Abstract
Estradiol (E2) regulates a wide range of neural functions, many of which require activation of estrogen receptor α (ERα) and/or ERβ, ligand-gated transcriptional regulators. Surprisingly, very few neural gene targets of ERs have been identified, and these cannot easily explain the myriad effects of E2. GABA regulates most of the same neural functions as E2, and GABAergic neurons throughout the brain contain ER. Therefore, we examined whether E2 directly regulates expression of glutamic acid decarboxylase 2 (gad2), the enzyme primarily responsible for GABA synthesis for synaptic release. Using dual luciferase assays, we found that E2, but not other gonadal steroids, stimulated the activity of a 2691 bp rat gad2 promoter reporter construct. Activation required either ERα or ERβ, and ERβ did not repress ERα-mediated transactivation. Site-directed mutagenesis studies identified three estrogen response elements (EREs) with cell-specific functions. An ERE at −711 upstream of the gad2 translational start site was essential for transactivation in both MCF-7 breast cancer cells and SN56.B5.G4 neural cells, but an ERE at −546 enhanced transcription only in neural cells. A third ERE at −1958 was inactive in neural cells but exerted potent transcriptional repression in E2-treated MCF-7 cells. Chromatin immunoprecipitation assays in mouse GABAergic N42 cells confirmed that E2 induced ERα binding to a DNA fragment containing sequences corresponding to the −546 and −711 EREs of the rat promoter. Based on these data, we propose that direct transcriptional regulation of gad2 may explain, at least in part, the ability of E2 to impact such a diverse array of neural functions.
Introduction
Estradiol (E2) is best known as a regulator of the female reproductive tract and breast tissue, but it also affects neural functions. Some functions are directly related to reproduction (Mong and Pfaff, 2003; Petersen et al., 2003), but E2 also modulates autonomic responses (Du et al., 1995), sensory processing (Papka and Mowa, 2003), arousal (Morgan et al., 2004), spatial learning (Fugger et al., 1998), and mood and cognition (Miller, 2003; Shively and Bethea, 2004). Many of these E2 effects depend on activation of estrogen receptor α (ERα) or ERβ, ligand-gated transcription factors, but few neural transcriptional targets have been identified. Therefore, our ability to use genetic tools to understand the etiology and pathology of neurological disease, particularly in women, is limited.
Evidence suggests that genes related to GABA synthesis may be E2 targets in the nervous system. GABA is the most abundant inhibitory neurotransmitter in the brain, and it impacts most neural functions. In addition, GABAergic neurons throughout the brain contain ERα and/or ERβ (Flügge et al., 1986; Herbison et al., 1993; Blurton-Jones and Tuszynski, 2002; Ottem et al., 2004). Therefore, it is not surprising that E2 increases GABA release (Elkind-Hirsch et al., 1992; Fabre-Nys et al., 1994; Jarry et al., 1995; Luine et al., 1999; Mitsushima et al., 2002; Saleh and Connell, 2003), possibly by regulating glutamic acid decarboxylase (GAD) levels.
There are two GAD isoforms, GAD65 (encoded by the gad2 gene) and GAD67 (encoded by gad1) (Erlander et al., 1991). Both convert glutamate to GABA (Martin et al., 1991), but GAD65 is the isoform thought to regulate synaptic release of GABA (Kaufman et al., 1991; Martin and Rimvall, 1993). The other isoform, GAD67, is distributed throughout the neuron (Kaufman et al., 1991) and may support nonvesicular GABA release (Soghomonian and Martin, 1998). Importantly, E2 consistently elevates GAD65 (Weiland, 1992; McCarthy et al., 1995; Bosma et al., 2001; Curran-Rauhut and Petersen, 2002), but not GAD67 mRNA levels. In addition, the rat gad1 proximal promoter region contains no likely estrogen response elements (EREs) (Pedersen et al., 2001), whereas the gad2 promoter region has three putative EREs (Skak and Michelsen, 1999). It is unclear whether ERE sequences found in the gad2 promoter region are functional or what roles ERα or ERβ play in gad2 gene expression.
To address these issues, we examined the effects of E2 and other steroids on gad2 proximal promoter activity. We also compared the abilities of ERα and ERβ, alone or in combination, to mediate E2 induction of gad2 promoter activity. Finally, we used site-directed mutagenesis and chromatin immunoprecipitation (ChIP) to evaluate putative ERE sequences in the gad2 promoter.
Materials and Methods
General methods
Cell lines and culturing methods.
All cells were maintained at 37°C and 5% CO2 in DMEM supplemented with 10% (v/v) fetal bovine serum (FBS; Hyclone), 100 U/ml penicillin, 100 mg/ml streptomycin, and 2 mm l-glutamine (PS-Gln; Invitrogen, BRL). The medium for MCF-7 breast cancer cells (American Type Culture Collection) was further supplemented with 1 mg/ml insulin. Medium for SN56.B5.G4 cells derived from septal neurons (provided by Dr. Jan Krysztof Blusztajn, Boston University, Boston, MA) and N42 immortalized hypothalamic neurons (Cellutions Biosystems) was supplemented with 1 mm sodium pyruvate. COS-7 green monkey kidney cells (American Type Culture Collection) received no media supplements. In all cases, medium was changed every 48 h.
To verify that SN56.B5.G4 cells lose E2 responsiveness after the tenth passage (Watters and Dorsa, 1998), we examined E2 effects on the activity of a pGL3 ERE–Luc reporter vector (provided by Sallie Smith Schneider, University of Massachusetts Amherst, Amherst, MA) transiently transfected into SN56.B5.G4 cells of a passage number >20. We also used PCR and Western blots to verify ER status and E2 responsiveness of MCF-7 and N42 cells.
gad2 promoter construct.
We used reverse transcriptase-PCR and rat hypothalamic DNA to generate previously described promoter regions of the rat gad2 gene (accession #AF090195; bases 18–2708) (Skak and Michelsen, 1999). The forward primer was 5′-CCGGTACCGAGATTT-CTGGGTGGCGTGAA, and the reverse primer was 5′-TAGTCTGG-CGCTGGTCGC. The resulting 2691 bp fragment of the proximal gad2 promoter region was cloned into the pGL3 Basic vector (Promega).
Site-directed mutagenesis.
To determine the sites through which ERα and ERβ activate gad2 promoter activity, we performed site-directed mutagenesis. We searched the sequences 3000 bp upstream from the transcriptional start site for known EREs or ERE half-sites and then confirmed our results using the online tools Dragon ERE (Bajic et al., 2003) and Match (Biobase Biological Databases) (Goessling et al., 2001). We found putative ERE sequences at positions 776, 2023, and 2190 [National Center for Biotechnology Information (NCBI) accession #AF090195]. The gad2 promoter contains several transcriptional start sites (Skak and Michelsen, 1999), so to avoid ambiguity, we used the translational start site as the reference position (+1). Thus, the putative EREs were at −1958, −711, and −546 upstream from the translational start site (Fig. 1). We designed a series of oligonucleotide primers to specifically disrupt putative EREs at each of these positions. Mutations were as follows (mutations underlined): AGTTAAGGCGTAGGGATCCGATCTCTACCTTCCCTCAACTGC (for the −1958 site), CCTGGGCTCTTGAGGATCCAGAGATCTCCGCACGGGTTTGG (−711 site), and GTGCCCAGCTGCACCGAGGCCAGC (−546 site).
Figure 1.
Diagram of relative locations of three putative EREs in rat gad2 promoter sequence used in these studies. Site locations are mapped based on the location of the translational start site (Trl).
