevalence of clinically relevant CYP2C19*2 polymorphisms in Puerto Ricans, it is necessary to educate pharmacists and healthcare providers in CYP2C19-related pharmacogenetics in order to achieve better outcomes by developing DNA-guided individual (personalized) CYP2C19 substrates dosing regimens.
Acknowledgements
This investigation was supported in part by a Research Centers in Minority Institution Award G12RR-03051 from the National Center for Research Resources; a Clinical Research Center Infrastructure Initiative Pilot Projects Award (RCRII) Grant No. 5P20RR011126; the NIH grants R25GM61838 and by the Puerto Rico Newborn Screening Program and Genomas internal research and development funds. The authors want to thank Yolanda Rodríguez for her support in collecting the samples for this survey. We also like to thank the Bio-Minds and MBRS-RISE program. This study (protocol A4070107) was exempt from IRB review under FDA and OHRP guidelines based on category 4 45CFR46.118 for proposed research that does not plan a human subject involvement. The authors have no potential conflicts of interest to disclose. A glossary of genetic terminology is maintained by the National Human Genome Research Institute at www.genome.gov/glossary.cfm.
Contributor Information
Jorge Duconge, School of Pharmacy, Department of Pharmaceutical Sciences, Room 420, School of Pharmacy, Medical Sciences Campus, University of Puerto Rico, PO Box 365067, San Juan, PR 00936-5067.
Carmen L. Cadilla, School of Medicine, Laboratory of Molecular Genetics.
Jessica Y. Renta, School of Medicine, Laboratory of Molecular Genetics.
Pamela Silén-Rivera, School of Medicine, Laboratory of Molecular Genetics.
Paola Piovanetti, School of Medicine, Laboratory of Molecular Genetics.
Rafael García-Berdecía, School of Pharmacy, Department of Pharmaceutical Sciences.
Liza M. Castro-Rosario, School of Medicine, Laboratory of Molecular Genetics
Shalom Monzón, School of Pharmacy, Department of Pharmaceutical Sciences.
Linet Vélez, School of Pharmacy, Department of Pharmaceutical Sciences.
Guillermo Rosas, School of Pharmacy, Department of Pharmaceutical Sciences.
Jhon A. Guerra, Puerto Rico Newborn Screening Program; University of Puerto Rico, School of Medicine’s Pediatrics Department, Pediatric Hospital, San Juan PR 00936.
Pedro J. Santiago-Borrero, Puerto Rico Newborn Screening Program; University of Puerto Rico, School of Medicine’s Pediatrics Department, Pediatric Hospital, San Juan PR 00936.
References
- 1.Streetman DS, Bertino JS, Jr, Nafziger AN. Phenotyping of drug-metabolizing enzymes in adults: a review of in vivo cytochrome P450 phenotyping probes. Pharmacogenetics. 2000;10:187–216. doi: 10.1097/00008571-200004000-00001. [DOI] [PubMed] [Google Scholar]
- 2.De Morais SM, Wilkinson GR, Blaisdell J, Meyer UA, Nakamura K, Goldstein JA. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol. 1994;46:594–598. [PubMed] [Google Scholar]
- 3.De Morais SM, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem. 1994;269:15419–15422. [PubMed] [Google Scholar]
- 4.Ruaño G, Windemuth A, Holford T. Physiogenomics: Integrating Systems Engineering and Nanotechnology for Personalized Medicine. In: Bronzino JD, editor. The Biomedical Engineering Hand-book. 3rd edition CRC Press; 2005. [Google Scholar]
- 5.Holford TR, Windemuth A, Ruaño G. Designing Physiogenomic studies. Pharmacogenomics. 2006;7:157–158. doi: 10.2217/14622416.7.2.157. [DOI] [PubMed] [Google Scholar]
- 6.Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. BeadArray technology: enabling an accurate, cost-effective approach to high- throughput genotyping. Biotechniques. 2002;(Suppl):56–61. [PubMed] [Google Scholar]
- 7.Shu Y, Zhou HH. Individual and ethnic differences in CYP2C19 activity in Chinese populations. Acta Pharmacol Sin. 2000;21:193–199. [PubMed] [Google Scholar]
- 8.Shi WS, Chen SQ. Frequencies of poor metabolizers of cytochrome P450 2C19 in esophagus cancer, stomach cancer, lung cancer and bladder cancer in Chinese population. World J Gastroenterology. 2004;10:1961–1963. doi: 10.3748/wjg.v10.i13.463. [DOI] [PMC free article] [PubMed] [Google Scholar]


