Abstract
Background
Human Herpesvirus-6 (HHV-6) has been associated with a diverse spectrum of central nervous system (CNS) diseases and reported glial tropism.
Objective
To determine if HHV-6 is present in a series of pediatric brain tumors.
Study Design
Pediatric gliomas from 88 untreated patients represented in a tissue microarray (TMA) were screened for HHV-6 by nested polymerase chain reaction (PCR), in situ hybridization (ISH), and immunohistochemistry (IHC) and compared to non glial tumors (N=22) and control brain (N=32). Results were correlated with tumor grade and overall survival.
Results
HHV-6 U57 was detected by nested PCR in 68/120 (57%) tumors and 7/32 (22%) age-matched non-tumor brain (P=0.001). HHV-6 U31 was positive in 73/120 (61%) tumors and 11/32 (34%) controls (P=0.019). Seventy-two percent (43/60) of tumors were HHV-6 Variant A. HHV-6 U57 was confirmed by ISH in 83/150 (54%) tumors and 10/32 (31%) controls (P=0.021), revealing a non-lymphocytic origin of HHV-6. HHV-6A/B gp116/64/54 late antigen was detected by IHC in 50/124 (40%) tumors and 6/32 (18%) controls (P=0.013). Interestingly, 58% of low grade gliomas (N=67) were IHC positive compared to 19% of high grade gliomas (N=21, P=0.002) and 25% of non gliomas (N=36; P=0.001). HHV-6A/B gp116/64/54 antigen co-localized with glial fibrillary acidic protein, confirming the astrocytic origin of antigen. Overall, there was no primary association between HHV-6A/B gp116/64/54 antigen detection and survival (P=0.861).
Conclusions
We provide the first reported series of HHV-6 detection in pediatric brain tumors. The predominance of HHV-6 in glial tumors warrants further investigation into potential neurooncologic disease mechanisms.
Keywords: human herpesvirus-6, pediatric brain tumor, tissue microarray
1. Background
Human herpesvirus-6 (HHV-6), a member of the beta herpesvirus family, has been associated with several CNS diseases1-3. HHV-6 has been shown to infect human oligodendrocytes and astrocytes in vitro and in vivo, altering cytokine production, cellular proliferation, and differentiation4-7. There are two known variants of HHV-6 (A and B); each is believed to exhibit differential tropism and pathogenicity despite greater than 90% sequence homology8-9. Regarding CNS tumors, HHV-6 variants have been reported in 8-37% of primary adult brain tumors10-12; however, it is not known whether these observations represent active infection since only one study reported HHV-6 p41 early antigen in 4/5 adult PCR positive brain tumors12. In children, two cases of glial tumors in association with HHV-6 have been reported13-14. Since HHV-6 is commonly acquired during early childhood and exhibits neurotropism in vivo, we sought to determine the prevalence of HHV-6 DNA and late antigen in a cohort of pediatric glial and non glial tumors and whether there is any effect on survival.
2. Materials and Methods
2.1 Pediatric brain tumor and control tissue
Brain tumor tissue from pediatric patients ages 4 months to 18 years (112 gliomas, 10 medulloblastomas, 10 meningiomas, 2 germinomas, 10 oligodendrogliomas) and non tumor control autopsy brain tissue (ages 2months to 15 years) was provided by Children's National Medical Center, Washington D.C., following Institutional Review Board approval. The non tumor control tissue was obtained from pediatric autopsy cases of non neurological causes of death with normal brain histology. Positive control HHV-6 encephalitis tissue was provided by the National Institute of Neurological Disorders and Stroke, Division of Immunology (Dr. Steven Jacobson).
2.2 Pediatric Brain Tumor Tissue Microarrays
For HHV-6 in-situ hybridization (ISH) and immunohistochemistry (IHC) experiments, a pediatric glioma tissue microarray (TMA) was probed, consisting of 88 untreated patients with glial tumors of varying WHO grades (42 Grade I, 24 Grade II, 13 Grade III and 9 Grade IV) with known clinical information (Ages 4months to 18 years, Mean 10.4y) as previously described.15 A separate cohort of pediatric autopsy brains and non glial tumors were used in comparison. Due to TMA sectioning and processing, not all of the tumors were represented in duplicate and explains why there are differences in the total number of tumors tested by ISH and IHC.
2.3 HHV-6 Nested PCR and Subtype Analysis
Five hundred nanograms of DNA (100ng for fresh frozen tumors) were used for nested PCR using U57 Major Capsid Protein (MCP) and U31 Large Tegument Protein (LTP)16-20. U251 cell lines infected with HHV-6A (U1102 strain) or SupT1 cells infected with HHV-6B (Z29 strain) served as positive controls. Great care was taken to avoid contamination of HHV-6 nested PCR reactions. All DNA was extracted in a facility that does not handle HHV-6 (CNMC). Fresh gloves and blades were used for each extraction. Negative controls were run for all PCR reactions. PCR was performed in a separate room in a sterile hood following UV irradiation.