Each mutation introduced a new restriction site to aid in identification. Reagents and protocols from the GeneEditor in vitro Site-Directed Mutagenesis Kit (Promega) were used in conjunction with these oligonucleotides to introduce ERE mutations in the pGL3/gad2 promoter construct. This was accomplished by annealing kinased mutagenic oligonucleotides to the plasmid along with a kit-supplied oligonucleotide designed to alter antibiotic resistance. After mutant strand synthesis (using T4 DNA polymerase) and ligation, resultant plasmids were introduced into Escherichia coli and transformants selected using the novel antibiotic resistance. Further screening involved restriction analysis to assay for the mutant sites.
Transient transfections.
Using Superfect reagent (Qiagen), and following the manufacturer's protocols, we transiently transfected MCF-7, COS-7, and SN56.B5.G4 cells with pGL3 Basic vectors containing various gad2 promoter constructs. Twenty-four hours before transfection, cells were plated to produce a density of ∼80% confluence. A ratio of 5 μg pGL3 vector with promoter construct to 0.16 μg cytomegalovirus (CMV) renilla luciferase internal control plasmid (pRL–CMV; Superfect) was optimal for transfecting MCF-7 and COS-7 cells grown in 48-well plates. For SN56.B5.G4 cells, the optimal ratio was 0.68 μg reporter construct to 0.04 μg pRL–CMV plasmid. Two to three hours later, cells were rinsed with PBS and transferred to phenol red-free DMEM with 10% FBS (charcoal-stripped) and PS-Gln. Transfected cells were incubated for 18–24 h before receiving steroid treatments as described below. All transfections and assays were performed in triplicate and replicated at least three times.
Dual-luciferase assays.
Activity of the gad2 promoter reporter constructs was determined in lysates and analyzed using the Dual Luciferase Assay kit (Promega) and a TD 20/20 luminometer (Turner Designs), according to manufacturer's instructions.
Specific studies
Steroid regulation of gad2 promoter activity.
We performed initial studies of steroid effects on gad2 promoter activity in MCF-7 breast cancer cells. These cells contain androgen receptors (Andò et al., 2002), progestin receptors (Horwitz et al., 1978), and ERα but little or no ERβ (Chang et al., 2008). We performed a dose–response study of E2 effects on gad2 promoter activity using vehicle (ethanol), 0.01, 0.1, 1.0, and 10 nm E2 and lysing cells 24 h after steroid addition. We then performed a time course study wherein we collected lysates at 0, 4, 8, 12, 16, 24, and 48 h after treatment with 10 nm E2. Next, we compared effects of E2 on gad2 promoter activity with those of other gonadal steroids using 10 nm E2, testosterone (T), progesterone (P4), or combined E2 and P4. Similar studies were done in COS-7 cells as negative controls because these cells have no steroid receptors. MCF-7 cells containing the gad2 promoter construct were treated also with the pure antiestrogen, ICI 182,780 (100 nm) or vehicle. Six hours later, 10 nm E2 or vehicle was added, and cultures were incubated 42 h before lysates were collected.
ERα and ERβ regulation of gad2 promoter activity.
The gad2 gene is most commonly expressed in the nervous system; therefore, we followed up positive findings from studies of MCF-7 cells with studies in a mouse neural cell line, SN56.B5.G4 cells (described above). E2 did not activate a pGL3 ERE–Luc reporter construct in these cells without cotransfection with ER expression vectors. This allowed us to use SN56.B5.G4 cells to assess the roles of ERα and ERβ in gad2 promoter regulation. We performed a series of dose–response (1 nm through 100 nm) studies of gad2 promoter activity in cells cotransfected with pRST7–hERα and/or pRST7–hERβ expression vectors (provided by Julie Hall, Duke University Medical Center, Durham, NC).
Identification of functional EREs in gad2 promoter.
In MCF-7 cells, we compared the effects of 10 nm E2 on the reporter activity of wild-type and mutated gad2 promoter sequences prepared as described above. Mutations were at putative EREs found at −1958, −711, and −546 upstream from the translational start site. We performed similar studies in SN56.B5.G4 cells cotransfected with pRST7–hERα and/or pRST7–hERβ expression vectors.
We also investigated whether the three ERE sequences in the rat gad2 promoter were found in the gad2 promoters of other animal species for which there was data in the NCBI database. For these studies, we used BLAST (NCBI) (Altschul et al., 1990) to find the ERE sequences, then aligned identified ERE sequences with the rat promoter using CLUSTALW (1.81) (http://align.genome.jp) (Larkin et al., 2007).
ChIP assays.
To determine whether ERα binds to the ERE identified as a binding target in mutagenesis studies, we used N42 hypothalamic cells after verifying with PCR that they contain endogenous GAD2 mRNA (our unpublished observations). We focused on ERα binding to verify the importance of the ERE, because at the time of the studies, we were unable to find appropriate ERβ antibodies validated for ChIP, a problem described previously (Weitsman et al., 2006). Cells were plated at 1.90 × 106 density in 10 cm tissue culture dishes in DMEM without phenol red and supplemented with PS-Gln and 10% FBS overnight at 37°C with 5% CO2. The following day, N42 cells were treated with 10 nm E2 or vehicle and incubated for 2 h, after which they were treated with 1% formaldehyde in PBS for 10 min at room temperature on a platform shaker. The crosslinking reaction was stopped by adding glycine at a final concentration of 125 mm. Cells were rinsed twice with cold PBS before harvesting then resuspended in cell lysis buffer (5 mm Tris-HCl, pH 8.0, and 85 mm KCl) with 0.5% NP-40 and Protease Inhibitor Cocktail III (Calbiochem). After a 10 min incubation on ice, we collected nuclei by centrifugation and resuspended them in nuclear lysis buffer (50 mm Tris-HCl, pH 8.1, 10 mm EDTA, 1% SDS and Protease Inhibitor Cocktail III). Cell lysates were incubated on ice for 10 min then sonicated seven times for 10 s at 15% amplitude (Branson Ultrasonic Corporation) followed by centrifugation at 4°C. Sonicated cell lysate (30 μl) was used to verify that the average size of DNA fragments was in the range of 100–500 bp. DNA/protein complex (4 μg) was diluted in 0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.0, and 167 mm NaCl) to a final volume of 400 μl. The samples were next precleared with 40 μl 50% salmon sperm DNA/protein G bead slurry (Millipore Bioscience Research Reagents) for 30 min at 4°C. Supernatant (30 μl) was saved for total input control after preclearing. Immunoprecipitation was performed by adding 1 μg ERα antibody (Abcam) and then incubating at 4°C overnight with rotation. Samples with no antibody were run in parallel as negative controls.
The following day, 40 μl 50% salmon sperm DNA/protein G beads slurry was added and incubated for 2 h at 4°C to capture the immune complexes. Proteins and DNA nonspecifically associated with the beads were removed by sequentially washing with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.0, 150 mm NaCl), high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.0, 500 mm NaCl), and LiCl washing buffer (0.25 m LiCl; 1% deoxycholate sodium salt, 1 mm EDTA, 10 mm Tris-HCl, pH 8.0, and 1% IGEPAL-CA630; MP Biomedicals), and twice with Tris-EDTA buffer (10 mm Tris-HCl, 1 mm EDTA, pH 8.0) at 4°C for 12 min each wash. The immunoprecipitated DNA–protein complexes were eluted from beads with 1% SDS in 0.1 m NaHCO3. Then the DNA–protein complexes were incubated in 0.2 m NaCl at 65°C overnight followed by proteinase K digestion for 2 h at 45°C to reverse cross linking. After protein removal, DNA was purified by phenol/chloroform extraction followed by EtOH precipitation.
Quantitative PCR analysis of ChIP assays.
To quantify ChIP DNA, we used a MX3500P real-time quantitative PCR (QPCR) system (Stratagene) and designed to flank the putative ERE of the mouse gad2 promoter (accession #AB032757) at −693 that align with the rat −711 ERE. The primers were 5′-ATGAGTTCGTTGGTGTGGAAG (forward) and 5′-AGTGCTGAGGTCGCTGTG (reverse). A second set of primers was designed to flank the putative ERE at −532 of the mouse promoter that aligns with the rat −546 ERE. The sequences of those primers were 5′-GCCAAGGTCTCCCTGTCC (forward) and 5′-TGCCTGCGTA-GTGCTGAC. In addition, negative control primers were designed for a part of the coding region of the mouse gad2 gene devoid of an identifiable ERE (accession #NM_008078, bases 2402–2610). The sequences of these primers were 5′-TCAAAACCAACAGGAAACATCA (forward) and 5′-TTCAAGGGTGATTTGGCACT (reverse).
Each 15 μl DNA amplification reaction contained 5 mm Tris HCl, 5 mm Tris-base, 50 mm KCl, 3 mm MgCl2, 0.01% Tween 20, 0.8% glycerol, 40,000-fold dilution of SYBR Green I (Invitrogen), 200 μm dNTPs, 50 nm ROX reference dye (Stratagene), 5 pmol of each primer, 2 μl of DNA, and 1.0 U Platinum TaqDNA polymerase (Invitrogen). The QPCR conditions were 95°C (5 min), followed by 40 cycles of 95°C (15 s), 55–67°C (30 s) for annealing and 72°C (30 s) for extension. We included one sample without cDNA template to establish the specificity of target cDNA amplification, and we verified identity of products by electrophoresis on 2% agarose gels and sequence confirmation. In these studies, we ran three separate ChIP reactions in duplicate for each of the three targets (negative control, −693 and −532), as well as a sample with no antibody and one for total input for each target.
Statistical analyses.
We first obtained means for each treatment from at least three replicates of three or more independent runs for each promoter reporter assay. Treatment effects were evaluated using one-way ANOVA or two-way ANOVA, depending on the number of independent variables. We used Bonferroni t tests for post hoc analyses to probe treatment effects when a significant interaction of main effects was detected by two-way ANOVA.
We analyzed QPCR data for ChIP studies using the ΔΔCt method (Livak and Schmittgen, 2001). For these analyses, duplicates for each sample were averaged and corrected for background by subtracting values of the no antibody controls. Fold enrichment was determined for each sample by comparing with total input controls. To facilitate comparisons, data were expressed as fold induction over the vehicle controls. Finally, means of the vehicle group and E2-treated group for each DNA target were compared using Student's t tests.
Results
E2, but not other gonadal steroids, stimulated gad2 promoter activity in vitro
MCF-7 cells
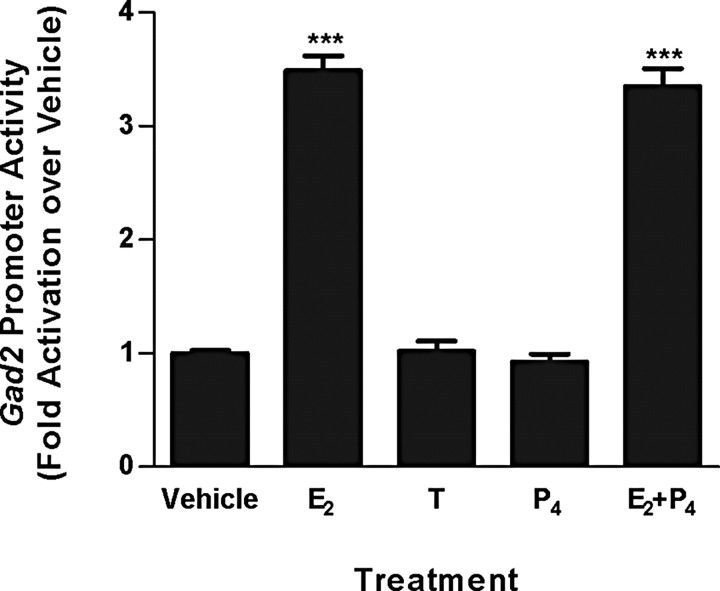
E2, but not T or P4, increased gad2 promoter reporter activity when the reporter construct was expressed in MCF-7 cells (Fig. 2). Results of dose–response studies showed that E2 increased gad2 promoter activity in a dose-dependent manner, reaching saturation at 10−8 m (data not shown). Time course studies showed that promoter activity peaked 12 h after initiation of E2 treatment and remained elevated for at least 48 h (data not shown). The stimulatory effect of E2 on gad2 promoter activity was blocked by pretreatment with the ER antagonist, ICI 182,780 (data not shown).
Figure 2.
Effects of 10 nm E2, T, P4, or E2 plus P4 (E2 pretreatment followed 6 h later with P4) on rat gad2 promoter activity. gad2 promoter reporter construct was transiently transfected into MCF-7 cells, and cells were treated for 24 h with steroids after 18 h incubation in steroid-free media. Reporter activity was measured using a dual-luciferase assay with normalization to a renilla luciferase control. Each bar represents treatment mean ± SEM of at least nine samples. ***Significantly different from vehicle; p < 0.0001.
SN56.B5.G4 cells
As reported previously (Watters and Dorsa, 1998), SN56.B5.G4 cells were unresponsive to E2 unless transfected with ER expression vectors. E2 had no effect on reporter activity in the absence of ER, and the stimulatory effect of E2 was completely blocked by pretreatment with ICI 182,780 (data not shown). In log dose–response studies (10−12 to 10−7 m; data not shown), we found that E2-induced gad2 promoter reporter activity peaked at 10−8 m, regardless of whether cells were transfected with hERα, hERβ, or both expression vectors; however, activity was generally higher when both receptors were present.
EREs are important for cell-specific regulation of the gad2 promoter
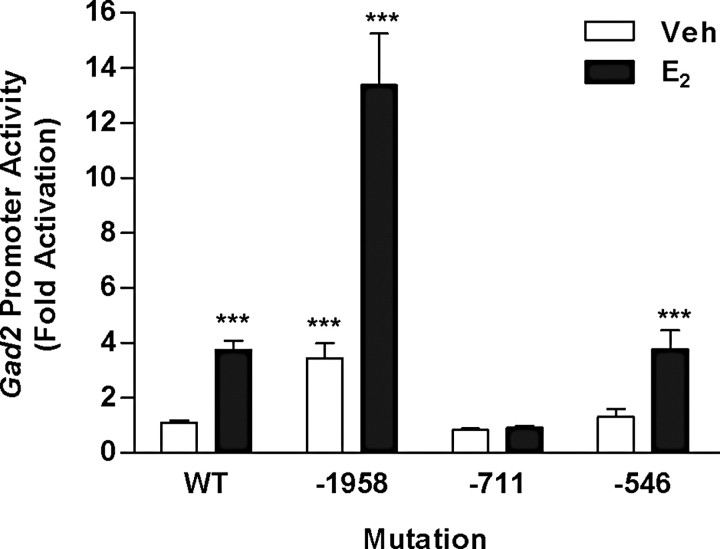
Mutations of putative EREs in the gad2 promoter resulted in different responses to E2 in MCF-7 breast cancer cells compared with SN56.B5.G4 neural cells. In MCF-7 cells, only mutation of the −711 site abrogated transactivational effects of E2 (Fig. 3). Importantly, mutation of the −1958 ERE increased basal transcriptional levels significantly and also increased E2 responsiveness from less than fourfold to over 13-fold.
Figure 3.
E2 activation of gad2 wild-type (WT) promoter and promoter constructs with mutated EREs. Activation was assessed using dual-luciferase assays. Designation of constructs is based on the positions of the mutated ERE upstream of the translational start site (see Materials and Methods). Constructs were transiently transfected into MCF-7 cells and treated with ethanol vehicle (Veh) or 10 nm E2 for 24 h. Values are expressed relative to the wild-type construct in cells treated with vehicle. Each bar represents the mean ± SEM of values from at least nine independent samples. ***Significantly different from WT, vehicle control (p < 0.001).