2.4 In Situ Hybridization (ISH)
HHV-6 biotinylated DNA oligonucleotide probes that recognize DNA/RNA corresponding to U57 ORF4L (Major Capsid Protein) were used on TMAs, non glial tumors, and control brain according to the manufacturer's protocol (Maxim Biotech, Bethesda MD). An HHV-6 encephalitis PCR positive sample served as positive control for in situ hybridization experiments. Herpes simplex virus (HSV1/2) oligonucleotide probe (Maxim Biotech, Bethesda MD) was used as the negative control (data not shown).
2.5 Immunohistochemistry (IHC) and Immunofluorescence Microscopy
Pediatric tumor and non tumor controls were probed with HHV6A/B late antigen gp116/64/54 mouse monoclonal antibody (Advanced Biotechnologies, Columbia Maryland) as previously described17,19-21. PCR positive HHV6 encephalitis samples served as the positive controls16,19. For co-localization studies, rabbit anti-human GFAP FITC conjugated antibody (Sigma, St Louis Missouri) was used at a dilution of 1:250 for 1 hour at room temperature following addition of primary antibody (HHV6A/B gp116/54/64 at 1:50 dilution) and goat anti-mouse IgG Texas-red conjugated secondary antibody at 1:200 dilution (Sigma, St Louis Missouri). Light and Fluorescence microscopy were performed using Ziess Axioskop with Axiocam Imaging Software (Chester, VA). IHC and ISH samples were graded in a blinded fashion as positive or negative based on a well demarcated cytosolic/perinuclear signal observed in greater than 10% of tumor cells at 20× magnification in each of the 3 different fields.
2.6 Statistical Analysis
Fisher's exact test, ANOVA, and Kaplan Meier progression free survival were performed using GraphPad5 software (GraphPad Inc. San Diego CA). Error bars on column graphs represent standard error. A P value of ≤ 0.05 was considered statistically significant.
3. Results
3.1 Detection of Human herpesvirus- 6 (HHV-6) Variants in Pediatric Brain Tumors and Age-Matched Control Brain by In Situ Hybridization (ISH) and Nested PCR
We screened a series of more than 120 pediatric gliomas from 88 untreated patients represented in a tissue microarray (TMA), 22 non glial tumors, and 32 pediatric non tumor autopsy controls for HHV-6 by in situ hybridization (ISH) (Fig. 1A,Table 1). Treatment with a biotinylated oligonucleotide DNA probe corresponding to ORF4L Major Capsid Protein (MCP) of HHV-6 revealed a perinuclear/cytosplasmic signal in 57% (73/128) of glial tumors (66/114 glioma, 5/10 oligodendroglioma, 2/4 ganglioglioma) compared to 31% (10/32) of non tumor control brain tissue (P=0.0138). Of 22 non-glial tumors (10 medulloblastoma, 10 meningioma, 2 germinoma), 45% (10/24) were positive. HHV-6 was detected by ISH more in tumor tissue than in adjacent normal appearing brain (Fig. 1B).
Figure 1. Detection of HHV-6 by In Situ Hybridization in Pediatric Brain Tumors and Non Tumor Brain.
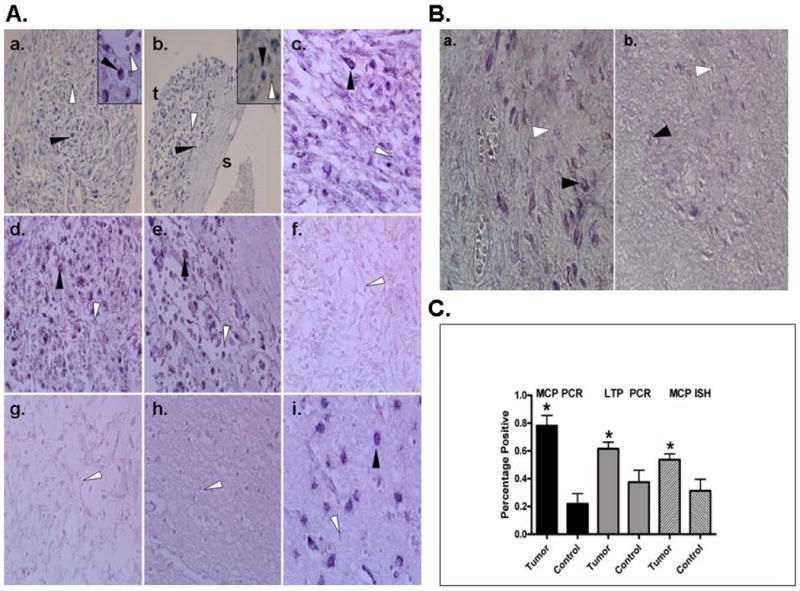
A. Nested HHV-6 MCP PCR positive (a-e,i) and negative (f-g) paraffin-embedded pediatric brain tumors were probed with biotinylated oligonucleotide probe corresponding to HHV-6 Major Capsid Protein. At 20× magnification (a,b) perinuclear cytoplasmic staining is observed diffusely throughout the tumor (t) but is not observed in surrounding stromal tissue (s) as shown in b (insets 40× magnification). Areas of representative HHV-6 positive ISH signal is indicated by black arrows while negative signal is shown by white arrows. No staining was observed in PCR negative tumors (f-g) or non tumor brain (h) compared to HHV-6 encephalitis positive control (i).