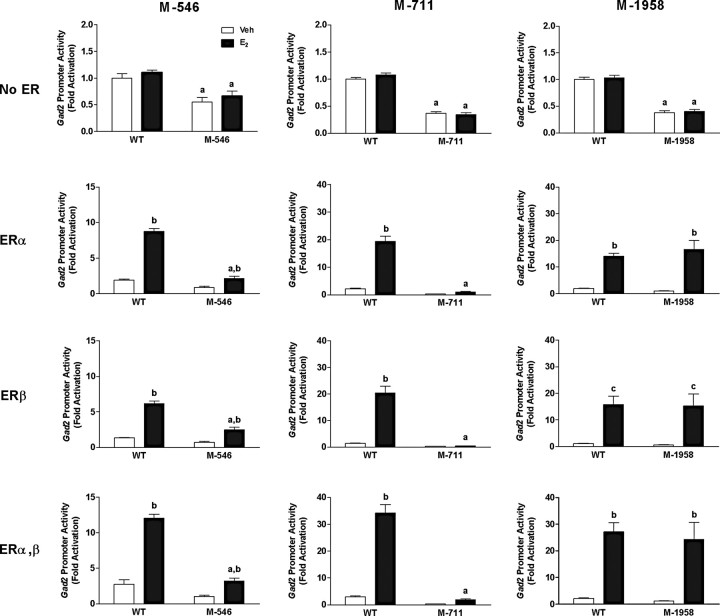
Unlike responses seen in MCF-7 cells, we found that in SN56.B5.G4 cells, mutation of the −546 site repressed E2 transactivation when hERα, hERβ, or both expression vectors were present (Fig. 4, left column). However, mutation of the −711 site completely abolished E2 responsiveness of the gad2 promoter when the construct was expressed in SN56.B5.G4 cells (Fig. 4, middle column), as it did when tested in MCF-7 cells (Fig. 3). Unlike the dramatic stimulatory effect seen in MCF-7 cells when the −1958 site was mutated in MCF7 cells, the same mutation of the gad2 promoter had no effect on E2 transactivation in SN56.B5.G4 neural cells (Fig. 4, right column). Finally, in contrast to the responses observed in MCF-7 cells, basal promoter activity (no ER) decreased in SN56.B5.G4 cells when any of the three EREs was mutated (Fig. 4, top row).
Figure 4.
Effects of E2 on reporter activity of a wild-type (WT) gad2 promoter construct or constructs with mutations of ERE sequences at −549, −711, and −1958 upstream of the gad2 translational start site (M-549, left column; M-711, middle column; and −1958, right column; for details, see Materials and Methods). Constructs were expressed in SN56.B5.G4 neural cells that were transiently transfected with empty vector (No ER, top row), hERα (second row), hERβ (third row), or both hERα and hERβ (ERαβ, bottom row). Cells were treated with 10 nm E2 or ethanol vehicle (Veh) for 24 h before assaying reporter activity using dual-luciferase assays. The induction of promoter activity was expressed as fold activation compared with vehicle control. Each bar represents the mean ± SEM of values from at least nine independent samples. aSignificantly different from corresponding WT (p < 0.001); bsignificantly different from corresponding Veh control (p < 0.001); csignificantly different from corresponding Veh control (p < 0.01).
Rat gad2–711 ERE is similar to the consensus ERE and aligns with similar sites in several species
The structures of the various imperfect EREs found in the gad2 promoter, as well as the vitellogenin and consensus EREs, are shown in Table 1. Database analysis of the rat gad2 promoter (AF090195) showed that the sequence of the ERE at −711 differs from the consensus ERE at the +1 position, the −546 site differs at the +3 position, and the −1958 site at the −5 position.
Table 1.
Structures of imperfect EREs found in the rat gad2 DNA sequence between +1 and −3000 bp upstream of the translational start site
| −5 | −4 | −3 | −2 | −1 | N | N | N | +1 | +2 | +3 | +4 | +5 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −546 | 5′ | G | G | T | C | A | C | C | G | A | G | G | C | C | 3′ |
| −711 | G | G | T | C | A | C | A | G | C | G | A | C | C | ||
| −1958 | G | G | T | C | A | A | G | A | T | G | A | C | T | ||
| Vitellogenin A2 | G | G | T | C | A | C | A | G | T | G | A | C | C | ||
| Consensus ERE | G | G | T | C | A | N | N | N | T | G | A | C | C |
The first column denotes the site of the ERE in the promoter region. The consensus and vitellogenin A2 ERE sequences are shown in the last two rows for comparison. Bold indicates differences between gad2 and consensus EREs. N, Any nucleotide.
Alignment of rat, mouse (Rattus norvegicus, Mus musculus; AB032757), dog (Canis lupus familiaris; NW_876290), horse (Equus caballus; NW_001867401), rhesus monkey (Macaca mulatta; NW_001124107), chimpanzee (Pan troglodytes; NW_001220649), and human (Homo sapiens; NT_008705) gad2 promoter regions are shown in Figure 5. Numbering of sites is relative to the known translational start sites for rat, mouse, and human and to predicted translational start sites for dog, horse, rhesus monkey, and chimpanzee. The site we found most important for E2 induction of gad2 promoter activity, the rat −711 site, is identical with sites at −693 in mouse, −752 in dog, and −730 in horse. A site at −713 of the rhesus monkey differs from the rat sequence by one base (A to G at −1 position). Sites at −702 of the chimpanzee and −708 in human were identical to one another and differed from the rat sequence by two bases (A to G at −1; C to T at +1). The reverse palindrome sequences of the rat −546 and −1958 EREs are similar to sites at −532 and −1838 of the mouse gad2 promoter, and each differs by only one base in the intervening 3-base sequence. Sequences of dog and horse that align with the rat −546 site may be ERE half-sites, but the sites in monkey, chimpanzee, and human do not appear to be either EREs or ERE half-sites. BLAST analysis showed that the region containing the rat −1958 ERE was not sufficiently homologous to nonrodent species to allow sequence alignment.
Figure 5.
Promoter sequence alignment for gad2 promoters of rat (AF090195), mouse (AB032757), dog (NW_876290), horse (NW_001867401), rhesus monkey (NW_001124107), chimpanzee (NW_001220649), and human (NT_008705AD2) using CLUSTALW (1.81) multiple alignment software. Designations of −1958, −711, and −546 refer to positions of putative EREs of rat GAD2 promoter relative to the translational start site. Shading of bases denotes identity with bases of the rat sequence, and asterisks indicate sites conserved among all species compared. BLAST analysis found insufficient homology to allow alignment between rodent and nonrodent species in the region containing the rat −1958 site.
E2 increases ERα binding to DNA fragments containing ERE sequences
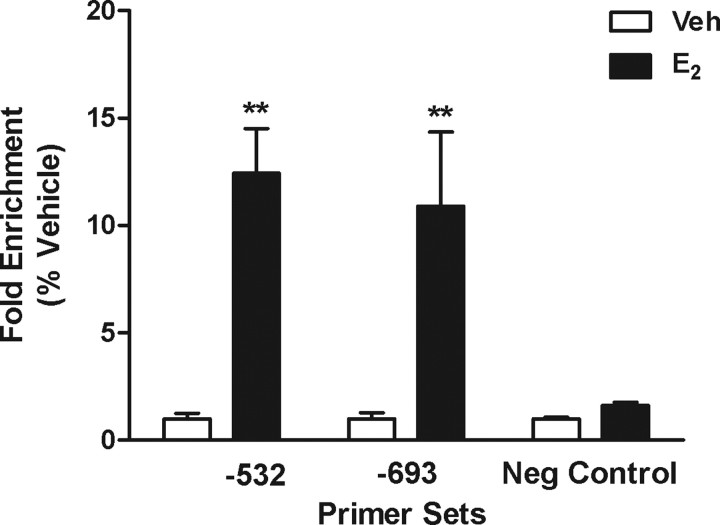
Results of ChIP assays using ERα antibody showed that fragments containing the −532 and −693 ERE sequences were significantly enriched in DNA samples from E2-treated N42 cells compared with DNA from vehicle-treated cells (Fig. 6). No enrichment was seen using negative control primers.
Figure 6.
Results of ChIP using ERα antibody and DNA isolated from N42 cells. Cells were treated with ethanol vehicle (Veh) or E2 for 2 h before they were harvested for ChIP assays. Samples were assayed using primers flanking the −532 ERE and the −693 ERE or a site with no ERE consensus sequences (for primer sequences, see Materials and Methods). Each data point represents the mean ± SEM of three independent samples, each analyzed in duplicate. **Significantly different from corresponding vehicle control; p < 0.01. Neg, Negative.
Discussion
Our findings show that the gad2 promoter is a direct target of E2 and activated through the canonical pathway by ERα and ERβ. We identified three novel EREs within the first 3000 bp upstream of the translational start site of the gad2 promoter. Mutational and ChIP analyses showed that two of these sites were important for promoter activation by E2 in neural cells. Importantly, a third ERE was a strong repressor of E2 transactivation in breast cancer cells. Although either ERα or ERβ could mediate transactivation of the gad2 promoter, we found no evidence for ERβ repression of ERα-mediated transcription. Together, these findings show that the gad2 gene is a direct transcriptional target of E2 regulated, at least in part, by the canonical ER pathway in a tissue-specific manner. In view of the widespread colocalization of ER and GAD2, our findings may explain, at least in part, the effects of E2 on diverse neural functions.
Consistent with our in vitro findings, previous studies showed that E2 increases GAD2 mRNA levels in vivo, specifically in neuronal populations that contain ER. For example, in the rat CA1, an ER-containing region of the hippocampus, GAD2 mRNA levels decline after ovariectomy and then increase after E2 replacement (Nakamura et al., 2004). Another intriguing report suggests that estrogen synthesized locally also upregulates gad2 gene expression in the hippocampus (Ikeda et al., 2006). Similarly, in the rat anteroventral periventricular nucleus (AVPV), nearly all GABAergic neurons contain ERα mRNA and many also contain ERβ mRNA (Ottem et al., 2004). E2 consistently elevates GAD2 mRNA levels in the ER-rich AVPV but not in a more rostral preoptic region where ER expression is found only in scattered GABAergic cells (Curran-Rauhut and Petersen, 2002). These in vivo findings support our contention that the rat gad2 promoter region is a direct target of ERs.
Results of our mutational and ChIP analyses indicate that E2 regulates the gad2 promoter through the classical pathway wherein ER binds to ERE sequences (Matthews and Gustafsson, 2003; Dietz and Carroll, 2008), although nonclassical mechanisms also may be involved in the expression of the gene in vivo. The ERE most important for regulating gad2 promoter activity was −711 upstream of the translational start site of the rat promoter and identical to ones in the mouse, as well as the predicted promoters of dog and horse. Alignment studies also detected a similar site in the predicted gad2 promoters of rhesus monkey, chimpanzee, and in the human promoter. The ERE in the rhesus monkey promoter differed from the rat sequence by one base and the sites in chimpanzee and human by two bases. Notably, differences among species were at either the −1 or +1 position. We detected no likely ERE sequences that aligned with either the rat −546 or −1958 sites in any of the nonrodent animals. These findings provide further evidence that the highly conserved site at −711 of the rat is critical to E2 regulation of the gad2 promoter.
The other ERE active in the rat gad2 promoter was at −546 and identical to an ERE at −532 in the mouse gene. ChIP assays in mouse N42 cells showed E2 induction of ERα binding to DNA fragments quantified using QPCR and primers flanking either the −532 or the more highly conserved −693 ERE. Considering the proximity of these sequences and the size of fragments used in the ChIP assays, it is not possible to determine whether binding differs between the −693 and −532 sites. However, EREs separated by >2000 base pairs can synergistically activate a promoter (Sathya et al., 1997), so both sites may contribute to activation of the native rodent gad2 promoter.
Despite the extensive colocalization of ERα and ERβ, little is known about the relative contributions of the two isoforms in regulating neural gene expression. Some have suggested that ERβ acts primarily as a dominant repressor of ERα transactivation (Pettersson et al., 2000; Liu et al., 2002), particularly when E2 levels are not saturating (Hall and McDonnell, 1999). In contrast, we found that ERβ stimulated, rather than repressed, gad2 gene expression. Consistent with our findings, microarray studies from other laboratories identified genes activated solely by ERβ and others in which ERβ enhances ERα-induced expression (Monroe et al., 2005; Chang et al., 2006). These data support our findings that ERβ can potently stimulate gene transcription independent of ERα.
E2 induction of gad2 promoter activity was significantly lower in MCF-7 breast cancer cells than in SN56.B5.G4 neural cells. MCF-7 cells lack ERβ, but the differences persist even when we compare MCF-7 cells with SN56.B5.G4 cells lacking ERβ. It may be that the tissue-specific responses are attributable to differential utilization of EREs or to the complement of ERα in the two cell lines. Interestingly, the ERE at −1958 was inactive in SN56.B5.G4 cells but served as a potent repressor in MCF-7 cells. In fact, after mutating the −1958 site, E2 stimulated gad2 promoter activity in MCF-7 cells to nearly the same extent as wild-type promoter activation in SN56.B5.G4 neural cells. Future studies will evaluate possible mechanisms underlying the ability of the −1958 site to repress gad2 gene transcription in MCF-7 cells.
E2 alters expression of many neural genes, but few are known to be regulated directly through EREs in the promoter region. Until recently, it was assumed that such genes had not yet been identified, but this view is changing, as investigators interrogate the entire genome for ER binding sites. Researchers using whole-genome ChIP-based strategies made the surprising discovery that only 4% of ERα binding sites map to the proximal promoter region 0–1 kb upstream of the transcriptional start site (Carroll et al., 2006) or 5% to the 0–5 kb upstream region (Lin et al., 2007). Interestingly, EREs adjacent to E2-upregulated, but not E2-downregulated genes, are enriched in the proximal promoter region, particularly around the transcriptional start site (Lin et al., 2007). If the distribution of EREs in the neural transcriptome is similar, our finding that the gad2 proximal promoter has two stimulatory EREs would suggest that the gad2 gene is an unusual and important neural target of E2.
The finding that E2 directly targeted the gad2 gene is notable, because GAD65 maintains levels of GABA used in vesicular release (Soghomonian and Martin, 1998). Considering the ubiquitous distribution and myriad functions of GABA, regulation of gad2 gene expression may explain how E2 is able to regulate such a wide array of neural functions. Two regions in particular, the AVPV and bed nucleus of the stria terminalis (BNST), have among the highest concentrations of ERα- and ERβ-containing neurons in the brain (Simerly et al., 1990; Shughrue et al., 1997). In the AVPV, virtually all ER expression is in GABAergic neurons (Ottem et al., 2004), and E2 tonically elevates gad2 gene expression in these cells (Curran-Rauhut and Petersen, 2002). In turn, tonic elevation of GABA release plays a key role in regulating E2-dependent gonadotropin release during the estrous cycle (Herbison, 1998; Petersen et al., 2003). E2 also regulates gad2 gene expression (Pompolo et al., 2002) in the BNST, a nucleus important for stress responses. Disruption of gad2 expression results in anxiety-like behavior (Stork et al., 2000), whereas E2 attenuates stress responses (Gerrits et al., 2005). The underlying mechanism may be through E2 activation of GABA release from BNST neurons that project to corticotrophin-releasing hormone neurons in the medial parvocellullar subdivision of the paraventricular nucleus (Miklos and Kovacs, 2002).