B. HHV-6 MCP ISH of a pediatric glioma containing tumor (a.) and adjacent non-tumor (b.) reveals an increased perinuclear/cytoplasmic signal in the tumor containing region compared to adjacent normal apprearing brain. C. Statistical analysis of HHV-6 PCR and ISH findings show a significance between tumor samples and control brain for MCP PCR (P<0.001), LTP PCR (P<0.019) and MCP ISH (P<0.021).
Table 1. Summary of HHV-6 PCR, In Situ Hybridization, and Immunohistochemistry in Pediatric Brain Tumors and Control Non Tumor Brain.
| Pediatric Samples | HHV-6 PCR | HHV-6 ISH | HHV-6 IHC | ||
|---|---|---|---|---|---|
| MCP(%) | LTP(%) | HHV6 Subtype | MCP(%) | gp116/54/64 (%) | |
| TUMORS | |||||
| Paraffin-Embedded | 36/64 (56) | 43/64(67) | 31A 12B | 83/150 (55) | 50/124 (40) |
| Glioma | 27/41 (66) | 29/41 (71) | 23A 6B | 66/114 (58) | 41/88 (47) |
| Grade 1 | 10/18 (56) | 13/18 (72) | 30/46 (65) | 19/43 (44) | |
| Grade 2 | 2/2 (100) | 2/2 (100) | 16/29 (55) | 9/24 (38) | |
| Grade 3 | 6/7 (86) | 4/7 (57) | 13/24 (54) | 2/14 (14) | |
| Grade 4 | 9/14 (64) | 10/14 (71) | 7/15 (47) | 2/7 (29) | |
| Oligodendroglioma | 1/1 (100) | 1/1 (100) | 0A 2B | 5/10 (50) | 5/10 (50) |
| Ganglioglioma | N/A | N/A | N/A | 2/4 (50) | 2/4 (50) |
| Meningioma | 5/10 (50) | 7/10 (70) | 3A 4B | 5/10 (50) | 0/10 (0) |
| Medulloblastoma | 2/10 (20) | 4/10 (40) | 4A 0B | 4/10 (40) | 2/10 (20) |
| Germinoma | ½ (50) | 2/2 (100) | 1A 0B | ½ (50) | 0/2 (0) |
| Fresh Frozen | 32/56 (57) | 30/56 (54) | 12A 5B | N/A | N/A |
| Glioma | 14/24 (58) | 17/24 (71) | 12A 5B | N/A | N/A |
| Medulloblastoma | 14/24 (58) | 11/24 (46) | NP | N/A | N/A |
| Meningioma | 4/8 (50) | 2/8 (25) | NP | N/A | N/A |
| NON TUMOR CONTROLS | |||||
| Paraffin-Embedded Non-Tumor Brain | 7/32 (22) | 11/32 (34) | 4A 7B | 10/32 (31) | 6/32 (18) |
Abbreviations: HHV-6, Human Herpesvirus-6; MCP, Major Capsid Protein; LTP, Large Tegument Protein; ISH, In situ hybridization; IHC, Immunohistochemistry; NA not applicable; NP, not performed.