In addition to the AVPV and BNST, ER and GAD2 are coexpressed in neurons throughout the hypothalamus, as well as in regions of the hippocampus, amygdala, and brainstem. In these regions, GABA and E2 modulate such diverse functions as feeding behavior (Butera et al., 1990; Meister, 2007), spatial memory (Spencer et al., 2008), emotional responses (Krezel et al., 2001), and autonomic functions (Saleh and Connell, 2003). Finally, gad2 gene expression is found in ER-containing cells of non-neural tissues, including β-cells of the pancreas (Franklin and Wollheim, 2004; Alonso-Magdalena et al., 2008), epiphyseal chondrocytes (Nilsson et al., 1999; Tamayama et al., 2001), and oviductal epithelial cells (Tillakaratne et al., 1992; Wang et al., 2000). Thus, it is tempting to speculate that the relationship between ER and GAD2 is widespread and important both inside and outside the nervous system.
In summary, these findings show that E2 directly stimulates gad2 promoter activity through the canonical ER pathway. Transcriptional activation in neural cells required ERα or ERβ, as well as two EREs located within the proximal promoter region. Importantly, a third ERE was inactive in neural cells but had potent repressor activity in breast cancer cell lines. Considering the codistribution patterns of GAD2 and ERs in the brain, direct regulation of the gad2 gene by E2 may explain the wide range of neural functions modulated by this steroid.
Footnotes
This research was funded by National Institutes of Health Grant HD027305 to S.L.P.
References
- Alonso-Magdalena et al., 2008.Alonso-Magdalena P, Ropero AB, Carrera MP, Cederroth CR, Baquié M, Gauthier BR, Nef S, Stefani E, Nadal A. Pancreatic insulin content regulation by the estrogen receptor ER alpha. PLoS ONE. 2008;3:e2069. doi: 10.1371/journal.pone.0002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul et al., 1990.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andò et al., 2002.Andò S, De Amicis F, Rago V, Carpino A, Maggiolini M, Panno ML, Lanzino M. Breast cancer: from estrogen to androgen receptor. Mol Cell Endocrinol. 2002;193:121–128. doi: 10.1016/s0303-7207(02)00105-3. [DOI] [PubMed] [Google Scholar]
- Bajic et al., 2003.Bajic VB, Tan SL, Chong A, Tang S, Ström A, Gustafsson JA, Lin CY, Liu ET. Dragon ERE finder version 2: a tool for accurate detection and analysis of estrogen response elements in vertebrate genomes. Nucleic Acids Res. 2003;31:3605–3607. doi: 10.1093/nar/gkg517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blurton-Jones and Tuszynski, 2002.Blurton-Jones M, Tuszynski MH. Estrogen receptor-beta colocalizes extensively with parvalbumin-labeled inhibitory neurons in the cortex, amygdala, basal forebrain, and hippocampal formation of intact and ovariectomized adult rats. J Comp Neurol. 2002;452:276–287. doi: 10.1002/cne.10393. [DOI] [PubMed] [Google Scholar]
- Bosma et al., 2001.Bosma PT, Blázquez M, Fraser EJ, Schulz RW, Docherty K, Trudeau VL. Sex steroid regulation of glutamate decarboxylase mRNA expression in goldfish brain is sexually dimorphic. J Neurochem. 2001;76:945–956. doi: 10.1046/j.1471-4159.2001.00086.x. [DOI] [PubMed] [Google Scholar]
- Butera et al., 1990.Butera PC, Beikirch RJ, Willard DM. Changes in ingestive behaviors and body weight following intracranial application of 17 alpha-estradiol. Physiol Behav. 1990;47:1291–1293. doi: 10.1016/0031-9384(90)90386-i. [DOI] [PubMed] [Google Scholar]
- Carroll et al., 2006.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Chang et al., 2006.Chang EC, Frasor J, Komm B, Katzenellenbogen BS. Impact of estrogen receptor beta on gene networks regulated by estrogen receptor alpha in breast cancer cells. Endocrinology. 2006;147:4831–4842. doi: 10.1210/en.2006-0563. [DOI] [PubMed] [Google Scholar]
- Chang et al., 2008.Chang EC, Charn TH, Park SH, Helferich WG, Komm B, Katzenellenbogen JA, Katzenellenbogen BS. Estrogen receptors alpha and beta as determinants of gene expression: influence of ligand, dose, and chromatin binding. Mol Endocrinol. 2008;22:1032–1043. doi: 10.1210/me.2007-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran-Rauhut and Petersen, 2002.Curran-Rauhut MA, Petersen SL. Regulation of glutamic acid decarboxylase 65 and 67 gene expression by ovarian steroids: identification of two functionally distinct populations of GABA neurons in the preoptic area. J Neuroendocrinol. 2002;14:310–317. doi: 10.1046/j.1365-2826.2002.00780.x. [DOI] [PubMed] [Google Scholar]
- Dietz and Carroll, 2008.Dietz SC, Carroll JS. Interrogating the genome to understand oestrogen-receptor-mediated transcription. Expert Rev Mol Med. 2008;10:e10. doi: 10.1017/S1462399408000653. [DOI] [PubMed] [Google Scholar]
- Du et al., 1995.Du XJ, Riemersma RA, Dart AM. Cardiovascular protection by oestrogen is partly mediated through modulation of autonomic nervous function. Cardiovasc Res. 1995;30:161–165. [PubMed] [Google Scholar]
- Elkind-Hirsch et al., 1992.Elkind-Hirsch KE, Wallace E, Stach BA, Jerger JF. Cyclic steroid replacement alters auditory brainstem responses in young women with premature ovarian failure. Hear Res. 1992;64:93–98. doi: 10.1016/0378-5955(92)90171-i. [DOI] [PubMed] [Google Scholar]
- Erlander et al., 1991.Erlander MG, Tillakaratne NJ, Feldblum S, Patel N, Tobin AJ. Two genes encode distinct glutamate decarboxylases. Neuron. 1991;7:91–100. doi: 10.1016/0896-6273(91)90077-d. [DOI] [PubMed] [Google Scholar]
- Fabre-Nys et al., 1994.Fabre-Nys C, Blache D, Hinton MR, Goode JA, Kendrick KM. Microdialysis measurement of neurochemical changes in the mediobasal hypothalamus of ovariectomized ewes during oestrus. Brain Res. 1994;649:282–296. doi: 10.1016/0006-8993(94)91075-8. [DOI] [PubMed] [Google Scholar]
- Flügge et al., 1986.Flügge G, Oertel WH, Wuttke W. Evidence for estrogen-receptive GABAergic neurones in the preoptic/anterior hypothalamic area of the rat brain. Neuroendocrinology. 1986;43:1–5. doi: 10.1159/000124500. [DOI] [PubMed] [Google Scholar]
- Franklin and Wollheim, 2004.Franklin IK, Wollheim CB. GABA in the endocrine pancreas: its putative role as an islet cell paracrine-signalling molecule. J Gen Physiol. 2004;123:185–190. doi: 10.1085/jgp.200409016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugger et al., 1998.Fugger HN, Cunningham SG, Rissman EF, Foster TC. Sex differences in the activational effect of ERalpha on spatial learning. Horm Behav. 1998;34:163–170. doi: 10.1006/hbeh.1998.1475. [DOI] [PubMed] [Google Scholar]
- Gerrits et al., 2005.Gerrits M, Grootkarijn A, Bekkering BF, Bruinsma M, Den Boer JA, Ter Horst GJ. Cyclic estradiol replacement attenuates stress-induced c-Fos expression in the PVN of ovariectomized rats. Brain Res Bull. 2005;67:147–155. doi: 10.1016/j.brainresbull.2005.06.021. [DOI] [PubMed] [Google Scholar]
- Goessling et al., 2001.Goessling E, Okel-Margoulis OV, Kel AE, Wingender E. MATCHTM–a tool for searching transcription factor binding sites in DNA sequences. Applications for the analysis of human chromosomes. Paper presented at the German Conference on Bioinformatics; October; Braunschweig, Germany. 2001. [Google Scholar]
- Hall and McDonnell, 1999.Hall JM, McDonnell DP. The estrogen receptor beta-isoform (ERbeta) of the human estrogen receptor modulates ERalpha transcriptional activity and is a key regulator of the cellular response to estrogens and antiestrogens. Endocrinology. 1999;140:5566–5578. doi: 10.1210/endo.140.12.7179. [DOI] [PubMed] [Google Scholar]
- Herbison, 1998.Herbison AE. Multimodal influence of estrogen upon gonadotropin-releasing hormone neurons. Endocr Rev. 1998;19:302–330. doi: 10.1210/edrv.19.3.0332. [DOI] [PubMed] [Google Scholar]
- Herbison et al., 1993.Herbison AE, Robinson JE, Skinner DC. Distribution of estrogen receptor-immunoreactive cells in the preoptic area of the ewe: co-localisation with glutamic acid decarboxylase but not luteinizing hormone-releasing hormone. Neuroendocrinology. 1993;57:751–759. doi: 10.1159/000126433. [DOI] [PubMed] [Google Scholar]
- Horwitz et al., 1978.Horwitz KB, Koseki Y, McGuire WL. Estrogen control of progesterone receptor in human breast cancer: role of estradiol and antiestrogen. Endocrinology. 1978;103:1742–1751. doi: 10.1210/endo-103-5-1742. [DOI] [PubMed] [Google Scholar]
- Ikeda et al., 2006.Ikeda T, Matsuki N, Yamada MK. Estrogen produced in cultured hippocampal neurons is a functional regulator of a GABAergic machinery. J Neurosci Res. 2006;84:1771–1777. doi: 10.1002/jnr.21083. [DOI] [PubMed] [Google Scholar]
- Jarry et al., 1995.Jarry H, Leonhardt S, Schwarze T, Wuttke W. Preoptic rather than mediobasal hypothalamic amino acid neurotransmitter release regulates GnRH secretion during the estrogen-induced LH surge in the ovariectomized rat. Neuroendocrinology. 1995;62:479–486. doi: 10.1159/000127037. [DOI] [PubMed] [Google Scholar]
- Kaufman et al., 1991.Kaufman DL, Houser CR, Tobin AJ. Two forms of the gamma-aminobutyric acid synthetic enzyme glutamate decarboxylase have distinct intraneuronal distributions and cofactor interactions. J Neurochem. 1991;56:720–723. doi: 10.1111/j.1471-4159.1991.tb08211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krezel et al., 2001.Krezel W, Dupont S, Krust A, Chambon P, Chapman PF. Increased anxiety and synaptic plasticity in estrogen receptor beta-deficient mice. Proc Natl Acad Sci U S A. 2001;98:12278–12282. doi: 10.1073/pnas.221451898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin et al., 2007.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lin et al., 2007.Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, Yeo A, George J, Kuznetsov VA, Lee YK, Charn TH, Palanisamy N, Miller LD, Cheung E, Katzenellenbogen BS, Ruan Y, et al. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al., 2002.Liu MM, Albanese C, Anderson CM, Hilty K, Webb P, Uht RM, Price RH, Jr, Pestell RG, Kushner PJ. Opposing action of estrogen receptors alpha and beta on cyclin D1 gene expression. J Biol Chem. 2002;277:24353–24360. doi: 10.1074/jbc.M201829200. [DOI] [PubMed] [Google Scholar]
- Livak and Schmittgen, 2001.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luine et al., 1999.Luine VN, Wu V, Hoffman CS, Renner KJ. GABAergic regulation of lordosis: influence of gonadal hormones on turnover of GABA and interaction of GABA with 5-HT. Neuroendocrinology. 1999;69:438–445. doi: 10.1159/000054447. [DOI] [PubMed] [Google Scholar]
- Martin and Rimvall, 1993.Martin DL, Rimvall K. Regulation of gamma-aminobutyric acid synthesis in the brain. J Neurochem. 1993;60:395–407. doi: 10.1111/j.1471-4159.1993.tb03165.x. [DOI] [PubMed] [Google Scholar]
- Martin et al., 1991.Martin DL, Martin SB, Wu SJ, Espina N. Regulatory properties of brain glutamate decarboxylase (GAD): the apoenzyme of GAD is present principally as the smaller of two molecular forms of GAD in brain. J Neurosci. 1991;11:2725–2731. doi: 10.1523/JNEUROSCI.11-09-02725.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews and Gustafsson, 2003.Matthews J, Gustafsson JA. Estrogen signaling: a subtle balance between ER alpha and ER beta. Mol Interv. 2003;3:281–292. doi: 10.1124/mi.3.5.281. [DOI] [PubMed] [Google Scholar]
- McCarthy et al., 1995.McCarthy MM, Kaufman LC, Brooks PJ, Pfaff DW, Schwartz-Giblin S. Estrogen modulation of mRNA levels for the two forms of glutamic acid decarboxylase (GAD) in female rat brain. J Comp Neurol. 1995;360:685–697. doi: 10.1002/cne.903600412. [DOI] [PubMed] [Google Scholar]
- Meister, 2007.Meister B. Neurotransmitters in key neurons of the hypothalamus that regulate feeding behavior and body weight. Physiol Behav. 2007;92:263–271. doi: 10.1016/j.physbeh.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Miklos and Kovacs, 2002.Miklos IH, Kovacs KJ. GABAergic innervation of corticotropin-releasing hormone (CRH)-secreting parvocellular neurons and its plasticity as demonstrated by quantitative immunoelectron microscopy. Neuroscience. 2002;113:581–592. doi: 10.1016/s0306-4522(02)00147-1. [DOI] [PubMed] [Google Scholar]
- Miller, 2003.Miller KJ. The other side of estrogen replacement therapy: outcome study results of mood improvement in estrogen users and nonusers. Curr Psychiatry Rep. 2003;5:439–444. doi: 10.1007/s11920-003-0082-5. [DOI] [PubMed] [Google Scholar]
- Mitsushima et al., 2002.Mitsushima D, Shwe TT, Funabashi T, Shinohara K, Kimura F. GABA release in the medial preoptic area of cyclic female rats. Neuroscience. 2002;113:109–114. doi: 10.1016/s0306-4522(02)00160-4. [DOI] [PubMed] [Google Scholar]
- Mong and Pfaff, 2003.Mong JA, Pfaff DW. Hormonal and genetic influences underlying arousal as it drives sex and aggression in animal and human brains. Neurobiol Aging. 2003;24(Suppl 1):S83–S88. doi: 10.1016/s0197-4580(03)00053-8. discussion S91–S82. [DOI] [PubMed] [Google Scholar]
- Monroe et al., 2005.Monroe DG, Secreto FJ, Subramaniam M, Getz BJ, Khosla S, Spelsberg TC. Estrogen receptor alpha and beta heterodimers exert unique effects on estrogen- and tamoxifen-dependent gene expression in human U2OS osteosarcoma cells. Mol Endocrinol. 2005;19:1555–1568. doi: 10.1210/me.2004-0381. [DOI] [PubMed] [Google Scholar]