To confirm the specificity of HHV-6 ISH, a subset of formalin-fixed paraffin-embedded glial tumors (N=42), non glial tumors (N=22), and control non tumor brain (N=32) were subjected to nested PCR using HHV-6 U57 Major Capsid Protein (MCP) and U31 Large Tegument Protein (LTP) gene-specific primers. Fifty-six percent (36/64) of paraffin-embedded tumors were HHV-6 MCP nested PCR positive compared to 22% (7/32) of non tumor controls (P=0.001) (Fig. 1C, Table 1). HHV-6 LTP nested PCR revealed 67% (43/64) positive compared to 34% (11/32) of non tumor controls (P=0.019). There was no difference between non glial tumors and controls by either MCP (P=0.18) or LTP PCR (P=0.09). There was no difference between PCR positivity and tumor grade (P=0.221). Overall, LTP was more sensitive than both MCP PCR and ISH in detecting HHV-6 DNA. Five of 41 PCR positive tumors were not detected by ISH and no tumor was negative by PCR and positive by ISH. ANOVA analysis revealed no differences in sensitivity among the PCR or ISH assays to detect HHV-6 DNA among paraffin embedded tumors (P=0.249) or non tumor controls (P=0.527). To validate PCR results on formalin fixed paraffin embedded tumor material, a separate cohort of fresh frozen glial (N=24) and non glial tumors (N=32) were subject to nested PCR. Fifty-eight percent of fresh frozen gliomas (14/24) were HHV-6 MCP PCR positive compared to 71% (17/24) for LTP. HHV-6 subtype analysis revealed 72% (43/60) HHV6 Variant A in all pediatric tumors compared to 33% (4/11) in non tumor brain (P=0.025).
3.2 Detection of HHV-6A/B gp116/64/54 Late Antigen in Pediatric Brain Tumors and Age-Matched Control Brain by Immunohistochemistry and Immunoflourescence Microscopy
We used an HHV-6 monoclonal antibody gp116/64/54 that recognizes both A and B variants which was previously demonstrated on paraffin embedded brain tissue16,19,22 to screen both tumor and non tumor tissue for the presence of late antigen, suggestive of active infection. Figure 2A reveals a cytoplasmic signal in the HHV-6 PCR positive gliomas and the encephalitis control but not in the PCR negative gliomas. Overall, 41 of 88 (47%) gliomas were positive compared to 18% of non tumor control brain (P=0.003). Immunoreactivity was seen in 5/10 oligodendrogliomas, while none of the non-glial tumors (germinomas N=2, medulloblastomas N=10, meningiomas N=10) were positive (Fig. 2B). HHV-6A/B gp116/64/54 IHC was validated by immunofluorescence microscopy (Fig. 2C) in a subset of tumors and controls. No sample positive by immunofluorescence microscopy was negative by IHC. Similar to the ISH findings, HHV-6A/B gp116/64/54 was not specific for tumor tissue and was seen, albeit to a lesser extent, in areas of adjacent normal appearing brain (Fig. 2D).
Figure 2. Detection of HHV-6A/B gp116/64/54 Late Antigen by Immunohistochemistry and Immunofluorescence microscopy in Pediatric Brain Tumors and Control Brain.
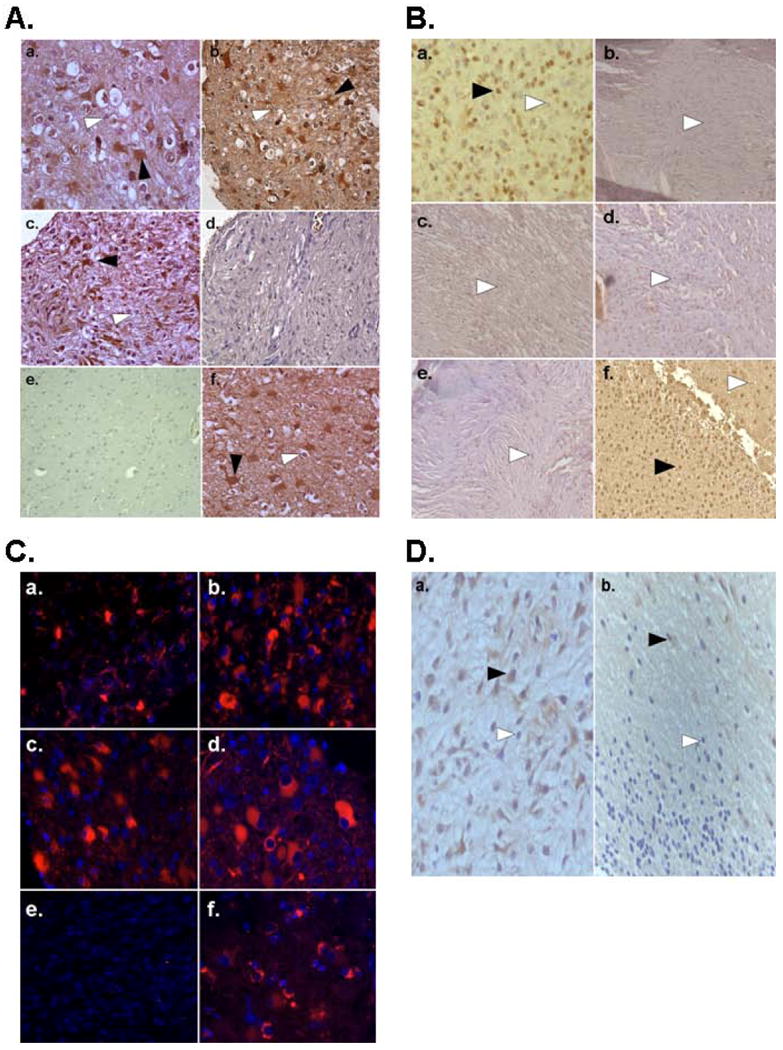
A. Immunohistochemistry using HHV-6A/B gp116/64/54 late antigen antibody of PCR positive pediatric tissue microarray gliomas (a-d) at 20× magnification reveals both positive (black arrows) and negative (white arrows) containing regions. Staining is preferentially observed in lower grade gliomas (a-c) and HHV-6 encephalitis positive control (f) compared to high grade glioma (d) or non tumor control tissue (e). B. HHV-6A/B gp116/64/54 late antigen staining is observed in an oligodendroglioma (a) but not in other non glial tumor subtypes meningioma (b,c), medulloblastoma (d), or germinoma (e) compared to positive control HHV-6 encephalitis (f). C. Immunofluorescence microscopy using HHV-6 A/B gp116/64/54 late antigen further reveals cytosolic signal in PCR positive gliomas (a-d) but not age-matched control brain (e) compared to HHV-6 encephalitis positive control (f). D. HHV-6A/B gp116/64/54 IHC of a pediatric glioma containing tumor (a) and adjacent non tumor (b) areas reveals increased signal in the tumor containing region.
3.3 Preferential Detection of HHV6A/B gp116/64/54 Late Antigen in Low Grade Glial Tumors and Co-Localization with Glial Fibrillary Acidic Protein
To determine the cellular origin of HHV-6A/B gp116/64/54, co-localization experiments were performed using the astrocyte marker glial fibrillary acidic protein. HHV-6A/B gp116/64/54 late antigen co-localized with glial fibrillary acidic protein in both tumor (ac) and encephalitis (d-f) biopsy (Fig. 3A), confirming the glial distribution of HHV-6. Overall, there was no difference in immunopositivity between non glial tumors and control brain (P=0.853). In contrast, glial tumors demonstrated increased frequency of immunopositivity for HHV-6A/B gp116/64/54 late antigen compared to non glial tumors (P=0.003) (Fig. 3B). In addition, a significant difference in immunopositivity was observed between lower grade (WHO Grade I-II) and higher grade (WHO grade III-IV) gliomas (42 and 19% positive, respectively) (P=0.002) (Fig. 3C).
Figure 3. Detection of HHV-6A/B gp116/64/54 Late Antigen in Cells of Glial Origin and Correlation between Tumor Type, Grade, and Progression Free Survival.
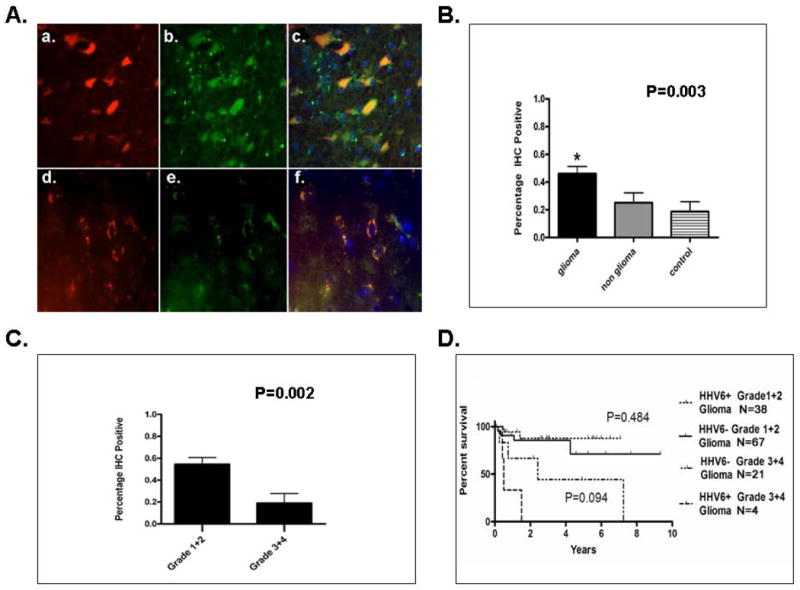
A. Immunofluorescence microscopy of pediatric glioma (a-c) and HHV-6 control encephalitis (d-f) paraffin embedded samples dual stained with HHV-6A/B gp116/64/54 late antigen(a,d) and glial fibrillary acidic protein (b,e) reveals colocalization (c,f) indicative of HHV-6 antigen in cells of glial origin. B. Statistical analysis of gp116/64/54 IHC results shows predilection of immunopositivity in glial tumors compared to non glial tumors (P<0.003). C. HHV-6A/B late antigen gp116/64/54 is preferentially detected in lower grade (1-2) gliomas compared to higher grade gliomas (3-4) (P<0.002). D. Kaplan Meier progression free survival analysis stratified by HHV-6A/B gp116/64/54 IHC status and tumor grade demonstrates no significance among IHC positive or negative lower grade (P=0.484) or higher grade (P=0.094) gliomas.