- Morgan et al., 2004.Morgan MA, Schulkin J, Pfaff DW. Estrogens and non-reproductive behaviors related to activity and fear. Neurosci Biobehav Rev. 2004;28:55–63. doi: 10.1016/j.neubiorev.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Nakamura et al., 2004.Nakamura NH, Rosell DR, Akama KT, McEwen BS. Estrogen and ovariectomy regulate mRNA and protein of glutamic acid decarboxylases and cation-chloride cotransporters in the adult rat hippocampus. Neuroendocrinology. 2004;80:308–323. doi: 10.1159/000083657. [DOI] [PubMed] [Google Scholar]
- Nilsson et al., 1999.Nilsson LO, Boman A, Sävendahl L, Grigelioniene G, Ohlsson C, Ritzén EM, Wroblewski J. Demonstration of estrogen receptor-beta immunoreactivity in human growth plate cartilage. J Clin Endocrinol Metab. 1999;84:370–373. doi: 10.1210/jcem.84.1.5531. [DOI] [PubMed] [Google Scholar]
- Ottem et al., 2004.Ottem EN, Godwin JG, Krishnan S, Petersen SL. Dual-phenotype GABA/glutamate neurons in adult preoptic area: sexual dimorphism and function. J Neurosci. 2004;24:8097–8105. doi: 10.1523/JNEUROSCI.2267-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papka and Mowa, 2003.Papka RE, Mowa CN. Estrogen receptors in the spinal cord, sensory ganglia, and pelvic autonomic ganglia. Int Rev Cytol. 2003;231:91–127. doi: 10.1016/s0074-7696(03)31003-4. [DOI] [PubMed] [Google Scholar]
- Pedersen et al., 2001.Pedersen AA, Videbaek N, Skak K, Petersen HV, Michelsen BK. Characterization of the rat GAD67 gene promoter reveals elements important for basal transcription and glucose responsiveness. DNA Seq. 2001;11:485–499. doi: 10.3109/10425170109041332. [DOI] [PubMed] [Google Scholar]
- Petersen et al., 2003.Petersen SL, Ottem EN, Carpenter CD. Direct and indirect regulation of gonadotropin-releasing hormone neurons by estradiol. Biol Reprod. 2003;69:1771–1778. doi: 10.1095/biolreprod.103.019745. [DOI] [PubMed] [Google Scholar]
- Pettersson et al., 2000.Pettersson K, Delaunay F, Gustafsson JA. Estrogen receptor beta acts as a dominant regulator of estrogen signaling. Oncogene. 2000;19:4970–4978. doi: 10.1038/sj.onc.1203828. [DOI] [PubMed] [Google Scholar]
- Pompolo et al., 2002.Pompolo S, Scott CJ, Clarke IJ. Selective regulation of glutamic decarboxylase isoform 65, but not isoform 67, in the bed nucleus of the stria terminalis and the preoptic area of the ewe brain across the estrous cycle. Endocrinology. 2002;143:544–550. doi: 10.1210/endo.143.2.8610. [DOI] [PubMed] [Google Scholar]
- Saleh and Connell, 2003.Saleh TM, Connell BJ. Estrogen-induced autonomic effects are mediated by NMDA and GABAA receptors in the parabrachial nucleus. Brain Res. 2003;973:161–170. doi: 10.1016/s0006-8993(03)02432-6. [DOI] [PubMed] [Google Scholar]
- Sathya et al., 1997.Sathya G, Li W, Klinge CM, Anolik JH, Hilf R, Bambara RA. Effects of multiple estrogen responsive elements, their spacing, and location on estrogen response of reporter genes. Mol Endocrinol. 1997;11:1994–2003. doi: 10.1210/mend.11.13.0039. [DOI] [PubMed] [Google Scholar]
- Shively and Bethea, 2004.Shively CA, Bethea CL. Cognition, mood disorders, and sex hormones. ILAR J. 2004;45:189–199. doi: 10.1093/ilar.45.2.189. [DOI] [PubMed] [Google Scholar]
- Shughrue et al., 1997.Shughrue PJ, Lane MV, Merchenthaler I. Regulation of progesterone receptor messenger ribonucleic acid in the rat medial preoptic nucleus by estrogenic and antiestrogenic compounds: an in situ hybridization study. Endocrinology. 1997;138:5476–5484. doi: 10.1210/endo.138.12.5595. [DOI] [PubMed] [Google Scholar]
- Simerly et al., 1990.Simerly RB, Chang C, Muramatsu M, Swanson LW. Distribution of androgen and estrogen receptor mRNA-containing cells in the rat brain: an in situ hybridization study. J Comp Neurol. 1990;294:76–95. doi: 10.1002/cne.902940107. [DOI] [PubMed] [Google Scholar]
- Skak and Michelsen, 1999.Skak K, Michelsen BK. The TATA-less rat GAD65 promoter can be activated by Sp1 through non-consensus elements. Gene. 1999;236:231–241. doi: 10.1016/s0378-1119(99)00286-3. [DOI] [PubMed] [Google Scholar]
- Soghomonian and Martin, 1998.Soghomonian JJ, Martin DL. Two isoforms of glutamate decarboxylase: why? Trends Pharmacol Sci. 1998;19:500–505. doi: 10.1016/s0165-6147(98)01270-x. [DOI] [PubMed] [Google Scholar]
- Spencer et al., 2008.Spencer JL, Waters EM, Romeo RD, Wood GE, Milner TA, McEwen BS. Uncovering the mechanisms of estrogen effects on hippocampal function. Front Neuroendocrinol. 2008;29:219–237. doi: 10.1016/j.yfrne.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork et al., 2000.Stork O, Ji FY, Kaneko K, Stork S, Yoshinobu Y, Moriya T, Shibata S, Obata K. Postnatal development of a GABA deficit and disturbance of neural functions in mice lacking GAD65. Brain Res. 2000;865:45–58. doi: 10.1016/s0006-8993(00)02206-x. [DOI] [PubMed] [Google Scholar]
- Tamayama et al., 2001.Tamayama T, Kanbara K, Maemura K, Kuno M, Watanabe M. Localization of GABA, GAD65 and GAD67 in rat epiphyseal growth plate chondrocytes. Acta Histochem Cytochem. 2001;34:201–206. doi: 10.1078/0065-1281-00604. [DOI] [PubMed] [Google Scholar]
- Tillakaratne et al., 1992.Tillakaratne NJ, Erlander MG, Collard MW, Greif KF, Tobin AJ. Glutamate decarboxylases in nonneural cells of rat testis and oviduct: differential expression of GAD65 and GAD67. J Neurochem. 1992;58:618–627. doi: 10.1111/j.1471-4159.1992.tb09763.x. [DOI] [PubMed] [Google Scholar]
- Wang et al., 2000.Wang H, Eriksson H, Sahlin L. Estrogen receptors alpha and beta in the female reproductive tract of the rat during the estrous cycle. Biol Reprod. 2000;63:1331–1340. doi: 10.1095/biolreprod63.5.1331. [DOI] [PubMed] [Google Scholar]
- Watters and Dorsa, 1998.Watters JJ, Dorsa DM. Transcriptional effects of estrogen on neuronal neurotensin gene expression involve cAMP/protein kinase A-dependent signaling mechanisms. J Neurosci. 1998;18:6672–6680. doi: 10.1523/JNEUROSCI.18-17-06672.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiland, 1992.Weiland NG. Glutamic acid decarboxylase messenger ribonucleic acid is regulated by estradiol and progesterone in the hippocampus. Endocrinology. 1992;131:2697–2702. doi: 10.1210/endo.131.6.1446611. [DOI] [PubMed] [Google Scholar]
- Weitsman et al., 2006.Weitsman GE, Skliris G, Ung K, Peng B, Younes M, Watson PH, Murphy LC. Assessment of multiple different estrogen receptor-beta antibodies for their ability to immunoprecipitate under chromatin immunoprecipitation conditions. Breast Cancer Res Treat. 2006;100:23–31. doi: 10.1007/s10549-006-9229-5. [DOI] [PubMed] [Google Scholar]