3.4 Lack of Association between HHV6A/B gp116/64/54 Late Antigen Detection and Survival in Pediatric Gliomas
IHC results were correlated with known clinical data (age at diagnosis, time to disease progression, overall survival) to determine if HHV6A/B gp116/64/54 positivity correlated with glial tumor survival. Overall, survival was 3.08 years (0.6-15.9y; 3 deaths) for HHV-6A/B gp116/64/54 IHC positive glioma patients compared to 3.25 years for IHC negative patients (0.1-10.9y; 8 deaths) (P=0.245). Progression free survival was 2.67 years (0.42-7.1y) for HHV-6A/B gp116/64/54 IHC positive glioma patients and 2.25 years (0.1-9.3) for IHC negative patients (P=0.653). Stratifying according to HHV-6A/B gp116/64/54 IHC status and glioma grade, revealed no differences in survival among lower grade (WHO grade 1-2) HHV-6A/B gp116/64/54 IHC positive and negative (P=0.484) patients. Among higher grade gliomas (WHO grade 3-4), there was a tendency for HHV-6A/B gp116/64/54 positive patients to have a shorter survival; however, this did not reach statistical significance (P=0.094) (Figure 3D).
4. Conclusions
Our results are consistent with prior studies demonstrating HHV-6 by PCR in both non tumor (32-67%) and malignant (8.2-37%) adult brain tissue 11-12, 24-25. Recently, Neves et al. reported no detectable HHV-6 DNA by real time PCR in 35 low grade astrocytomas26, compared to our finding of 44 to 72% positive by nested PCR, ISH, or IHC. This discrepancy can be explained in part by methodology. In a recent multicenter study, up to 100-fold changes in sensitivity have been reported among various HHV-6 PCR assays used for HHV-6 detection in serum27. Since the detection limit of HHV-6 MCP and LTP by primary PCR is 105 and 104 copies/million cells, respectively (data not shown), the amount of detectable HHV-6 by nested PCR reported in this study are low compared to immunocompromised patients with active disease17-18, 22. Others have similarly demonstrated nested PCR is necessary for detection in other HHV-6-associated diseases21, 23, 28. The requirement of nested PCR for detection of HHV-6 in our series is not consistent with chromosomally integrated HHV-629-34. The minimum copy number of HHV-6 to confer CNS disease is not known and it is possible that low copy number could cause significant changes at the cellular level to ultimately modify disease. Since the tumors in this study were from previously untreated patients, it is less likely that HHV-6 detection is due to viral reactivation as is seen in immunocompromised hosts. However, we cannot be certain whether detection of HHV-6 is due to primary infection or reactivation due to a localized tumor-induced dysfunction of normal immunosurveillance mechanisms and enhanced glial cell proliferation.
While overall and progression free survival did not correlate with HHV-6A/B gp116/54/64 late antigen detection, this does not exclude the possibility that HHV-6 may play an epigenetic role in glial tumor biology that may affect clinical course, given our relatively small numbers studied.
The distinction between viral disease causation and association is an important one to address. Our results do not suggest HHV-6 as a direct cause of brain tumors and should not be over interpreted. Only through the development of an animal model system, together with more reliable testing for viral macromolecules, will these questions be answerable. We feel that the association of HHV-6 in brain tumors, glial tumors in particular, is an important observation and worth further investigation. Our lab is currently working on a rigorous genomic analysis of HHV-6- induced changes in malignant astrocytes to more fully understand potential oncogenic and neuroimmune dysregulatory mechanisms of HHV-6 CNS infection.
Acknowledgments
The authors would like to thank the Friends of Ian Foundation for their generous support of the pediatric tissue microarray construction. We thank Dr. Julie Fotheringham (NINDS) for offering technical expertise in PCR and IHC experiments. This work was supported by the National Institutes of Health Neurological Sciences Academic Development Award (NSADA) K12NS052159-01A1.
Abbreviations
- HHV-6
Human herpesvirus-6
- PCR
Polymerase Chain Reaction
- ISH
In Situ Hybridization
- IHC
Immunohistochemistry
- TMA
Tissue Microarray
- MCP
Major Capsid Protein
- LTP
Large Tegument Protein
Footnotes
Disclosure: The authors have reported no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Caserta MT. Human Herpesvirus 6 Infection of the Central Nervous System. Curr Infect Dis Rep. 2004;6:316–321. doi: 10.1007/s11908-004-0054-x. [DOI] [PubMed] [Google Scholar]
- 2.De Bolle L, Naesens L, De Clercq E. Update on human herpesvirus 6 biology, clinical features, and therapy. Clinical Microbiology Reviews. 2005;18:217–245. doi: 10.1128/CMR.18.1.217-245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dockrell DH. Human herpesvirus 6: molecular biology and clinical features. J Med Microbiol. 2003;52:5–18. doi: 10.1099/jmm.0.05074-0. [DOI] [PubMed] [Google Scholar]
- 4.Albright AV, Lavi E, Black JB, Goldberg S, O'Connor MJ, Gonzalez-Scarano F. The effect of human herpesvirus-6 (HHV-6) on cultured human neural cells: oligodendrocytes and microglia. J Neurovirol. 1998;4:486–494. doi: 10.3109/13550289809113493. [DOI] [PubMed] [Google Scholar]
- 5.Kong H, Baerbig Q, Duncan L, Shepel N, Mayne M. Human herpesvirus type 6 indirectly enhances oligodendrocyte cell death. J Neurovirol. 2003;9:539–550. doi: 10.1080/13550280390241241. [DOI] [PubMed] [Google Scholar]
- 6.Dietrich J, Blumberg BM, Roshal M, et al. Infection with an endemic human herpesvirus disrupts critical glial precursor cell properties. J Neurosci. 2004;24:4875–4883. doi: 10.1523/JNEUROSCI.5584-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arena A, Liberto MC, Iannello D, Capozza AB, Foca A. Altered cytokine production after human herpes virus type 6 infection. New Microbiol. 1999;22:293–300. [PubMed] [Google Scholar]
- 8.De Bolle L, Van Loon J, De Clercq E, Naesens L. Quantitative analysis of human herpesvirus 6 cell tropism. J Med Virol. 2005;75:76–85. doi: 10.1002/jmv.20240. [DOI] [PubMed] [Google Scholar]
- 9.Donati D, Martinelli E, Cassiani-Ingoni R, et al. Variant-specific tropism of human herpesvirus 6 in human astrocytes. J Virol. 2005;79:9439–9448. doi: 10.1128/JVI.79.15.9439-9448.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan PK, Ng HK, Cheng AF. Detection of human herpesviruses 6 and 7 genomic sequences in brain tumours. J Clin Pathol. 1999;52:620–623. doi: 10.1136/jcp.52.8.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luppi M, Barozzi P, Maiorana A, et al. Human herpesvirus-6: a survey of presence and distribution of genomic sequences in normal brain and neuroglial tumors. J Med Virol. 1995;47:105–111. doi: 10.1002/jmv.1890470119. [DOI] [PubMed] [Google Scholar]
- 12.Cuomo L, Trivedi P, Cardillo MR, et al. Human herpesvirus 6 infection in neoplastic and normal brain tissue. J Med Virol. 2001;63:45–51. [PubMed] [Google Scholar]
- 13.Rantala H, Mannonen L, Ahtiluoto S, et al. Human herpesvirus-6 associated encephalitis with subsequent infantile spasms and cerebellar astrocytoma. Dev Med Child Neurol. 2000;42:418–421. doi: 10.1017/s0012162200000773. [DOI] [PubMed] [Google Scholar]
- 14.Stödberg T, Deniz Y, Esteitie N, et al. A case of diffuse leptomeningeal oligodendrogliomatosis associated with HHV-6 variant A. Neuropediatrics. 2002;33:266–270. doi: 10.1055/s-2002-36739. [DOI] [PubMed] [Google Scholar]
- 15.Thorarinsdottir HK, Santi MR, McCarter R, et al. Expression of PDGFR Correlates with Malignant Histology and PTEN with Survival in Childhood Gliomas. Clin Cancer Res. 2008;14:3386–3394. doi: 10.1158/1078-0432.CCR-07-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fotheringham J, Akhyani N, Vortmeyer A, et al. Detection of active human herpesvirus-6 infection in the brain: correlation with polymerase chain reaction detection in cerebrospinal fluid. J Infect Dis. 2007;195:450–4. doi: 10.1086/510757. [DOI] [PubMed] [Google Scholar]
- 17.Yao K, Gagnon S, Akhyani N, et al. Reactivation of human herpesvirus-6 in natalizumab treated multiple sclerosis patients. PLos ONE. 2008;3:e2028. doi: 10.1371/journal.pone.0002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akhyani N, Berti R, Brennan MB. Tissue distribution and variant characterization of human herpesvirus (HHV)-6: increased prevalence of HHV-6A in patients with multiple sclerosis. J Infect Dis. 2000;182:1321–5. doi: 10.1086/315893. [DOI] [PubMed] [Google Scholar]
- 19.Crawford JR, Kadom N, Santi MR, Mariani B, Lavenstein BL. Human Herpesvirus 6 Rhombencephalitis in Immunocompetent Children. J Child Neurol. 2007;22:1260–8.18. doi: 10.1177/0883073807307086. [DOI] [PubMed] [Google Scholar]
- 20.Lyall EGH, Cubie HA. Human Herpesvirus-6 DNA in the Saliva of Paediatric Oncology Patients and Controls. J Med Virol. 1995;47:317–322. doi: 10.1002/jmv.1890470405. [DOI] [PubMed] [Google Scholar]
- 21.Donati D, Akhyani N, Fogdell-Hahn A, et al. Detection of human herpesvirus-6 in mesial temporal lobe epilepsy surgical brain resections. Neurology. 2003;61:1405–1411. doi: 10.1212/01.wnl.0000094357.10782.f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fotheringham J, Akhyani N, Vortmeyer A, et al. Detection of active human herpesvirus-6 infection in the brain: correlation with polymerase chain reaction detection in cerebrospinal fluid. J Infect Dis. 2007;195:450–4. doi: 10.1086/510757. [DOI] [PubMed] [Google Scholar]
- 23.Fotheringham J, Donati D, Akhyani N, et al. Association of human herpesvirus-6B with mesial temporal lobe epilepsy. PLos Med. 2007;4:e180. doi: 10.1371/journal.pmed.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liedtke W, Trubner K, Schwechheimer K. On the role of human herpesvirus 6 in viral latency in nervous tissue and in cerebral lymphoma. J Neurol Sci. 1995;134:184–188. doi: 10.1016/0022-510x(95)00248-6. [DOI] [PubMed] [Google Scholar]
- 25.Paulus W, Jellinger K, Hallas C, Ott G, Muller-Hermelink HK. Human herpesvirus-6 and Epstein-Barr virus genome in primary cerebral lymphomas. Neurology. 1993;43:1591–1593. doi: 10.1212/wnl.43.8.1591. [DOI] [PubMed] [Google Scholar]
- 26.Neves AM, Thompson G, Carvalheira J, et al. Detection and quantitative analysis of human herpesvirus in pilocytic astrocytoma. Brain Res. 2008;1221:108–14. doi: 10.1016/j.brainres.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Flamand L, Gravel A, Boutolleau D, et al. Multicenter comparison of PCR assays for detection of human herpesvirus 6 DNA in serum. J Clin Microbiol. 2008;46:2700–6. doi: 10.1128/JCM.00370-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akhyani N, Berti R, Brennan MB. Tissue distribution and variant characterization of human herpesvirus (HHV)-6: increased prevalence of HHV-6A in patients with multiple sclerosis. J Infect Dis. 2000;182(5):1321–5. doi: 10.1086/315893. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka-Taya K, Sashihara J, Kurahashi H, et al. Human herpesvirus 6 (HHV-6) is transmitted from parent to child in an integrated form and characterization of cases with chromosomally integrated HHV-6 DNA. J Med Virol. 2004;73:465–73. doi: 10.1002/jmv.20113. [DOI] [PubMed] [Google Scholar]
- 30.Daibata M, Taguchi T, Nemoto Y, Taguchi H, Miyoshi I. Inheritance of chromosomally integrated human herpesvirus 6 DNA. Blood. 1999;94:1545–9. [PubMed] [Google Scholar]
- 31.Kamble RT, Clark DA, Leong HN, Heslop HE, Brenner MK, Carrum G. Transmission of integrated human herpesvirus-6 in allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant. 2007;40:563–6. doi: 10.1038/sj.bmt.1705780. [DOI] [PubMed] [Google Scholar]
- 32.Leong HN, Tuke PW, Tedder RS, Khanom AB, Eglin RP, Atkinson CE, Ward KN, Griffiths PD, Clark DA. The prevalence of chromosomally integrated human herpesvirus 6 genomes in the blood of UK blood donors. J Med Virol. 2007;79:45–51. doi: 10.1002/jmv.20760. [DOI] [PubMed] [Google Scholar]
- 33.Clark DA, Nacheva EP, Leong HN, et al. Transmission of integrated human herpesvirus 6 through stem cell transplantation: implications for laboratory diagnosis. J Infect Dis. 2006;193:912–6. doi: 10.1086/500838. [DOI] [PubMed] [Google Scholar]
- 34.Hall CB, Caserta MT, Schnabel K, et al. Chromosomal integration of human herpesvirus 6 is the major mode of congenital human herpesvirus 6 infection. Pediatrics. 2008;122:513–20. doi: 10.1542/peds.2007-2838. [DOI] [PubMed] [Google Scholar]


